ABSTRACT
Kisspeptin, produced from the brain and peripheral tissues, may constitute an important link in metabolic regulation in response to external cues, such as diet. The kisspeptin system is well described in the brain. However, its function and regulation in the peripheral tissues, especially in relation to metabolic disease and sex differences, remain to be elucidated. As Kiss1 and Kiss1r, encoding for kisspeptin and kisspeptin receptors, respectively, are altered by overnutrition/fasting and regulated by DNA methylation during puberty and cancer, epigenetic mechanisms in metabolic disorders are highly probable. In the present study, we experimentally induced type 2 diabetes mellitus (DM2) in female Wistar rats using high-fat diet/streptozocin. We analysed expression and DNA methylation of Kiss1 and Kiss1r in the peripheral tissues, using quantitative-reverse-transcription PCR (qRT-PCR) and pyrosequencing. We discovered differential expression of Kiss1 and Kiss1r in peripheral organs in DM2 females, as compared with healthy controls, and the profile differed from patterns reported earlier in males. DM2 in females was linked to the increased Kiss1 mRNA in the liver and increased Kiss1r mRNA in the liver and adipose tissue. However, Kiss1r promoter was hypermethylated in the liver, suggesting gene silencing. Indeed, the increase in DNA methylation of Kiss1r promoter was accompanied by a reduction in Kiss1r protein, implying epigenetic or translational gene repression. Our results deliver novel evidence for tissue-specific differences in Kiss1 and Kiss1r expression in peripheral organs in DM2 females and suggest DNA methylation as a player in regulation of the hepatic kisspeptin system in DM2.
Introduction
Kisspeptin (KP), encoded by the KISS1 gene, was discovered in 1996 as a peptide with anti-metastatic properties and named metastin Citation1. Later in 2006, two independent groups of researchers proved that the absence of KP or its functional receptor, KISS1R, resulted in hypogonadotropic hypogonadism (HH), a condition characterized by low sex steroids and gonadotropin levels [Citation2,Citation3]. Since that time, the crucial role of this peptide in regulating the hypothalamic-pituitary-gonadal (HPG) axis, which governs reproduction, was confirmed by many studies [Citation4–6]. KP is now considered the most potent stimulator of gonadotropin-releasing hormone (GnRH), which regulates the HPG axis [Citation4–6]. In addition to the importance in reproduction and puberty, KP was found to play a role in sending a cue to the HPG axis about the metabolic status of the organism [Citation7,Citation8]. This is possible due to the presence of KP and KP receptors in the brain, including the arcuate nucleus (ARC) that is a central side of the metabolic regulation, as well as in the peripheral tissues that control metabolic functions (i.e., pancreas, liver, and adipose tissue) [Citation7,Citation8]. Indeed, levels of the KP system in the brain have been shown to be altered in metabolic imbalances, e.g., obesity, type 2 diabetes mellitus (DM2), and starvation [Citation9,Citation10]. In addition, Kiss1r knockout (KO) mice had altered metabolic phenotypes with increased adiposity and reduced energy expenditure [Citation10].
While the central role of KP at the brain level is relatively well described, the function of the KP system in the peripheral tissues and KP-mediated regulation of metabolism at the periphery remain largely unknown. A piece of evidence comes from a study by Song et al., which reports on the tri-hormonal endocrine circuit between the pancreatic islets and the liver [Citation11]. The findings of this study suggest that glucagon produced by α-cells in the pancreas stimulates the cAMP/PKA/CREB signalling pathway in the liver, which upregulates expression of gluconeogenic genes and KISS1. These events lead to an increase in the production of glucose and KP in the liver. Glucose released into the bloodstream stimulates insulin release from the pancreatic β-cells, whereas KP released by the liver works in the opposite way and suppresses glucose-stimulated insulin secretion. This is a well-designed way of KP-mediated regulation of blood glucose levels and endocrine pancreatic functions. In terms of expression of the KP system at the periphery, we have recently reported tissue-specific KP alterations in male rats with experimentally induced DM2 [Citation12]. The study included only male rats, and thus, it remains unknown whether and how the KP system in peripheral tissue changes in DM2 females. There is only one report on the levels of KP in the adipose tissue in response to nutritional status where males and females were considered separately. Brown et al. observed a decrease in fat Kiss1 mRNA in Sprague-Dawley rats fed a high-fat diet (HFD) and in obese Zucker rats in both males and females [Citation13].
