ABSTRACT
The endoplasmic reticulum (ER) is the largest membranous organelle, and its turnover ensures cellular homeostasis. The selective macroautophagy/autophagy of the ER (reticulophagy) guarantees the balance of ER turnover. However, the mechanism and physiological relevance of reticulophagy is largely unknown. Recently, we identified ATL3 (atlastin GTPase 3), generally considered to mediate ER fusion, as a receptor for reticulophagy. ATL3 specifically interacts with the GABARAP subfamily proteins of the Atg8-family, and this association is crucial for ATL3’s role as a receptor for reticulophagy. Moreover, 2 hereditary sensory and autonomic neuropathies type 1 (HSANI)-associated mutations of ATL3 (Tyr192Cys and Pro338Arg) impair ATL3’s binding to GABARAP and function in reticulophagy. Therefore, we illuminate a new function of ATL3 in reticulophagy and the potential physiological relevance of reticulophagy in neurodegenerative diseases.
The endoplasmic reticulum (ER), the largest membranous organelle, is a vital component in protein synthesis and secretion, lipid metabolism, Ca2+ homeostasis, and organelle communication. Except for the well-known unfolded protein response (UPR) and ER-associated protein degradation (ERAD), reticulophagy is another major quality control pathway involved in ER turnover and homeostasis. In recent years, our understanding of reticulophagy is growing rapidly as the discovery of several reticulophagy receptors, including RETREG1/FAM134B, SEC62, the long splice variant of RTN3 (RTN3L) and CCPG1. In the view of the specificity of reticulophagy receptors in different ER subdomains or tissue and physiological or pathological conditions, undiscovered reticulophagy receptors are supposed to exist.
Figure 1. Model of ATL3 in reticulophagy. To drive tubular ER turnover by reticulophagy, ATL3 specifically binds to GABARAPs via 2 GIMs, which leads to the recruitment of the phagophore, the sequestration of tubular ER within autophagosomes and the final degradation by a lysosome.
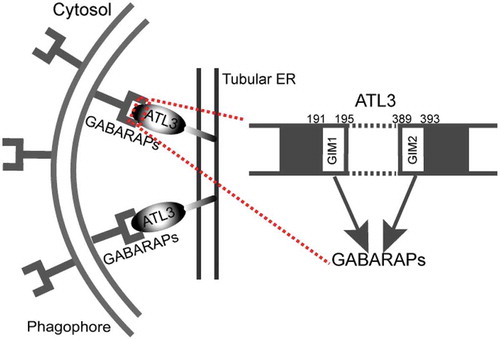
ATL3, belonging to a class of dynamin-like GTPase, was reported to mediate ER fusion. Recently, we identified ATL3 as a tubular reticulophagy receptor [Citation1]. Upon Earle’s balanced salt solution starvation, ATL3 traffics to autophagosomes and knockout of ATL3 inhibits tubular ER degradation. ATL3 specifically interacts with GABARAP subfamily proteins through 2 GABARAP-interacting motifs (GIMs) (). These 2 GIMs are required for ATL3 to function as an autophagy receptor for tubular ER.
RTN3L was also identified as a tubular reticulophagy receptor and this creates a question: Why are two receptors employed to mediate selective autophagy of tubular ER? Coincidentally, we use COS-7 cells as the experimental model and RTN3L is barely detectable in COS-7 cells. In fact, RTN3L expresses only in some tissues and organs, but is undetectable in others, such as kidney and heart. All these observations suggest that reticulophagy may be regulated by different autophagy receptors in different cell types or tissues. Thus, we propose that ATL3 is the main tubular reticulophagy receptor in cells lacking RTN3L. Moreover, the ER is the largest membrane organelle with complex morphology and function, and several reticulophagy receptors are reported to function in different ER subdomains and regulate their turnover. For instance, RETREG1/FAM134B and RTN3 or ATL3 regulate ER sheets and tubular ER turnover, respectively. Based on current research, different physiological and pathological conditions-induced reticulophagy may employ different reticulophagy receptors. For example, RETREG1/FAM134B, RTN3 and ATL3 or CCPG1 and SEC62 function in reticulophagy caused by nutritional deficiencies or ER stress, respectively. In short, multiple autophagy receptors are involved in reticulophagy.
The study of reticulophagy is mainly at the stage of identifying reticulophagy receptors; however, the physiological functions of reticulophagy need more attention. Right now, there are only two pieces of direct evidence of reticulophagy in physiological functions, which showed that retreg1/fam134b knockout mice display peripheral sensory neuropathy and Ccpg1 mutant mice exhibit significant accumulation of insoluble protein and metabolic disorder in the exocrine pancreas. It is worth noting that except for peripheral sensory neurons and pancreas, almost all other tissues and organs are normal in Retreg1/Fam134b- or Ccpg1-deficient mice. In our recent study, 2 disease-related mutations of ATL3 (Y192C and P338R), identified in patients with HSANI, were shown to weaken the interaction between ATL3 and GABARAP and impair ATL3’s role as a reticulophagy receptor [Citation1]. Combined with the results in retreg1/fam134b knockout mice, reticulophagy exhibits an important role in maintenance of peripheral sensory neuronal homeostasis.
In brief, ATL3 functions as a reticulophagy receptor through specific binding to GABARAP subfamily proteins (). The physiological importance of ATL3 requires further investigation. Besides, the ATL3 protein level is upregulated at the early stage of starvation, thus, the regulatory mechanism of transcriptional and post-translational modifications in reticulophagy is also a field worth studying.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
Reference
- Chen QZ, Xiao Y, Chai PY, et al. ATL3 is a tubular ER-phagy receptor for GABARAP-mediated selective autophagy. Curr Biol. 2019 Mar 4;29(5):846–855.e6. PubMed PMID: 30773365.
