Abstract
Hypermethylation is an important mechanism for the dynamic regulation of gene expression, necessary for metastasizing tumour cells. Our aim is to identify methylation tumour markers that have a predictive value for the presence of regional lymph node metastases in patients with oral and oropharyngeal squamous cell carcinoma (OOSCC). Significantly differentially expressed genes were retrieved from four reported microarray expression profiles comparing pN0 and pN+ head-neck tumours, and one expression array identifying functionally hypermethylated genes. Additional metastasis-associated genes were included from the literature. Thus genes were selected that influence the development of nodal metastases and might be regulated by methylation. Methylation-specific PCR (MSP) primers were designed and tested on 8 head-neck squamous cell carcinoma cell lines and technically validated on 10 formalin-fixed paraffin-embedded (FFPE) OOSCC cases. Predictive value was assessed in a clinical series of 70 FFPE OOSCC with pathologically determined nodal status. Five out of 28 methylation markers (OCLN, CDKN2A, MGMT, MLH1 and DAPK1) were frequently differentially methylated in OOSCC. Of these, MGMT methylation was associated with pN0 status (P = 0.02) and with lower immunoexpression (P = 0.02). DAPK1 methylation was associated with pN+ status (P = 0.008) but did not associate with protein expression. In conclusion, out of 28 candidate genes, two (7%) showed a predictive value for the pN status. Both genes, DAPK1 and MGMT, have predictive value for nodal metastasis in a clinical group of OOSCC. Therefore DNA methylation markers are capable of contributing to diagnosis and treatment selection in OOSCC. To efficiently identify additional new methylation markers, genome-wide methods are needed.
Abbreviations
| FFPE | = | formalin-fixed paraffin-embedded |
| OOSCC | = | oral and oropharyngeal squamous cell carcinoma |
| pN status | = | pathologically determined nodal status |
| (Q)MSP | = | (quantitative) methylation-specific PCR |
Introduction
Oral and oropharyngeal squamous cell carcinomas (OOSCC) compose the largest subgroup of head and neck cancer, and are estimated to have caused over 42,000 new cases in the United States in 2014.Citation1 OOSCC are characterized by regional metastatic spread to the lymph nodes of the neck in an early stage. Patients with regional lymph node metastases are generally treated with curative intent. When regional metastases are not adequately treated, distant spread results, which is considered as incurable disease. Therefore, it is essential to make an accurate assessment of the nodal (N) status of the neck to adequately treat patients with OOSCC.Citation2 However, current imaging methods to assess the presence of metastases in the palpation-negative neck showed a sensitivity of 60–70%.Citation3 Sentinel lymph node biopsy, when performed intra-operatively on frozen sections, has a comparable sensitivity of 50–70%.Citation4,5
DNA hypermethylation is an important mechanism for the regulation of gene expression, in both physiological and pathological conditions.Citation6 DNA hypermethylation is a form of epigenetic regulation, in which the genetic sequence is not altered, but CH3-groups are added to the cytosine of CpG dinucleotides which, when present in the promoter region of a gene, leads to transcriptional repression of the associated protein. This process is reversible, and hypomethylation leads to reactivation of gene transcription.Citation7 Thus, hypermethylation of tumor suppressor genes and hypomethylation of oncogenes may contribute to carcinogenesis and cancer progression.Citation8 Because of its dynamic nature, methylation is a possible candidate mechanism for the dynamic regulation of gene expression during metastatic progression of OOSCC cells.Citation9
Moreover, several demethylating drugs have been developed and show that treatment results in re-expression of formerly hypermethylated genes. Decitabine and Azacytidine are therapeutic demethylating agents and have already been used in treatment of specific hematological malignancies.Citation10 Therefore, methylation can also be therapeutically targeted.Citation11
Methylation-specific PCR (MSP) is one of the most widely used methylation detection methods, because of its cost-effectiveness and high sensitivity.Citation12 The availability of such a sensitive detection method may allow methylation to become a prognostic or diagnostic tool in the clinical setting. For example, hypermethylation of MGMT in gliomas has been shown to predict patient response to alkylating chemotherapy.Citation13
Various studies have identified several genes that are frequently hypermethylated in OOSCC,Citation14,15 such as CDH1, CDKN2A, O-6-methylguanine-DNA methyltransferase (MGMT), death-associated protein kinase 1 (DAPK1), RARB, and RASSF1, but only few of those have been associated with metastasis.Citation16,17 In other cancers, various methylation markers have been associated with cell migration and invasion in vitroCitation18,19 and the presence of nodal metastasis.Citation19,20
In this study, we set out to identify novel methylation markers that are associated with the presence of lymph node metastases in patients with OOSCC. We selected candidate genes with a CpG island from the most differentially expressed genes, as reported in 4 published metastasis-associated gene profiles,Citation21-24 and the genes from these 4 profiles that were functionally methylated (showing increased expression after demethylating treatment), as determined in a previous study performed in our lab.Citation25 Additionally, we selected several genes that were reported to be associated with metastasis in previous studies in squamous cell carcinomas. These methylation markers were tested by MSP in a clinical series of OOSCC with pathologically determined N status for their predictive value for the presence of lymph node metastases.
Results
Candidate gene selection and initial testing
Using the strategy outlined in , 28 candidate genes were selected for analysis (). Two markers did not show any product during the optimization phase and were excluded. Of the 26 markers tested on the initial set of 5 N0 and 5 N+ formalin-fixed, paraffin-embedded (FFPE) OOSCC samples, 17 markers were methylated in none of the 10 OOSCC samples, 3 markers (PPT2, BTG2, CAV1) were methylated in only one sample, and one marker (TJP1) was methylated in all samples. Five markers showed methylation in 2 or more tumor samples and were considered eligible for further analysis (OCLN, CDKN2A, MGMT, MLH1, and DAPK1).
Figure 1. Flowchart for candidate gene selection and testing * TJP1 showed methylation in all samples and was therefore excluded.