In the present study, we have assessed the status of Kiss1 and Kiss1r in peripheral tissues that are involved in metabolic functions in female rats with DM2. We have detected an increase in mRNA levels of Kiss1r in the liver and fat tissue, and a higher mRNA expression of Kiss1 in the liver of female rats with HFD and streptozotocin (STZ)-induced DM2, as compared with healthy females on a chow diet. We next asked a question about the potential mechanism behind the observed changes in gene expression. Since DM2 is often aetiologically linked to lifestyle factors, which is also reflected in our HFD-related rat model, an epigenetic mechanism becomes an obvious suspect. Epigenetics refers to changes in gene expression without changes in the DNA sequence [Citation14]. Such changes are dynamic and responsive to environmental cues. The concept is comprised of DNA methylation, covalent histone modifications, chromatin remodelling complexes, and non-coding RNA mechanisms. Although histone modifications, together with DNA-binding complexes, may signal changes in chromatin structure, DNA methylation acts by eventually stabilizing gene transcription and providing stable long-term regulation [Citation15]. Moreover, studies in the last decade suggest that DNA methylation in the regulatory regions of Kiss1 and Kiss1r regulates the expression of these genes both in the context of cancer [Citation16] and during puberty [Citation17,Citation18]. Recent reports imply a similar epigenetic regulation in diabetes as well [Citation19,Citation20]. For these reasons, we have investigated the DNA methylation status of Kiss1 and Kiss1r promoters in DM2 female rats. Of note, DNA methylation within gene regulatory regions, such as promoters and enhancers, is associated with gene silencing. In contrast, the loss of DNA methylation within those regions leads to gene activation [Citation14]. Our findings demonstrate, for the first time, that DNA hypermethylation of Kiss1r promoter in the liver of DM2 females is associated with a reduction in Kiss1r protein level despite a detectable rise in Kiss1r mRNA level. This inconsistency between mRNA and protein level could be linked to a bias due to different transcript variants of Kiss1r or to a translational regulation of Kiss1r protein synthesis. Hence, our study opens the door to future investigations on the role of DNA methylation in both the regulation of Kiss1r expression at mRNA/protein level and sex-differences in Kiss1r expression in healthy and DM2 conditions.
Material and methods
Animals
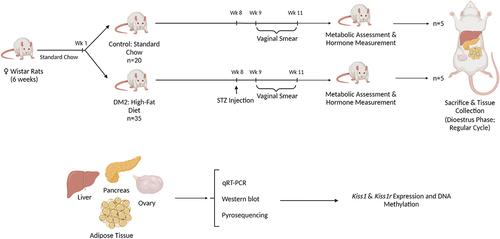
Female Wistar rats (weight: 160 ± 15 g) used in the experiment were born in an animal breeding house of the Department of Toxicology, Poznan University of Medical Sciences (Poznan, Poland). The animals were six weeks old at the beginning of our study. Rats were housed in cages (two rats per cage) in a temperature-controlled (22°C) room under 12 h light/12 h dark cycle. The animals were fed a laboratory chow diet (Labofeed B, Morawski, Kcynia, Poland); food and water were freely available. All procedures and experimental protocols followed European and Polish legislation and were approved by the local Animal Ethics Committee in Poznan (Poznan University of Life Sciences Poland). After 1-week of the acclimatization period, female rats were divided randomly into two groups: 1. Control fed with a regular lab chow diet (n = 20); 2. diabetic state (diabetes type 2; DM2) (n = 35) fed with a high fat diet providing 50% of energy from fat (HFD, Morawski, Kcynia, Poland) (please refer to the workflow, ). In week 8, DM2 group received i.p. injections of streptozotocin (STZ, 3 × 25 mg kg‐1 body weight [bw]; Sigma‐Aldrich, St Louis, MO, USA). Streptozotocin was dissolved in 0.1 mol L‐1 citric buffer (pH 4.4). STZ is toxic to the beta cells of the pancreas and, in small doses, was efficacious at inducing DM2. Diabetes was verified by measuring non-fasting glucose levels using a Roche AccuChek Active device (Roche Diabetes Care, Warsaw, Poland) and serum insulin concentrations using a commercially available radioimmunoassay (RIA) kit [as published in our previous work [Citation21]]. Non-fasting glucose concentrations were measured at the same time in all experimental groups [Citation21]. The study lasted 11 weeks. The animals were weighed weekly during the experiment.
Figure 1. Workflow of the animal study. Female Wistar rats were randomly divided into two groups, control and type 2 diabetes mellitus (DM2). Control rats were fed chow diet whereas DM2 rats were on high-fat-diet (HFD) containing 50% calories from fat. In week (wk) 8, three i.p. injections of streptozotocin (STZ) were applied to induce diabetes in DM2 group. Vaginal smears were collected throughout 10 d in week 9–11 to determine regularity of the oestrous cycle. At the end of the experimental period (week 11), one more vaginal smear was collected and only females in dioestrus phase were killed and subjected to tissue collection and molecular analyses. The drawings prepared using Biorender.
Assessment of the oestrous cycle
As the KP system is regulated by sex steroids, e.g., oestrogen and progesterone, we controlled rat’s oestrous cycle. We selected females in the dioestrus stage when the level of the hormones is low (n = 5 per each group, control and DM2).
To determine the stages of the oestrous cycle, vaginal smears were collected between 8 am and 11 am daily for 10 consecutive days and assessed using a DM500 light microscope (Leica, Wetzlar, Germany). For the evaluation of regular oestrous cycles, we used criteria established by Goldman et al. [Citation22]. The length of each phase in the cycle of 4–5 d was considered: pro‐oestrus: 1 d; oestrus: 1‐2 d; dioestrus: 2‐3 d. Additionally, we considered the determination of the time span for each phase of the oestrous cycle according to Paccola et al. [Citation23], namely pro‐oestrus and oestrus last for 12 hours each, and dioestrus lasts for 78 hours. Female rats that have the same phase of the oestrous cycle for 3 d or more were considered as those with irregular cycles and were excluded from the experiment.
