ABSTRACT
Macroautophagy/autophagy, a physiological process that is involved in tumorigenesis, is regulated at genetic and epigenetic levels. Emerging reports suggest that aberrant RNA modifications cause dysregulated autophagy and affect tumorigenesis, while the role of RNA modifications in the regulation of autophagy in cancers remains unclear. In a recent study, we describe a new role for the tRNA m7G methyltransferase complex components METTL1 and WDR4 as negative regulators of MTORC1-mediated autophagy in esophageal squamous cell carcinoma (ESCC). METTL1 and WDR4 show abnormally high expression in ESCC tissues, and are associated with poor ESCC prognosis. Targeting METTL1 or WDR4 leads to decreased expression of m7G-modified tRNAs and reduces the translation of a subset of oncogenic transcripts, including the genes related to the MTOR signaling pathway and negative regulators of autophagy in an m7G-related codon-dependent manner, thereby resulting in hyperactivated MTORC1-mediated autophagy via dephosphorylation of ULK1 and finally causes cell death in ESCC. Our findings provide a new layer of translation regulation mechanism mediated by tRNA m7G modification, link translational machinery with autophagic machinery, and suggest that METTL1 and its downstream signaling axis could be potential therapeutic targets for ESCC treatment.
Autophagy, a physiological process that clears damaged organelles or proteins via autophagosomes and autolysosomes, is associated with metabolism, development, immunity, and tumorigenesis. Some reports claim that autophagy promotes tumor cell migration and invasion, while others suggest that autophagy negatively regulates tumorigenesis by inducing cell death. Autophagy is well-regulated at genetic and epigenetic levels and could be a potential target for cancer treatment. Post-transcriptional modifications and post-translational modifications, such as RNA methylation or protein phosphorylation, are directly or indirectly involved in the regulation of autophagy. Notably, AMPK and MTOR (mechanistic target of rapamycin kinase) complex 1 (MTORC1)-mediated dynamic changes of phosphorylation on ULK1 play key roles in regulating the onset of autophagy. In addition, Upstream transcription factors and some non-coding RNAs such as microRNAs, circRNAs, and lncRNAs regulate autophagy by controlling the expression of autophagy-related proteins. In general, autophagy is regulated in cancers by the dynamic changes in the activity or gene expression of autophagic proteins or their regulators.
Dysregulated gene expression leads to aberrant autophagy in cancers. Gene expression is a complex multi-step process that is regulated at the DNA replication, gene transcription, mRNA translation, and post-translational levels. Emerging reports suggest that protein abundance is not proportional to mRNA abundance in cancers, and some oncogenes are abnormally overexpressed in cancers in a translationally regulated manner, which is closely related to the oncogenic processes in cancers. mRNA translation requires the participation of translational machinery, among which tRNAs are important adaptors to recognize codons on mRNAs and bridge amino acids for mRNA translation. tRNAs are subjected to diverse modifications, the roles and mechanisms of which in gene expression regulation and cancer progression remain poorly understood. tRNA N7-methylguanosine (m7G) modification is a common modification in tRNA. Mutations or dysregulation of m7G methyltransferase complex components METTL1 and WDR4 are closely associated with developmental disorders and cancers.
In our recent study [Citation1], we demonstrated that the m7G tRNA methyltransferase METTL1 shows significantly higher expression in esophageal squamous cell carcinoma (ESCC) tumor tissues than that in normal tissues, and the elevated METTL1 is associated with poor prognosis in ESCC patients. We further observed significantly reduced proliferative capacity, inhibited in vivo tumorigenic capacity, and increased apoptotic rate upon the m7G tRNA methyltransferase complex components METTL1 or WDR4 depletion in ESCC cells. Moreover, we established ESCC initiation and progression mouse models and further demonstrated that mettl1 conditional knockout or Mettl1 conditional knockin significantly impairs in vivo ESCC initiation and progression, strongly and directly confirming the critical role of METTL1 and tRNA m7G modifications in ESCC regulation.