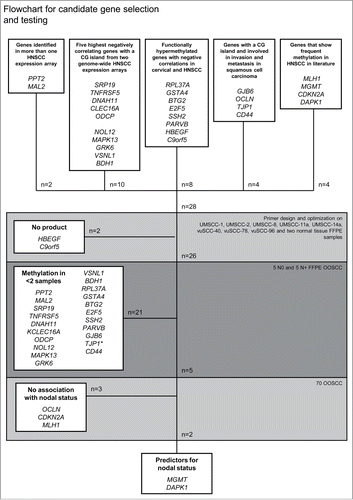
Table 1. Selected candidate genes
Predictor gene identification
OCLN, CDKN2A, MGMT, MLH1, and DAPK1 were tested on 32 pN0 and 38 pN+ cases (). MGMT was methylated in 13/32 (41%) of pN0 and 6/38 (16%) of pN+ cases and showed a significant association with nodal status (P = 0.02). DAPK1 methylation was also significantly associated with nodal status (P = 0.008); however, in contrast to MGMT, DAPK1 was more frequently methylated in pN+ (10/38, 26%) than in pN0 cases (1/32, 3%). OCLN, CDKN2A, and MLH1 showed more methylation in pN+ tumors also, but the difference was not statistically significant ().
Table 2. Clinicopathological characteristics
Table 3. Cross table analyses of the 5 genes eligible for testing on the patient series
MGMT had a predictive value of OR = 0.28 [95% confidence interval (CI): 0.09–0.84] and DAPK1 had an OR = 11.1 (95% CI: 1.33–92.1) for the pN status (). The wide 95% CI is probably attributable to the relatively small patient sample (n = 70) used in this study. Multivariate regression analysis revealed that both markers were not independent from currently used clinicopathological predictors, reflected in the cN status. However, the predictive values of MGMT and DAPK1 were independent from each other (). The combined regression model of MGMT and DAPK1 had a negative predictive value for the pN status of 76% ().
Table 4. Univariate and multiple logistic regression with pN status
Table 5. (A) Multiple logistic regression of DAPK1 and MGMT for pN status. (B) Crosstable for the DAPK1 and MGMT test combined vs. pN status
Immunohistochemistry
To assess if methylation of the 2 predictive markers MGMT and DAPK1 was associated with decreased expression, we performed immunohistochemistry on the available tumor tissue of the same cases that had been used to assess the predictive values of methylation. Because MGMTCitation26 and DAPK1,Citation27 in particular, are known to be heterogeneously expressed within the tumor, we investigated expression in the tumor center and tumor front separately in 66 OOSCC cases that were present on the tissue microarrays (). MGMT methylation was associated with low expression both in the tumor front (12% expression in methylated vs. 43% in unmethylated cases) and in the tumor center (26% in methylated vs. 36% in unmethylated cases), but this was only statistically significant in the tumor front (P = 0.02; ; ). For DAPK1 methylation, no associations were found with expression in tumor front (P = 1.0) or center (P = 0.14; ).
Figure 2. Representative examples of immunohistochemical staining. (A) DAPK1 low expression core, tumor center; (B) DAPK1 high expression, core tumor center; (C) DAPK1 low expression core, tumor front; (D) DAPK1 high expression core, tumor front; (E) MGMT low expression core, tumor center; (F) MGMT high expression core, tumor center; (G) MGMT low expression core, tumor front; (H) MGMT high expression core, tumor front.
Figure 3. Examples of 2 cases that showed MGMT methylation, associated with low expression in the invasive tumor front, but high expression in the tumor center. (A) MGMT methylation controls [pure water, leucocytes, and IV (in vitro SssI methylated leucocytes)] and 2 cases. (B) Low MGMT expression in the tumor invasive front (Case 1). (C) High MGMT expression in the tumor center (Case 1). (D) Low MGMT expression in the tumor invasive front (Case 2). (E) High MGMT expression in the tumor center (Case 2). U: unmethylated; M: methylated; Blanco: pure water control; Leuco: leucocytes; IV: in vitro SssI methylated leucocytes. T: tumor tissue. The border of the tumor area is indicated by a black line.
![Figure 3. Examples of 2 cases that showed MGMT methylation, associated with low expression in the invasive tumor front, but high expression in the tumor center. (A) MGMT methylation controls [pure water, leucocytes, and IV (in vitro SssI methylated leucocytes)] and 2 cases. (B) Low MGMT expression in the tumor invasive front (Case 1). (C) High MGMT expression in the tumor center (Case 1). (D) Low MGMT expression in the tumor invasive front (Case 2). (E) High MGMT expression in the tumor center (Case 2). U: unmethylated; M: methylated; Blanco: pure water control; Leuco: leucocytes; IV: in vitro SssI methylated leucocytes. T: tumor tissue. The border of the tumor area is indicated by a black line.](/cms/asset/7a2a2523-5d08-4944-9e99-b900ef426486/kepi_a_1075689_f0003_oc.gif)
Table 6. Associations between methylation and expression for MGMT and DAPK1
Discussion
The goal of our study was to identify novel methylation markers for the prediction of nodal metastasis. We selected 28 candidate genes, of which 2 (7%) showed a predictive value for the nodal (N) status. Both genes, DAPK1 and MGMT, have been described as frequently methylated in OOSCCCitation16,17 and other cancers.Citation28
Most candidate genes (12/28) were selected from the most differentially expressed genes in independent microarray studies of N0 vs. N+ HNSCC. We hypothesized that gene-specific promoter methylation lead to the observed gene silencing in N+ cases. However, none of these selected genes showed any methylation, indicating that other mechanisms are responsible for their downregulation. One explanation for the finding that the most differentially expressed genes lack promoter methylation is that our selection might have caused a bias toward genes downregulated by other mechanisms because methylation rarely causes complete transcriptional repression.Citation29
We also selected 8 genes that had predictive value in the metastatic gene profilesCitation21-24,30 and showed upregulation after demethylating treatment in cell lines.Citation25 However, the functional regulation of these genes by methylation in vitro might not apply to clinical tumor samples, due to (in vivo) intra-tumor heterogeneous methylation.Citation31 Additionally, genes selected from metastatic profiles reported in microarray studies do not accurately reflect the metastatic genotype, because these signatures are largely platform and analysis related and composition of predictive profiles varies enormously between different studies.Citation32 In fact, comparing the 4 microarray studies, shows that no single gene was reported in all 4 profiles.Citation21-24 This demonstrates that using expression profiles to identify new metastasis-specific OOSCC methylation markers is not effective.