Tissue collection
At the end of the 11 weeks of the experimental period, vaginal smears were collected from all rats, and only those in the dioestrus phase were killed (n = 5 per each group, control and DM2). Animals were deeply anaesthetized using ketamine/xylazine (40 mg kg‐[Citation1] bw and 5 mg kg‐[Citation1] bw, respectively), subjected to a target perfusion to perform brain immunohistochemistry for later experiments/projects, and then anaesthetized animals were killed by decapitation. Tissue samples (livers, pancreas, adipose tissue, ovaries) were dissected before perfusion, immediately frozen in the liquid nitrogen and then stored at −80°C until further analysis.
RNA isolation and quantitative reverse-transcription PCR (qRT-PCR)
TRIzol (Invitrogen) was used to isolate RNA from rat tissues. Total RNA was subsequently subjected to cDNA synthesis using AMV reverse transcriptase (Roche Diagnostics), according to the manufacturer’s protocol. Amplification of the target genes was performed in CFX96 Touch Real-Time PCR Detection System (Bio-Rad) using 2 µl of cDNA as a template, 400 nM forward and reverse primers (please see Supplementary Table S1A for sequences), and 10 µl of SsoFast EvaGreen Supermix (Bio-Rad) in a final volume of 20 µl. The amplification reaction included denaturation at 95°C for 10 min, amplification for 60 cycles at 95°C for 10 s, annealing temperature for 10 s, 72°C for 10 s, and final extension at 72°C for 10 min. The CFX Maestro Software (Bio-Rad) was used to quantify gene expression with a standard curve-based analysis. Quantitative RT-PCR data are presented as gene of interest/GAPDH (reference gene).
Western blots
The proteins were isolated from liver tissue. The total cellular proteins were extracted using a RIPA lysis buffer (25 mM Tris–HCl pH 7.6, 150 mM NaCl, 5 mM EDTA, 1% NP-40 or 1% Triton X-100, 1% sodium deoxycholate, 0.1% SDS) supplemented with a protease inhibitor cocktail (Roche Diagnostics, Penzberg, Germany). Then, samples (n = 5 per each group, control and DM2) were centrifuged, and intermediate-phase protein was collected and stored at −80°C. Twenty micrograms of total proteins were separated using SDS – polyacrylamide gel electrophoresis (SDS-PAGE). SDS-PAGE was performed in 10% polyacrylamide gel at 150 V, and then, proteins were transferred to the nitrocellulose membrane. The membranes were blocked with 3% BSA in TBS-Tween-20 (TBST) for 1 h at room temperature. Next, the membranes were incubated overnight at 4°C with primary, rabbit anti-GPR54 antibody (cat No 254,512, ABBIOTEC, San Diego, USA: dilution of 1:1000), in a TBST solution supplemented with 1% BSA. Membranes were washed in a TBST buffer and then incubated with anti-rabbit secondary antibody for 1 h at room temperature (dilution of 1:5000). The signals were visualized using an enhanced chemiluminescent reagent, and the expression of β-actin was used to normalize the results. The band density was analysed by TotalLab software.
DNA isolation and pyrosequencing
DNA was isolated from rat tissues using a standard phenol:chloroform extraction protocol. DNA was then subjected to bisulphite conversion, as previously described [Citation24]. Briefly, upon digestion of 2 mg of DNA with EcoRI for 3 h at 37°C, DNA was purified with the Quick Clean PCR Purification Kit (GenScript) and denatured with 3 M NaOH for 15 minutes at 37°C. Bisulphite conversion was performed with freshly prepared 3.6 M sodium bisulphite/1 mM hydroquinone mixture (pH 5.0) that was added to denatured DNA. The samples were incubated for 2 minutes at 95°C, 8 h at 55°C, and finally 2 minutes at 95°C and 2 h at 55°C, followed by second denaturation and purification (Quick Clean PCR Purification Kit, GenScript). DNA was precipitated with 95% ethanol and resuspended in TE buffer.
Following bisulphite conversion, DNA was amplified using HotStar Taq DNA polymerase (Qiagen) and biotinylated primers specific to target regions (please see Supplementary Table S1B for primer sequences). Sequencing by synthesis of the biotinylated DNA strands was performed in the PyroMark Q48 Autoprep instrument (Qiagen). We used PyroMark Q48 software to determine the percentage of DNA methylation at a single CpG site resolution.
Statistical analyses
An unpaired t-test with two-tailed distribution was used for statistical analyses of qRT-PCR, western blot, and pyrosequencing (Statistica, v. 13.1, StatSoft, Poland). All the results represent n = 5 rats per group. Results are presented as mean ± SEM. The results were considered statistically significant when P < 0.05.