Mechanistically, loss of METTL1 results in decreased m7G-modified tRNA expression, and defects of the translation machinery cause ribosome pausing at codons decoded by m7G tRNAs during translation, thereby inhibiting the translation efficiency of specific mRNAs in an m7G-related codon-dependent manner. The translation efficiency-decreased mRNAs are significantly enriched in the MTOR signaling pathway, which is involved in the negative regulation process of autophagy. We further found that RPTOR, one of the core components of MTORC1, is significantly downregulated in METTL1-knockdown ESCC cells. RPTOR phosphorylates and inactivates ULK1, a member of the autophagic machinery, thereby blocking the onset of autophagy, whereas downregulation of RPTOR dephosphorylates ULK1, unleashes the shackles of the autophagy machinery, and triggers the initiation of autophagy, which is consistent with the significantly enhanced autophagy we observe in METTL1-knockdown ESCC cells (). Additionally, exogenous expression of RPTOR or knockdown of ULK1 successfully reverses the decline in proliferation capacity and survival capacity of METTL1-knockdown ESCC cells, suggesting that METTL1-mediated m7G tRNA modification promotes ESCC progression via the RPTOR-ULK1 axis. In our study, we observed decreased translational efficiency of RPTOR and hyperactivated autophagy as well as decreased malignancy phenotype upon METTL1 knockdown in ESCC. Rescue assays suggested that hyperactivation of autophagy partially accounts for the impaired malignancy phenotype of METTL1-knockdown ESCC cells. Indeed, RPTOR and MTORC1 are involved in many biological processes and molecular functions, such as translational regulation, transcriptional regulation, and cell proliferation. Actually, we observed that phosphorylation of RPS6KB1/S6K1 and EIF4EBP1, two known downstream targets of RPTOR, is significantly decreased in METTL1-knockdown ESCC cells. To more comprehensively explain the METTL1-mediated regulation of ESCC initiation and progression, further investigations are required to characterize the potential effects of the METTL1-RPTOR axis in ESCC. Taken together, we found that METTL1-mediated m7G tRNA modification enhances the translation of MTOR signaling pathway components in an m7G-related codon-dependent manner, thus blocking hyperactivation of autophagy and promoting ESCC progression.
Figure 1. Mechanism of translational machinery in regulating autophagic machinery. METTL1-mediated m7G tRNA modification enhances the translation of MTORC1 components in an m7G-related codon-dependent manner to phosphorylates and inactivates ULK1, thus blocking hyperactivation of autophagy.
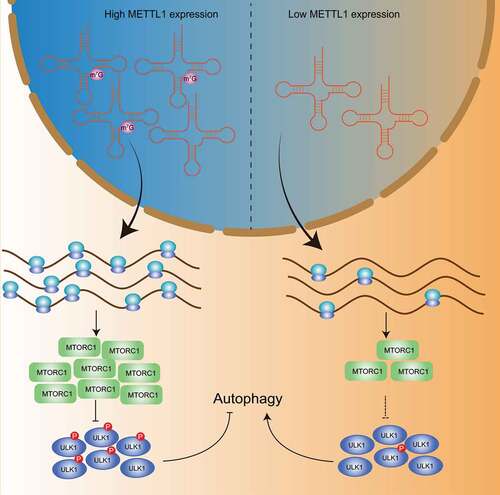
Autophagy is a conserved process that is essential for cell survival under various stress conditions. Previous studies uncovered that the epigenetic modifications in DNA, histone and mRNAs modulate dynamic gene expression and regulate autophagy under different cellular conditions. Using various in vitro and in vivo assays, we demonstrate that the elevated m7G modification on tRNA leads to increased translation of RPTOR mRNA and therefore further regulates the autophagy activity in ESCC cancer cells, which demonstrates a novel autophagy regulation mechanism at the post-transcriptional level. Our findings reveal the mechanism of translational machinery in regulating the autophagic machinery in ESCC, and suggest that METTL1 and its downstream autophagic signaling axis could be potential targets for ESCC treatment.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
Reference
- Han H, Yang C, Ma J, et al. N(7)-methylguanosine tRNA modification promotes esophageal squamous cell carcinoma tumorigenesis via the RPTOR/ULK1/autophagy axis. Nat Commun. 2022 Mar 18;13(1):1478.