Differentially hypermethylated regions (DMRs) in cancer are frequently found in or overlapping CpG islands (∼40% of hypermethylated DMRs). Another 30% of hypermethylated DMRs are located in a region of 500 bp flanking the CpG islands.Citation33 Our MSP primers were designed in the conventional areas [in CpG islands within −500 to +500 bp from the transcription start site (TSS)], which include 40–70% of the DMRs. However, it is possible that the regions most responsible for transcriptional regulation are located in specific regions outside these areas (CpG island shores).Citation33 The CpG island shores are not CG-rich and consequently not useful for optimal MSP primer design. Because we restricted our analysis to the CpG-rich regions close to the TSS to enable optimal MSP design, we cannot exclude that the differentially expressed genes are regulated by DNA methylation in other regions, such as CpG island shores, which contain ∼15% of the hypermethylated DMRs.
The selection of 4 genes that show frequent methylation in HNSCC produced the 2 methylation markers that were ultimately found to have predictive value for the presence of lymph node metastases (DAPK1 and MGMT).
DAPK1 is one of the most widely studied methylated genes. DAPK1 methylation is frequently found in a wide array of over 20 tumor types.Citation34 DAPK1 is a tumor suppressor gene, and methylation of this gene has been associated with shorter disease-free survival in surgically treated Stage I lung tumorsCitation34 and with metastasis in several tumor types including, head and neck tumors.Citation35 This latter study, which used similar primers, found comparable rates of DAPK1 methylation, 15/79 (19%) overall (compared to 16% in our study), and a significant association with N status (27% methylation in N+ group, compared to 26% in our study), confirming the results found in our study. In contrast to the studies in leiomyosarcoma and urothelial carcinoma that utilized the same immunohistochemical scoring method and found associations with methylation status,Citation36,37 we did not find an association between DAPK1 methylation and protein expression. Because this scoring method might not be reliable in OOSCC, we also analyzed high- and low-expression compared to the median (percentage of tumor cells having moderate or strong expression), and associated this with DAPK1 methylation. Again, no significant associations were found.DAPK1 is a serine/threonine kinase involved in several mechanisms linked to cell death and autophagy. It has pro-apoptotic activity by suppressing integrin-mediated survival signals, thus inducing a specific form of apoptosis, called anoikis. Tumor cells that have loss of anoikis by inactivated DAPK1 are more likely to survive during migration and, therefore, more likely to cause metastases.Citation38 Furthermore, DAPK1 has an antimigratory effect by blocking integrin-mediated cell polarization.Citation39 Therefore, DAPK1 downregulation by hypermethylation increases metastasis and tumor cell survival.
MGMT is a DNA repair enzyme. MGMT methylation is mostly known for being predictive for better response to alkylating chemotherapy in glioblastoma and, to a lesser extent, to radiotherapy.Citation40 In OOSCC, several studies assessing MGMT methylation using various techniques did not find associations with N status.Citation41,42 However, in a large study of >200 laryngeal and hypopharyngeal tumors, MGMT methylation was significantly associated with N0 status.Citation43 In that study, the same primers were used and a comparable MGMT methylation rate of 27% was found (also 27% in our study). How the higher methylation rates in pN0 cases affect the metastatic potential of OOSCC is not clear. Loss of the repair function of MGMT may increase the accumulation of mutations, especially in smoking-induced tumors, such as OOSCC. Because smoking is associated with higher methylation rates in generalCitation44 and methylation of MGMT specifically,Citation45 MGMT methylation might be a pseudo marker for smoking-induced tumors, rather than for HPV-associated tumors, which are more frequently pN+, according to some authors.Citation46 However, MGMT methylation was not associated with HPV status in our study (data not shown), nor in another study with more HPV-positive cases.Citation47 In our series, we show for the first time that in OOSCC, MGMT methylation is associated with a decreased expression in the invasive tumor front, but not in the tumor center (). This is in line with the reported heterogeneity of methylation markers and their associated proteins,Citation26,31 and with the fact that methylation is associated with heterogeneous rather than with overall low expression.Citation29
The negative predictive value (NPV) of the combined model of DAPK1 and MGMT methylation of 76% in the current study is even slightly better than the 72% found in a 696-gene expression signature.Citation48 However, a NPV of over 80% is needed to outperform current clinical nodal staging techniques,Citation33 including sentinel lymph node biopsy.Citation49 Obviously, further validation of the methylation markers, especially on the clinically most relevant subgroup of pT1-2cN0 cases, is needed. In the current study, both DAPK1 and MGMT were non-significant predictors in the pT1-2cN0 subgroup (n = 37; data not shown). Treatment of OOSCC patients using demethylating drugs may not be effective, as our study shows that demethylation of DAPK1 might be beneficial, but demethylation of MGMT might result in nodal disease.
MSP is not a quantitative technique. Although quantitative MSP for DAPK1 and MGMT enables specific cut-off values, thus customizing sensitivity and specificity, MSP is a more suitable technique for assessing a set of markers because it is a quick, low-cost and sensitive technique, able to detect a single methylated allele in a background of 1,000 unmethylated alleles.Citation50 However, selecting and testing of various possible methylation markers proved to be an inefficient method to identify new predictive markers. To improve marker selection efficiency, genome-wide methods are needed.Citation51
In conclusion, we analyzed 28 candidate methylation markers for their predictive value for N status by MSP on a large clinical group of OOSCC. MGMT and DAPK1 were identified as predictors of nodal metastasis in OOSCC with a high predictive value and specificity and sensitivity comparable to other markers previously reported. In addition, we showed for the first time that MGMT methylation is associated with a decreased expression in the invasive tumor front. This confirms the predictive value of methylation markers and the biological impact of methylation on the metastatic potential of OOSCC. In the future, DAPK1 and MGMT might be included in a panel of methylation markers that aid the clinician in the assessment of the N status, improving patient diagnosis and treatment selection.