Expression and DNA methylation microarray data from publicly available datasets are presented in boxplots and depict minimum, interquartile range (IQR), and maximum. GEO2R (http://www.ncbi.nlm.nih.gov/geo/geo2r/) was applied to determine the difference in KISS1 and KISS1R mRNA expression and DNA methylation between healthy or obese individuals and patients with DM2 in available microarray GEO series. GEO2R is an interactive web tool for comparing two groups of data to analyse any GEO series [Citation25]. In GEO2R, the adjusted P values (adj. P) using Benjamini and Hochberg (BH) false discovery rate (FDR) method by default are applied to correct for the occurrence of false-positive results. An adj. P < 0.05 and a |logFC| ≥1 are set as the cut-off criteria.
Results
Expression of Kiss1 and Kiss1r in peripheral tissues in DM2 and control female rats
We have employed a well-established rat model of DM2 experimentally induced by a high-fat diet and injections of streptozotocin (STZ), as shown in the workflow in [Citation26]. This toxin destroys the pancreas in rats, leading to severe metabolic alterations, including glucose intolerance, insulin resistance, increased cholesterol, triglyceride, and leptin levels, and abnormal functioning of the pancreatic islets [Citation27]. The model elegantly mimics the development and metabolic outcomes of DM2 in humans [Citation27].
In female Wistar rats used in our current study, DM2 state was confirmed based on the metabolic and hormonal data in our previously published work [Citation21], as shown in Supplementary Table S2. Briefly, DM2 females had increased glucose, triglycerides, cholesterol, and oestradiol levels compared to controls. Moreover, DM2 females had an impaired response to the intraperitoneal glucose tolerance test [Citation21].
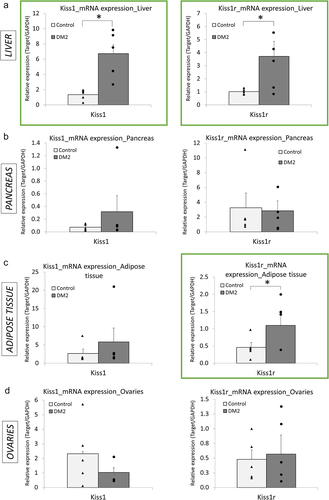
Using qRT-PCR, we have detected differential expression of the KP system in the peripheral tissues of the female rats with DM2 (). In the liver, both Kiss1 and Kiss1r were upregulated in DM2 females as compared with females on chow diet (healthy, control). Kiss1 showed fivefold increase in mRNA levels, whereas Kiss1r transcript was upregulated by 3.7-fold (). In the adipose tissue, Kiss1r was also upregulated with a 2.4-fold rise in mRNA level, whereas Kiss1 expression remained unchanged (). No statistically significant changes in Kiss1 and Kiss1r expression were detected in other tissues, including pancreas () and ovaries (). These findings are in contrast with our previous study on DM2 male rats where we reported an increase in mRNA levels of Kiss1 in the pancreas only with no alterations in Kiss1r expression in any of the organs, as compared with healthy males [Citation12].
Figure 2. Expression profile of Kiss1 and Kiss1r in peripheral tissues of female rats with type 2 diabetes mellitus (DM2), as compared with healthy (control) females. (a) Bar charts depicting expression of Kiss1 and Kiss1r in the liver (a), pancreas (b), adipose tissue (c), and ovaries (d) in female rats with DM2 caused by high-fat diet (HFD) and streptozotocin (STZ), as compared with healthy (control) females. Expression was measured using qRT-PCR. Charts in green squares represent changes that were statistically significant (P < 0.05). Results are expressed as mean ± SEM and points for each bar indicate values for individual rats; n = 5 rats per group. *P < 0.05.
DNA methylation state of Kiss1 and Kiss1r promoters in peripheral tissues in DM2 and control females
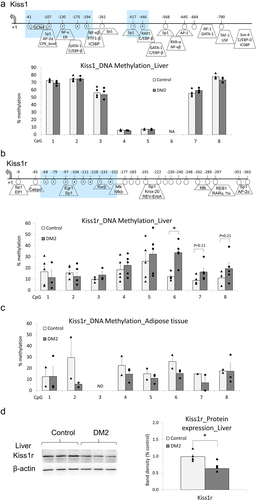
Using pyrosequencing, we have measured DNA methylation status of Kiss1 and Kiss1r promoters in the tissues of DM2 and control female rats, where altered expression at the mRNA level was detected. Although we did not observe changes in DNA methylation of the Kiss1 promoter in the liver (), Kiss1r DNA methylation levels were increased across several CpG sites with the highest nearly 22% increase at CpG #6 in DM2 females as compared with controls (healthy females) (). The same fragment of Kiss1r promoter was not differentially methylated in the adipose tissue implying tissue-specific epigenetic regulation ().
Figure 3. DNA methylation status of Kiss1 and Kiss1r promoters and a link between Kiss1r hypermethylation and Kiss1r downregulation at the protein level in females with type 2 diabetes mellitus (DM2), as compared with healthy (control) females. Average methylation status of CpG sites within promoters of Kiss1 in the liver (a), and Kiss1r in the liver (b) and in the adipose tissue (c), as determined by pyrosequencing in DM2 and healthy (control) females. Gene maps for Kiss1 (a) and Kiss1r (B) show transcription start site (TSS) indicated by +1, transcription factors predicted using Transfac, and pyrosequenced CpG sites that are circled and numbered. The blue-shaded fragment in the gene map refers to a region tested in pyrosequencing. CpG6 in Kiss1 promoter was not included in the region tested in pyrosequencing (NA). DNA methylation at CpG3 in Kiss1r promoter in the adipose tissue was not determined due to the lack of signal (ND). (d) Protein level of Kiss1r in the liver of DM2 and control females, as assessed by western blot. The band intensity was quantified and depicted in the bar chart. Results are expressed as mean ± SEM and points for each bar indicate values for individual rats; n = 5 rats per group. *P < 0.05.
DNA hypermethylation of Kiss1r promoter in the liver is accompanied by a decrease in Kiss1r protein level in DM2 females
DNA hypermethylation of Kiss1r promoter () would imply gene silencing since CpG methylation of gene regulatory regions such as promoters is linked to a condensed chromatin structure, blocking the region from binding of the transcriptional machinery, and to a transcriptional repression [Citation14]. However, we have detected an increase in Kiss1r transcript level in DM2 females as compared with controls (). If the gene is truly actively transcribed, we should also see an increase in Kiss1r receptor at the protein level. We therefore tested Kiss1r protein level in the liver of DM2 female rats, using western blot. In comparison with control females, the level of KP receptor was decreased by 36% in the liver of DM2 females ().
Differential expression and DNA methylation of KISS1 and KISS1R in peripheral tissues in humans with DM2 as compared with healthy or obese non-diabetic individuals
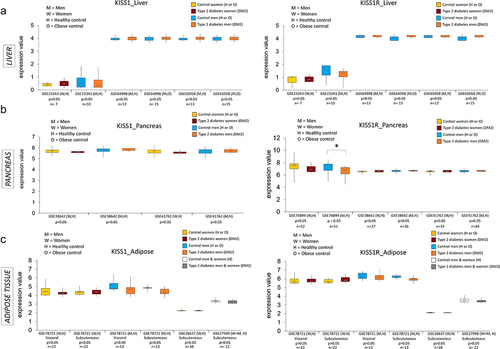
As being overweight or obese are the leading risk factors for the development of DM2 [Citation28], using publicly available data in the Gene Expression Omnibus (GEO), we have analysed expression and DNA methylation of KISS1 and KISS1R in peripheral tissues in diabetic patients and/or obese individuals. In GEO database, we used the following search terms: ‘type 2 diabetes,’ ‘expression’ (or ‘DNA methylation’ for DNA methylation analyses), and our tissue of interest (i.e., ‘liver,’ ‘pancreas,’ ‘adipose tissue’). We narrowed our search by only selecting the datasets pertaining to humans, reporting microarray data, and with cohorts of patients clinically diagnosed with DM2. The only exception was GSE76398 dataset where patients were categorized into insulin-resistant and insulin-sensitive based on homoeostatic model assessment for insulin resistance (HOMA-IR). Data for men and women were separately plotted whenever possible.
In both women and men, we have observed a higher median in KISS1 expression and a lower median in KISS1R expression in the liver of DM2 vs. healthy or obese individuals in several datasets analysed (). For instance, KISS1 median expression in the liver of DM2 women was 20% higher than that in healthy individuals in GSE23343 (, left panel). In men, we detected 4% increase in hepatic KISS1 median expression across all datasets except GSE23343. For KISS1R, 20% decrease in median expression in the liver of DM2 patients vs. healthy controls was found in both sexes in GSE23343 dataset (, right panel). However, the observed changes were not statistically significant, which could be due to a low number of DM2 patients and overall a low number of enrolled individuals (details on patient characteristics in Supplementary Table S3). The number of participants was substantially higher in the pancreas, where we found statistically significant 10% decrease in KISS1R expression in men and a 10% lower median expression of KISS1R in women, in the same dataset, GSE76894 (). Data in the adipose tissue were mixed and thus difficult to draw conclusions ().
Figure 4. Expression of KISS1 and KISS1R in human peripheral tissues from individuals with type 2 diabetes mellitus (DM2) or obesity and in healthy controls. KISS1 and KISS1R expression at the mRNA level in the liver (a), pancreas (b) and adipose tissue (c) of patients with DM2 vs. healthy individuals or individuals with obesity (controls), based on the microarray data from publicly available datasets in the Gene Expression Omnibus (GEO) database. Results expressed as min, IQR, and max of the array intensity (expression value); *P < 0.05. Patient characteristics corresponding to the data are depicted in Supplementary Table S3.
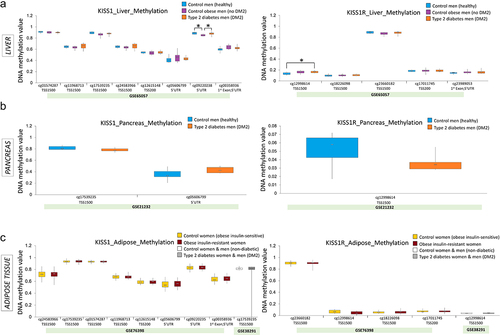
Interestingly, we detected statistically significant 15% hypermethylation of KISS1R promoter (, right panel, GSE65057, cg12998614), which is in agreement with a lower median in KISS1R expression in the liver of DM2 patients vs. healthy individuals (, right panel, GSE23343 dataset). On the contrary, KISS1 promoter was 5% hypomethylated in the liver of obese vs. healthy individuals (, left panel, GSE65057, cg09220235); however, the difference in the median dropped to 1% at the same CpG site when comparing healthy individuals to DM2 patients. No apparent changes in DNA methylation of KISS1 and KISS1R promoters were observed in other tissues ( b and c).