Materials and Methods
Selection of candidate genes
To select candidate genes that are regulated by methylation and associated with lymph node metastasis, we used reported microarray data from 4 independent studies in HNSCC.Citation21-24 All selected candidate genes should have a CpG island present in the promoter region of the gene, and a negative correlation with nodal metastases, as methylated genes have an associated downregulation on mRNA level. From these lists of genes we selected (): (1) all genes found in more than one of the 4 expression profilesCitation21-24; (2) the 5 highest ranking genes from the 2 studies that performed genome-wide arraysCitation21,24; (3) candidate genes that were reported in the 4 HNSCC expression profilesCitation21-24 and showed functional methylation (increased expression after treatment with 5-aza-2′-deoxycytidine (DAC)/ trichostatin A (TSA) in vitro and an association with lymph node metastasis in cervical squamous cell carcinoma, in a previous study performed in our lab.Citation25,30 Furthermore, 4 genes were selected that have been described to be associated with invasion and metastasis in squamous cell carcinoma: GJB6,Citation52 OCLN, Citation53 TJP1, Citation54 and CD44.Citation55
In this way, a total of 24 genes were selected that were not reported to be methylated in OOSCC and, consequently, were potential new candidate metastasis-associated genes whose expression might be regulated by methylation.
Four genes (MLH1, MGMT, CDKN2A, and DAPK1) were included that showed frequent methylation in HNSCC in the literatureCitation41,43,56,57 ().
MSP primer design
For optimal MSP primer design in a region with the highest chance of finding differentially methylated regions,Citation33 all candidate genes were checked for the presence of a CpG island in a range of −500 to +500 bp relative to the TSS, and primers were designed in this region using Methyl Primer Design software [Applied Biosystems, Foster City, CA, USA]. Primers that were selected generally had 3 CGs in their sequence. Maximum product size was set at 160 bp, due to working with DNA isolated from FFPE tissue. For MGMT, CDKN2A, and DAPK1, primer sequences from literature were usedCitation50,58,59 (see supplementary data 2 for all primer sequences).
Candidate gene testing strategy
Selected candidate genes were tested for optimal annealing temperature and MgCl2 concentration on a set of 8 HNSCC cell lines (UMSCC-1, UMSCC-2, UMSCC-8, UMSCC-11a, UMSCC-14a, vuSCC-40, vuSCC-78, vuSCC-96) and 2 normal tonsil FFPE samples. After optimization, MSPs were performed on a set of 5 N0 and 5 N+ tumors. All markers that showed methylation in 2 or more tumor samples were further tested on our total patient series (n = 70: 32 pN0 and 38 pN+; ). All tumor samples were tested twice in separate experiments. Samples with discordant results were tested for a third time.
Patient selection
From the database of the Netherlands Cancer Registry, all records with the following criteria were retrieved: oral or oropharyngeal primary tumor location (ICD-O-3 locations 00.3–6.9 and 9.0–10.9), histologically proven squamous cell carcinoma, diagnosed between 1997 and 2008, treated in the UMC Groningen, without prior head and neck or systemic oncological treatment, as reported previously.Citation60 For all tumors, information was collected regarding patient characteristics (e.g., previous cancer treatments, other diagnoses, last follow-up, recurrences, date, and cause of death), clinical tumor characteristics (e.g., localization, lateralization, synchronicity, cTNM, method of nodal diagnosis, and treatment), and pathological tumor characteristics (e.g., pTNM, histology, perineural and lymphovascular invasion status, margin status, nodal status, and infiltration depth). All FFPE tissue blocks and original hematoxylin and eosin (HE)-slides were retrieved from the archives of the Department of Pathology. The histopathological diagnoses were revised for all cases by an experienced head and neck pathologist. All patient tissues were coded. All data and tissues were treated according to the Code for Proper Secondary Use of Human Tissue in the Netherlands,Citation61 as well as to the relevant institutional and national guidelines.
Tissue microarrays (TMAs) were constructed as reported previously.Citation60 For the current study, we selected 2 TMAs that contained 70 randomly selected first primary tumors (32 pN0 and 38 pN+) that were treated by resection and neck dissection and for which tissue was available to perform MSP and immunohistochemistry (). HPV status was previously assessed for 64/70 (91%) cases using a triple algorithm, including p16 expression, HPV-PCR and HPV-BRISH, which identified high-risk HPV in 3/64 (5%).Citation62
DNA isolation
From the FFPE blocks of the tumors, 2 10-μm thick sections were cut and used for DNA extraction. Subsequently, a 3-μm thick section was cut and HE-stained to check if tumor load was sufficient through the sections (preferably >60%). After deparaffinization, DNA isolation was performed, using standard salt-chloroform extraction and ethanol precipitation.Citation62 For quality control, genomic DNA was amplified in a multiplex PCR containing a control gene primer set resulting in products of 100, 200 300, 400, and 600 bp, according to the BIOMED-2 protocol.Citation63 Only cases with products ≥200 bp were included for further analysis.
Bisulfite treatment and methylation-specific PCR (MSP)
Bisulfite-converted DNA (bisDNA) was made using the EZ DNA methylation kit according to the manufacturer's protocol (Zymo Research, Irvine, CA, USA). Methylation specific PCR (MSP) was performed using 20 ng bisDNA. All MSPs were run as follows: 10 min 95°C, 40 times (1 min 95°C, 1 min Ta, 1 min 72°C), 10 min 72°C, ∞ 4°C. Controls consisted of leukocyte DNA that was in vitro methylated by SssI methyltransferase (methylated control) or untreated leukocyte DNA (unmethylated control). Adequate bisulfite conversion was checked by β-actin MSP (Forward: 5′TAGGGAGTATATAGGTTGGGGAAGTT 3′; Reverse: 5′AACACACAATAACAAACACAAATTCAC 3′). A sample was considered methylated when the methylated product of the right size was visible. It was considered unmethylated when the unmethylated product of the right size was visible and no methylated product was visible. A sample was considered not assessable, when no unmethylated and methylated products of the right size were present. Methylation- and unmethylation-specific PCRs were performed in parallel, and performed at the same annealing temperature (Ta), on the same plate.
Immunohistochemistry
TMA sections were deparaffinized in xylene and rehydrated in a graded alcohol series. Antigen retrieval was performed by heating in a microwave oven for 15 min in either Tris/EDTA pH = 9.0 (for MGMT) or EDTA pH = 8.0 (for DAPK1). After antigen retrieval endogenous peroxide was blocked by incubating the slide in 0.3% peroxide solution. After one-hour incubation with anti-MGMT 1:100 (MT3.1, Millipore, Billerica, MA, USA) or anti-DAPK1 1:200 (D1319, Sigma-Aldrich, St. Louis MO, USA), a horseradish peroxidase-conjugated secondary antibody was used, followed by a horseradish conjugated tertiary antibody. Slides were developed with di-aminobenzidene chromogen solution, followed by hematoxylin counterstaining. In addition to the control tissues included on the TMA slide, full sections of the control tissue, specific for each staining, were also included (normal liver for MGMTCitation64; normal duodenum for DAPK1Citation65).
Analysis of immunohistochemistry
Cases were semi-quantitatively scored, assessing percentage of tumor cells stained and the intensity of staining (0, no staining; 1, weak; 2, moderate; 3, strong). Staining was scored by 2 observers, independently. Discordant results were discussed until consensus was reached. High MGMT expression was defined as moderate to strong nuclear expression in ≥10% of tumor cells, as reported previously.Citation66-68 For DAPK1, scores were given to cell proportion: 0: staining in <1% of tumor cells; 1: staining in 1–10%; 2: staining in 11–50%; and 3: staining in >50% of tumor cells. Intensity was then scored as 0: negative; 1: weak; 2: moderate; and 3: strong. The final score (ranging 0–9) was obtained by multiplying the cell proportion by the intensity. A final score of <4 was considered to indicate low expression, and ≥4 was considered high expression.Citation36,37
Statistical analysis
Statistical analysis was performed with SPSS version 20. Categorical data were compared using the Chi-square test, or Fisher's exact test, when appropriate. Univariate logistic regression was used to assess the relationship between predictor variables and the dichotomous pN status. All predictor variables with P < 0.10 in univariate logistic regression were entered in multiple logistic regression. All tests were performed 2-tailed. Results were considered significant when P < 0.05.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
1075689_Supplemental_Material.docx
Download MS Word (24.2 KB)Supplemental Material
Supplemental data for this article can be accessed on the publisher's website.
Funding
This work was partly funded by the CTMM Air Force consortium (http://www.ctmm.nl). CTMM pays for part of the salary of MJAMC and MFM and had no role in study design, data collection and analysis, decision to publish, and preparation of the manuscript.
References
- Siegel R, Ma J, Zou Z, Jemal A. Cancer statistics, 2014. CA Cancer J Clin 2014; 64:9-29; PMID:24399786; http://dx.doi.org/10.3322/caac.21208
- Arduino PG, Carrozzo M, Chiecchio A, Broccoletti R, Tirone F, Borra E, Bertolusso G, Gandolfo S. Clinical and histopathologic independent prognostic factors in oral squamous cell carcinoma: A retrospective study of 334 cases. J Oral Maxillofac Surg 2008; 66:1570-9.; http://dx.doi.org/10.1016/j.joms.2007.12.024
- Liao LJ, Lo WC, Hsu WL, Wang CT, Lai MS. Detection of cervical lymph node metastasis in head and neck cancer patients with clinically N0 neck-a meta-analysis comparing different imaging modalities. BMC Cancer 2012; 12:236, 2407-12-236; PMID:22691269; http://dx.doi.org/10.1186/1471-2407-12-236
- Vorburger MS, Broglie MA, Soltermann A, Haerle SK, Haile SR, Huber GF, Stoeckli SJ. Validity of frozen section in sentinel lymph node biopsy for the staging in oral and oropharyngeal squamous cell carcinoma. J Surg Oncol 2012; 106:816-9; PMID:22585742; http://dx.doi.org/10.1002/jso.23156
- Terada A, Hasegawa Y, Goto M, Sato E, Hyodo I, Ogawa T, Nakashima T, Yatabe Y. Sentinel lymph node radiolocalization in clinically negative neck oral cancer. Head Neck 2006; 28:114-20; PMID:16155916; http://dx.doi.org/10.1002/hed.20305
- Baylin SB, Jones PA. A decade of exploring the cancer epigenome - biological and translational implications. Nat Rev Cancer 2011; 11:726-34; PMID:21941284; http://dx.doi.org/10.1038/nrc3130
- Esteller M. Cancer epigenomics: DNA methylomes and histone-modification maps. Nat Rev Genet 2007; 8:286-98; PMID:17339880; http://dx.doi.org/10.1038/nrg2005
- Roossink F, de Jong S, Wisman GB, van der Zee AG, Schuuring E. DNA hypermethylation biomarkers to predict response to cisplatin treatment, radiotherapy or chemoradiation: The present state of art. Cell Oncol (Dordr) 2012; 35:231-41; PMID:22836879; http://dx.doi.org/10.1007/s13402-012-0091-7
- Smiraglia DJ, Smith LT, Lang JC, Rush LJ, Dai Z, Schuller DE, Plass C. Differential targets of CpG island hypermethylation in primary and metastatic head and neck squamous cell carcinoma (HNSCC). J Med Genet 2003; 40:25-33; PMID:12525538.; http://dx.doi.org/10.1136/jmg.40.1.25
- Rius M, Lyko F. Epigenetic cancer therapy: Rationales, targets and drugs. Oncogene 2012; 31:4257-65; PMID:22179827; http://dx.doi.org/10.1038/onc.2011.601