Figure 5. DNA methylation of KISS1 and KISS1R promoters in human peripheral tissues from individuals with type 2 diabetes mellitus (DM2) or obesity and in healthy controls. KISS1 and KISS1R DNA methylation levels at single CpG site in promoter regions in the liver (a), pancreas (b), and adipose tissue (c) of patients with DM2 vs. healthy individuals or individuals with obesity (controls), based on the DNA methylation Illumina microarray data from publicly available datasets in the Gene Expression Omnibus (GEO) database. Results expressed as min, IQR, and max of the array intensity (beta values = DNA methylation value)); *P < 0.05. Patient characteristics corresponding to the data are depicted in Supplementary Table S3.
Discussion
In the present study, we have investigated females that are largely understudied in numerous fields of science. For the first time, we have determined the profile of Kiss1 and Kiss1r expression in peripheral tissues in female rats with DM2 and explored the possibility that epigenetic mechanisms shape this profile of expression. Our results deliver novel evidence for differential expression of Kiss1 and Kiss1r in female peripheral organs and for DNA methylation as a player in regulation of Kiss1r expression in the liver of DM2 female rats.
Epigenetic regulation of Kiss1 and Kiss1r genes via DNA methylation has been described in different tissues, conditions, and experimental models [Citation16–20]. In the ARC region of the hypothalamus of C57BL/6 female mice, expression of Kiss1 during the initiation of puberty has been demonstrated to be driven by DNA demethylation of short interspersed nuclear elements (SINEs) within the Kiss1 enhancer [Citation29]. Another study on the brain tissue of female Sprague-Dawley rats showed that differential DNA methylation of Kiss1 and Kiss1r promoters at various CpG residues could be a mechanism that regulates differential expression of those genes before and after puberty [Citation30]. In cancer, high expression of KISS1R has been linked to higher survival in endometrial cancer patients, and DNA hypermethylation has been revealed as a mechanism of KISS1R silencing [Citation31]. Hypermethylated KISS1R has also been identified in genome-wide methylation studies in thyroid cancer [Citation32]. Furthermore, Kiss1r has been shown as one of the targets of the polycomb repressive complex-2 (PRC2) and Kiss1r promoter was hypermethylated with age in haematopoietic stem cells of female C57b/6 mice [Citation33]. Polycomb-group (PcG) proteins were also shown to repress the promoter activity of Kiss1 in hypothalamus, which was maintained by the interaction of PcG complexes with sirtuin 1, SIRT1, a fuel-sensing deacetylase [Citation34].
Although epigenetic alterations in genes involved in reproduction and energy homoeostasis have previously been demonstrated in metabolic disorders, such as obesity and diabetes, existing data on the epigenetic regulation of the KP system are very limited [Citation20]. In the present study, we found hypermethylation of Kiss1r promoter in the liver of DM2 female rats (), which corresponded with lower Kiss1r expression at the protein level (). Similarly, hypermethylation of Kiss1r has been reported in peripheral blood DNA of obese dogs [Citation19]. In the study on dogs, there was a trend towards more profound hypermethylation in male vs. female dogs, whereas we detected altered Kiss1r expression at mRNA and protein level in the liver of females, but not male rats. Along with our previous [Citation12] and present study, the findings imply that epigenetic regulation of Kiss1r occurs in a sex-dimorphic manner, which may explain, at least partially, sex-differences in the metabolic regulation by KP signalling. As previously reported by others, body weight and metabolic homoeostasis are regulated by KP signalling with a major role of gonadal hormone-dependent actions, and thus the regulation differs between sexes [Citation35]. Mice obesity triggered by Kiss1r KO [Citation10] was prevented by reinsertion of Kiss1r in GnRH neurons in female mice with gonadal functions preserved [Citation35]. However, the phenotype was not fully rescued in male animals [Citation35].
Among peripheral tissues, we have detected an increase in Kiss1 expression in the liver of DM2 females (). Indeed, increased levels of KP were reported in livers from patients with DM2 and in diabetic mouse models [Citation11]. According to the pancreas-liver hormonal endocrine circuit proposed by Song et al. [Citation11], hyperglucagonemia, which is often an early hallmark of DM2, could result in high hepatic KP. KP would then suppress glucose-stimulated insulin secretion and consequently lead to insufficient insulin levels [Citation11]. Thus, the liver appears to be essential in the KP-mediated reduction in glucose-stimulated insulin release. Blocking a hepatic KP production or antagonizing KP could be considered as a potential therapeutic approach to improve β-cell function in DM2 [Citation11]. Although we did not detect changes in DNA methylation of Kiss1 promoter, there may be another epigenetic mechanism involved to explain Kiss1 upregulation. Importantly, overnutrition and obesity have been shown to lead to downregulation of SIRT1 content in the hypothalamus, and consequently, the lack of SIRT1 at the Kiss1 promoter reduces the repressive action of PcG complexes and facilitates an increase in Kiss1 transcriptional activity [Citation34]. It is possible that such a mechanism could also exist at the periphery.