- Cock-Rada A, Weitzman JB. The methylation landscape of tumour metastasis. Biol Cell 2013; 105:73-90; PMID:23198959; http://dx.doi.org/10.1111/boc.201200029
- Kristensen LS, Hansen LL. PCR-based methods for detecting single-locus DNA methylation biomarkers in cancer diagnostics, prognostics, and response to treatment. Clin Chem 2009; 55:1471-83; PMID:19520761; http://dx.doi.org/10.1373/clinchem.2008.121962
- Jacinto FV, Esteller M. MGMT hypermethylation: A prognostic foe, a predictive friend. DNA Repair (Amst) 2007; 6:1155-60; PMID:17482895; http://dx.doi.org/10.1016/j.dnarep.2007.03.013
- Gasche JA, Goel A. Epigenetic mechanisms in oral carcinogenesis. Future Oncol 2012; 8:1407-25; PMID:23148615; http://dx.doi.org/10.2217/fon.12.138
- Jithesh PV, Risk JM, Schache AG, Dhanda J, Lane B, Liloglou T, Shaw RJ. The epigenetic landscape of oral squamous cell carcinoma. Br J Cancer 2013; 108:370-9; PMID:23287992; http://dx.doi.org/10.1038/bjc.2012.568
- Gonzalez-Ramirez I, Garcia-Cuellar C, Sanchez-Perez Y, Granados-Garcia M. DNA methylation in oral squamous cell carcinoma: Molecular mechanisms and clinical implications. Oral Dis 2011; 17:771-8; PMID:21781230; http://dx.doi.org/10.1111/j.1601-0825.2011.01833.x
- Mascolo M, Siano M, Ilardi G, Russo D, Merolla F, De Rosa G, Staibano S. Epigenetic disregulation in oral cancer. Int J Mol Sci 2012; 13:2331-53; PMID:22408457; http://dx.doi.org/10.3390/ijms13022331
- Borges S, Doppler H, Perez EA, Andorfer CA, Sun Z, Anastasiadis PZ, Thompson EA, Geiger XJ, Storz P. Pharmacologic reversion of epigenetic silencing of the PRKD1 promoter blocks breast tumor cell invasion and metastasis. Breast Cancer Res 2013; 15:R66; PMID:23971832; http://dx.doi.org/10.1186/bcr3460
- Li Y, Zhu CL, Nie CJ, Li JC, Zeng TT, Zhou J, Chen J, Chen K, Fu L, Liu H, et al. Investigation of tumor suppressing function of CACNA2D3 in esophageal squamous cell carcinoma. PLoS One 2013; 8:e60027; PMID:23560067; http://dx.doi.org/10.1371/journal.pone.0060027
- Xing X, Cai W, Shi H, Wang Y, Li M, Jiao J, Chen M. The prognostic value of CDKN2A hypermethylation in colorectal cancer: A meta-analysis. Br J Cancer 2013; 108:2542-8; PMID:23703248; http://dx.doi.org/10.1038/bjc.2013.251
- Roepman P, Wessels LF, Kettelarij N, Kemmeren P, Miles AJ, Lijnzaad P, Tilanus MG, Koole R, Hordijk GJ, van d V, et al. An expression profile for diagnosis of lymph node metastases from primary head and neck squamous cell carcinomas. Nat Genet 2005; 37:182-6.; http://dx.doi.org/10.1038/ng1502
- Schmalbach CE, Chepeha DB, Giordano TJ, Rubin MA, Teknos TN, Bradford CR, Wolf GT, Kuick R, Misek DE, Trask DK, et al. Molecular profiling and the identification of genes associated with metastatic oral cavity/pharynx squamous cell carcinoma. Arch Otolaryngol Head Neck Surg 2004; 130:295-302.; http://dx.doi.org/10.1001/archotol.130.3.295
- Mendez E, Houck JR, Doody DR, Fan W, Lohavanichbutr P, Rue TC, Yueh B, Futran ND, Upton MP, Farwell DG, et al. A genetic expression profile associated with oral cancer identifies a group of patients at high risk of poor survival. Clin Cancer Res 2009; 15:1353-61.; http://dx.doi.org/10.1158/1078-0432.CCR-08-1816
- O'Donnell RK, Kupferman M, Wei SJ, Singhal S, Weber R, O'Malley B, Cheng Y, Putt M, Feldman M, Ziober B, et al. Gene expression signature predicts lymphatic metastasis in squamous cell carcinoma of the oral cavity. Oncogene 2005; 24:1244-51.; http://dx.doi.org/10.1038/sj.onc.1208285
- Ongenaert M, Wisman GB, Volders HH, Koning AJ, van der Zee AG, van Criekinge W, Schuuring E. Discovery of DNA methylation markers in cervical cancer using relaxation ranking. BMC Med Genomics 2008; 1:57.; http://dx.doi.org/10.1186/1755-8794-1-57
- Cao VT, Jung TY, Jung S, Jin SG, Moon KS, Kim IY, Kang SS, Park CS, Lee KH, Chae HJ. The correlation and prognostic significance of MGMT promoter methylation and MGMT protein in glioblastomas. Neurosurgery 2009; 65:866, 75; discussion 875; PMID:19834398; http://dx.doi.org/10.1227/01.NEU.0000357325.90347.A1
- de Meneses IS, de Souza RR, Jeraldo Verónica de Lourdes Sierpe, Rodrigues D, Cavalcante R, Reis FP, de Albuquerque Júnior, Ricardo Luiz Cavalcanti, MENESES I, SOUZA R, JERALDO V. Death-associated protein kinase is underexpressed in high-grade oral squamous cell carcinoma. Int J Morphol 2010; 28:609-13.; http://dx.doi.org/10.4067/S0717-95022010000200044
- Esteller M, Corn PG, Baylin SB, Herman JG. A gene hypermethylation profile of human cancer. Cancer Res 2001; 61:3225-9; PMID:11309270.
- Ingold B, Schraml P, Heppner FL, Moch H. Homogeneous MGMT immunoreactivity correlates with an unmethylated MGMT promoter status in brain metastases of various solid tumors. PLoS One 2009; 4:e4775; PMID:19274096; http://dx.doi.org/10.1371/journal.pone.0004775
- Noordhuis MG, Fehrmann RS, Wisman GB, Nijhuis ER, van Zanden JJ, Moerland PD, Ver Loren van Themaat E, Volders HH, Kok M, ten Hoor KA, et al. Involvement of the TGF-beta and beta-catenin pathways in pelvic lymph node metastasis in early-stage cervical cancer. Clin Cancer Res 2011; 17:1317-30; PMID:21385933; http://dx.doi.org/10.1158/1078-0432.CCR-10-2320
- Rastetter M, Schagdarsurengin U, Lahtz C, Fiedler E, Marsch WC, Dammann R, Helmbold P. Frequent intra-tumoural heterogeneity of promoter hypermethylation in malignant melanoma. Histol Histopathol 2007; 22:1005-15; PMID:17523078.
- Braakhuis BJ, Brakenhoff RH, Leemans CR. Gene expression profiling in head and neck squamous cell carcinoma. Curr Opin Otolaryngol Head Neck Surg 2010; 18:67-71; PMID:20084003; http://dx.doi.org/10.1097/MOO.0b013e32833693ce
- Irizarry RA, Ladd-Acosta C, Wen B, Wu Z, Montano C, Onyango P, Cui H, Gabo K, Rongione M, Webster M, et al. The human colon cancer methylome shows similar hypo- and hypermethylation at conserved tissue-specific CpG island shores. Nat Genet 2009; 41:178-86; PMID:19151715; http://dx.doi.org/10.1038/ng.298
- Gozuacik D, Kimchi A. DAPk protein family and cancer. Autophagy 2006; 2:74-9; PMID:17139808.; http://dx.doi.org/10.4161/auto.2.2.2459
- Sanchez-Cespedes M, Esteller M, Wu L, Nawroz-Danish H, Yoo GH, Koch WM, Jen J, Herman JG, Sidransky D. Gene promoter hypermethylation in tumors and serum of head and neck cancer patients. Cancer Res 2000; 60:892-5.
- Kawaguchi K, Oda Y, Saito T, Yamamoto H, Takahira T, Tamiya S, Iwamoto Y, Tsuneyoshi M. Death-associated protein kinase (DAP kinase) alteration in soft tissue leiomyosarcoma: Promoter methylation or homozygous deletion is associated with a loss of DAP kinase expression. Hum Pathol 2004; 35:1266-71; PMID:15492995.; http://dx.doi.org/10.1016/j.humpath.2004.07.007
- Chen WT, Hung WC, Kang WY, Huang YC, Chai CY. Urothelial carcinomas arising in arsenic-contaminated areas are associated with hypermethylation of the gene promoter of the death-associated protein kinase. Histopathology 2007; 51:785-92; PMID:17953697; http://dx.doi.org/10.1111/j.1365-2559.2007.02871.x
- Wang WJ, Kuo JC, Yao CC, Chen RH. DAP-kinase induces apoptosis by suppressing integrin activity and disrupting matrix survival signals. J Cell Biol 2002; 159:169-79; PMID:12370243; http://dx.doi.org/10.1083/jcb.200204050
- Kuo JC, Wang WJ, Yao CC, Wu PR, Chen RH. The tumor suppressor DAPK inhibits cell motility by blocking the integrin-mediated polarity pathway. J Cell Biol 2006; 172:619-31; PMID:16476779; http://dx.doi.org/10.1083/jcb.200505138
- Olson RA, Brastianos PK, Palma DA. Prognostic and predictive value of epigenetic silencing of MGMT in patients with high grade gliomas: A systematic review and meta-analysis. J Neurooncol 2011; 105:325-35; PMID:21523485; http://dx.doi.org/10.1007/s11060-011-0594-5
- Supic G, Kozomara R, Brankovic-Magic M, Jovic N, Magic Z. Gene hypermethylation in tumor tissue of advanced oral squamous cell carcinoma patients. Oral Oncol 2009; 45:1051-7; PMID:19665921; http://dx.doi.org/10.1016/j.oraloncology.2009.07.007
- Su PF, Huang WL, Wu HT, Wu CH, Liu TY, Kao SY. p16(INK4A) promoter hypermethylation is associated with invasiveness and prognosis of oral squamous cell carcinoma in an age-dependent manner. Oral Oncol 2010; 46:734-9; PMID:20729138; http://dx.doi.org/10.1016/j.oraloncology.2010.07.002
- Dikshit RP, Gillio-Tos A, Brennan P, De Marco L, Fiano V, Martinez-Penuela JM, Boffetta P, Merletti F. Hypermethylation, risk factors, clinical characteristics, and survival in 235 patients with laryngeal and hypopharyngeal cancers. Cancer 2007; 110:1745-51; PMID:17786935; http://dx.doi.org/10.1002/cncr.22975
- Zeilinger S, Kuhnel B, Klopp N, Baurecht H, Kleinschmidt A, Gieger C, Weidinger S, Lattka E, Adamski J, Peters A, et al. Tobacco smoking leads to extensive genome-wide changes in DNA methylation. PLoS One 2013; 8:e63812; PMID:23691101; http://dx.doi.org/10.1371/journal.pone.0063812
- Christmann M, Kaina B. O(6)-methylguanine-DNA methyltransferase (MGMT): Impact on cancer risk in response to tobacco smoke. Mutat Res 2012; 736:64-74; PMID:21708177; http://dx.doi.org/10.1016/j.mrfmmm.2011.06.004
- Lassen P. The role of human papillomavirus in head and neck cancer and the impact on radiotherapy outcome. Radiother Oncol 2010; 95:371-80; PMID:20493569; http://dx.doi.org/10.1016/j.radonc.2010.04.022
- Weiss D, Basel T, Sachse F, Braeuninger A, Rudack C. Promoter methylation of cyclin A1 is associated with human papillomavirus 16 induced head and neck squamous cell carcinoma independently of p53 mutation. Mol Carcinog 2011; 50:680-8; PMID:21563216; http://dx.doi.org/10.1002/mc.20798
- van Hooff SR, Leusink FK, Roepman P, Baatenburg de Jong RJ, Speel EJ, van den Brekel MW, van Velthuysen ML, van Diest PJ, van Es RJ, Merkx MA, et al. Validation of a gene expression signature for assessment of lymph node metastasis in oral squamous cell carcinoma. J Clin Oncol 2012; 30:4104-10; PMID:23045589; http://dx.doi.org/10.1200/JCO.2011.40.4509
- Alkureishi LW, Ross GL, Shoaib T, Soutar DS, Robertson AG, Thompson R, Hunter KD, Sorensen JA, Thomsen J, Krogdahl A, et al. Sentinel node biopsy in head and neck squamous cell cancer: 5-year follow-up of a european multicenter trial. Ann Surg Oncol 2010; 17:2459-64; PMID:20552410; http://dx.doi.org/10.1245/s10434-010-1111-3