We have found an increase in mRNA levels of both Kiss1 and Kiss1r in the liver of DM2 females as compared with healthy (control) females (). However, only Kiss1r promoter displayed an altered DNA methylation status (). The consensus is that actively transcribed genes have hypomethylated promoters [Citation14]. Surprisingly, the increase in mRNA levels of Kiss1r was linked to promoter hypermethylation () and not, as we would expect, to promoter hypomethylation. This could potentially mean that the region we tested in pyrosequencing is actually a Kiss1r silencer. When a silencer regulatory region is methylated, the recruitment of transcriptional repressors is blocked due to the condensed chromatin structure and transcription of a given gene increase [Citation36]. Interestingly, such a mechanism was described for Kiss1 at the brain level [Citation17,Citation37]. In the anteroventral periventricular nucleus of the mouse hypothalamus, higher Kiss1 mRNA expression of female mice, as compared with males, was associated with higher CpG methylation of Kiss1 promoter [Citation37]. Furthermore, it is also possible that the observed hypermethylation actually reflects an increase in DNA hydroxymethylation. Since bisulphite sequencing such as pyrosequencing cannot discriminate 5-methyl-cytosine from 5-hydroxy-methyl-cytosine, the increase in 5-hydroxy-methyl-cytosine, which is linked to transcriptional activation, would be registered as DNA hypermethylation [Citation18].
Alternatively, the lack of correlation between changes in mRNA expression and DNA methylation of Kiss1r promoter could be due to the fact that there are different transcript variants of Kiss1r. We found two Kiss1r variants in the genome browser of the rn7 rat assembly. These isoforms have almost identical mRNA sequence and differ only with six nucleotides in exon 5. For this reason, qRT-PCR primers cannot distinguish between the isoforms which may introduce a bias. We therefore proceeded with western blot to assess Kiss1r at the protein level. Strikingly, we observed a decrease in Kiss1r protein level in the liver of DM2 females (). Thus, it becomes highly probable that mRNA readings are not reliable and Kiss1r promoter hypermethylation is indeed linked to transcriptional repression of the gene.
Another explanation for the detected discrepancy between increased mRNA and decreased protein of Kiss1r in the liver of DM2 female rats could lie in the aberrant translation from mRNA to protein. Many factors, including translation initiation and elongation factors, RNA-binding proteins, mRNA methylation (epitranscriptome), microRNAs and long non-coding RNAs, have been shown to contribute to translational regulation [Citation38–40]. Although Kiss1r may be prompted for activation at the promoter level (i.e., hypermethylation of a silencer; hydroxymethylation read as hypermethylation), production of Kiss1r protein may be disturbed by another mechanism at the translation step.
As the majority of studies have been performed on males, including our previous work [Citation12] and sex-specific effects in the development of DM2 have been more and more recognized [Citation41,Citation42], we have focused on females in the present study. Sex differences in the development of DM2 were shown in humans, with women favouring fat tissue storage and men tending to mobilize fat tissue burning [Citation41]. It was also found that women have greater insulin sensitivity than men. Additionally, sex differences in body fat distribution exist due to differences in sex hormones and the adipose tissue microenvironment between men and women [Citation42]. Of note, although KP neurons in the ARC are not sexually dimorphic, sex-dependent KP effects on metabolism were observed. Mature Kiss1r KO females had higher body weight and increased white adipose tissue mass [Citation43], as compared with Kiss1r KO males [Citation35]. Moreover, we have previously found sex differences in the central response of the KP neurons to experimentally induced DM2 in rats [Citation44,Citation45]. Contrary to males, DM2 female rats did not show an increased number of KP neurons in the ARC either in gonad-intact group [Citation21,Citation44] or after gonadectomy with/without sex hormone replacement [Citation45]. KP neurons in the ARC increased only in DM2 males.
As the KP system is implicated in the regulation of metabolism [Citation35,Citation43], and Kiss1/Kiss1r show sexually dimorphic expression in the brain [Citation21,Citation44,Citation45], it was not surprising for us to detect that the patterns of tissue-specific expression of Kiss1 and Kiss1r in peripheral organs of control and DM2 female rats are different from the patterns described in male rats in our previous study [Citation12]. DM2 in male rates was linked to increased expression of Kiss1 at mRNA level in the pancreas and at the protein level in the liver [Citation12], while DM2 females showed Kiss1 increase only in the liver (). Of interest, Kiss1r expression was altered in the pancreas and liver of DM2 females vs. control females (), while no changes in Kiss1r were reported in DM2 male rats vs. control males [Citation12]. However, we need to be cautious when comparing male and female data as the studies were not run simultaneously. Distinct phases in the development of DM2 between both sexes were the main reason for a separate design. In males, the induction of DM2 required a high-fat diet and a single dose of STZ (25 mg/kg body weight), which mimics an early stage of DM2 [Citation44,Citation45]. Females were more resistant to DM2 development and needed 3 i.p. injections of STZ (3 × 25 mg/kg body weight). As a result, DM2 in female rats would reflect the later stages of the disease [Citation21]. Hence, although our findings indicate possible sex-specific differences in DM2-mediated alterations in the KP system, further experiments are warranted to explore DM2 effects in males and females in parallel.