- Herman JG, Graff JR, Myohanen S, Nelkin BD, Baylin SB. Methylation-specific PCR: A novel PCR assay for methylation status of CpG islands. Proc Natl Acad Sci U S A 1996; 93:9821-6.; http://dx.doi.org/10.1073/pnas.93.18.9821
- Bock C, Tomazou EM, Brinkman AB, Muller F, Simmer F, Gu H, Jager N, Gnirke A, Stunnenberg HG, Meissner A. Quantitative comparison of genome-wide DNA methylation mapping technologies. Nat Biotechnol 2010; 28:1106-14; PMID:20852634; http://dx.doi.org/10.1038/nbt.1681
- Erdem NF, Carlson ER, Gerard DA. Characterization of gene expression profiles of 3 different human oral squamous cell carcinoma cell lines with different invasion and metastatic capacities. J Oral Maxillofac Surg 2008; 66:918-27; PMID:18423281; http://dx.doi.org/10.1016/j.joms.2007.12.036
- Martin TA, Mansel RE, Jiang WG. Loss of occludin leads to the progression of human breast cancer. Int J Mol Med 2010; 26:723-34; PMID:20878095.; http://dx.doi.org/10.3892/ijmm_00000519
- Blanco D, Vicent S, Elizegi E, Pino I, Fraga MF, Esteller M, Saffiotti U, Lecanda F, Montuenga LM. Altered expression of adhesion molecules and epithelial-mesenchymal transition in silica-induced rat lung carcinogenesis. Lab Invest 2004; 84:999-1012; PMID:15195114; http://dx.doi.org/10.1038/labinvest.3700129
- Perez A, Neskey DM, Wen J, Pereira L, Reategui EP, Goodwin WJ, Carraway KL, Franzmann EJ. CD44 interacts with EGFR and promotes head and neck squamous cell carcinoma initiation and progression. Oral Oncol 2013; 49:306-13; PMID:23265944; http://dx.doi.org/10.1016/j.oraloncology.2012.11.009
- Puri SK, Si L, Fan CY, Hanna E. Aberrant promoter hypermethylation of multiple genes in head and neck squamous cell carcinoma. Am J Otolaryngol 2005; 26:12-7; PMID:15635575.; http://dx.doi.org/10.1016/j.amjoto.2004.06.007
- Tawfik HM, El-Maqsoud NM, Hak BH, El-Sherbiny YM. Head and neck squamous cell carcinoma: Mismatch repair immunohistochemistry and promoter hypermethylation of hMLH1 gene. Am J Otolaryngol 2011; 32:528-36; PMID:21353335
- Rosas SL, Koch W, da Costa Carvalho MG, Wu L, Califano J, Westra W, Jen J, Sidransky D. Promoter hypermethylation patterns of p16, O6-methylguanine-DNA-methyltransferase, and death-associated protein kinase in tumors and saliva of head and neck cancer patients. Cancer Res 2001; 61:939-42; PMID:11221887.
- Esteller M, Hamilton SR, Burger PC, Baylin SB, Herman JG. Inactivation of the DNA repair gene O6-methylguanine-DNA methyltransferase by promoter hypermethylation is a common event in primary human neoplasia. Cancer Res 1999; 59:793-7; PMID:10029064.
- Melchers LJ, Bruine de Bruin L, Schnell U, Slagter-Menkema L, Mastik MF, de Bock GH, van Dijk BAC, Giepmans BNG, van der Laan BFAM, van der Wal JE, et al. Lack of claudin-7 is a strong predictor of regional recurrence in oral and oropharyngeal squamous cell carcinoma. Oral Oncol 2013; 49:998-1005; http://dx.doi.org/10.1016/j.oraloncology.2013.07.008
- Federation of Medical Scientific Societies. Code for proper secondary use of human tissue in the netherlands. 2011; 2012.
- Melchers L, Mastik M, Cameron BS, van Dijk B, de Bock G, van der Laan B, van der Vegt B, Speel E, Roodenburg J, Witjes M. Detection of HPV-associated oropharyngeal tumours in a 16-year cohort: More than meets the eye. Br J Cancer 2015; 112(8):1349-57; http://dx.doi.org/10.1038/bjc.2015.99
- van Dongen JJ, Langerak AW, Bruggemann M, Evans PA, Hummel M, Lavender FL, Delabesse E, Davi F, Schuuring E, Garcia-Sanz R, et al. Design and standardization of PCR primers and protocols for detection of clonal immunoglobulin and T-cell receptor gene recombinations in suspect lymphoproliferations: Report of the BIOMED-2 concerted action BMH4-CT98-3936. Leukemia 2003; 17:2257-317; PMID:14671650; http://dx.doi.org/10.1038/sj.leu.2403202
- Grafstrom RC, Pegg AE, Trump BF, Harris CC. O6-alkylguanine-DNA alkyltransferase activity in normal human tissues and cells. Cancer Res 1984; 44:2855-7; PMID:6722814.
- Santa Cruz Biotechnology I. Datasheet DAPK (H-300). 2013; 2013.
- Rodriguez MJ, Acha A, Ruesga MT, Rodriguez C, Rivera JM, Aguirre JM. Loss of expression of DNA repair enzyme MGMT in oral leukoplakia and early oral squamous cell carcinoma. A prognostic tool? Cancer Lett 2007; 245:263-8; PMID:16517062; http://dx.doi.org/10.1016/j.canlet.2006.01.015
- Sawhney M, Rohatgi N, Kaur J, Gupta SD, Deo SV, Shukla NK, Ralhan R. MGMT expression in oral precancerous and cancerous lesions: Correlation with progression, nodal metastasis and poor prognosis. Oral Oncol 2007; 43:515-22; PMID:16996781; http://dx.doi.org/10.1016/j.oraloncology.2006.05.007
- Huang SH, Lee HS, Mar K, Ji DD, Huang MS, Hsia KT. Loss expression of O6-methylguanine DNA methyltransferase by promoter hypermethylation and its relationship to betel quid chewing in oral squamous cell carcinoma. Oral Surg Oral Med Oral Pathol Oral Radiol Endod 2010; 109:883-9; PMID:20451846; http://dx.doi.org/10.1016/j.tripleo.2009.12.019