Although we did not observe statistically significant changes in Kiss1 and Kiss1r in the adipose tissue of female rats with DM2 vs. controls (), there was a slight decrease suggesting Kiss1 repression in DM2 group. The observation is consistent with existing data which show a decrease in fat Kiss1 mRNA in obese rats [Citation13]. On the other hand, a study on fat samples from humans demonstrated a positive correlation between KISS1 mRNA and body mass index in women [Citation46]. Such correlation was only registered for visceral, but not subcutaneous fat, which indicates differences in KP levels depending on the type of the adipose tissue.
Our findings prompted us to determine patterns of KISS1 and KISS1R expression and DNA methylation in peripheral tissues in humans with DM2, using publicly available datasets (). Although the results were mixed and statistically non-significant, we found a higher median in KISS1 expression and a lower median in KISS1R expression in the liver of DM2 men and women vs. healthy or obese individuals (). This finding is in agreement with the hormonal regulatory connection between the liver and pancreas, as described by Song et al. [Citation11], where increased production of hepatic KP in DM2 patients and animals leads to suppression of glucose-stimulated insulin secretion and thereby to low levels of insulin [Citation11]. Decrease in receptors for KP in the liver could be logical as the system is shifted towards KP action on the pancreas. The DNA methylation analysis showed hypermethylation of hepatic KISS1R promoter in DM2 vs. healthy individuals and hypomethylation of hepatic KISS1 promoter in obese vs. healthy individuals (). Our findings suggest that epigenetic mechanisms may be involved in regulation of KISS1 and KISS1R expression, and those mechanisms may be altered in obesity/DM2 resulting in aberrant gene expression. However, more comprehensive studies on gene expression and DNA methylation are needed with large homogenous cohorts where age, sex, treatment regimens, and other possible confounding factors are taken into account.
In summary, our study demonstrates differential expression of Kiss1 and Kiss1r in peripheral organs in DM2 females, as compared with healthy controls. We deliver novel findings on the regulation of the KP system in the peripheral tissues in female rats with DM2. We show that Kiss1 expression increases in the liver of DM2 female rats, which is consistent with existing data on the increased production of hepatic KP in DM2 patients. Our study also proposes an epigenetic mechanism, involving DNA methylation, for regulation of Kiss1r expression, which may be tissue-specific. Despite a detectable rise in mRNA of Kiss1r, we observe DNA hypermethylation of Kiss1r promoter which is associated with reduction in Kiss1r protein level. The discrepancy between the mRNA and protein level may result from different transcript variants of Kiss1r or a regulatory mechanism at the translation stage. Thus, our findings open the door to further research on Kiss1r regulation in DM2 and to potential elucidation of epigenetic therapeutics as a new KP-focused approach in DM2 therapy and presumably prevention. Our study also suggests that the regulation of the KP system at the periphery in DM2 may differ between sexes, as indicated by distinct patterns of Kiss1 and Kiss1r expression in DM2 vs. healthy controls in females in the current study and males in our previous work [Citation12]. Although sex differences need to be further confirmed in an experiment investigating females and males in parallel, there is a premise for the development of new sex-specific therapeutic strategies to face increasing rates of DM2 in humans.
Supplemental Material
Download Zip (54.9 KB)Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The Authors will share the data upon request.
Supplementary material
Supplemental data for this article can be accessed online at https://doi.org/10.1080/15592294.2022.2119120.
Additional information
Funding
References
- Lee JH, Miele ME, Hicks DJ, et al. KiSS-1, a novel human malignant melanoma metastasis-suppressor gene. J Natl Cancer Inst. 1996;88:1731–1737.
- de Roux N, Genin E, Carel JC, et al. Hypogonadotropic hypogonadism due to loss of function of the KiSS1-derived peptide receptor GPR54. Proc Natl Acad Sci U S A. 2003; 100:10972–10976. doi:10.1073/pnas.1834399100.
- Seminara SB, Messager S, Chatzidaki EE, et al. The GPR54 gene as a regulator of puberty. N Engl J Med. 2003;349:1614–1627.
- Lehman MN, Coolen LM, Goodman RL. Minireview: kisspeptin/neurokinin B/dynorphin (KNDy) cells of the arcuate nucleus: a central node in the control of gonadotropin-releasing hormone secretion. Endocrinology. 2010;151:3479–3489.
- Hrabovszky E. Neuroanatomy of the human hypothalamic kisspeptin system. Neuroendocrinology. 2014;99:33–48.
- Rometo AM, Krajewski SJ, Voytko ML, et al. Hypertrophy and increased kisspeptin gene expression in the hypothalamic infundibular nucleus of postmenopausal women and ovariectomized monkeys. J Clin Endocrinol Metab. 2007;92:2744–2750.
- Finley JC, Maderdrut JL, Roger LJ, et al. The immunocytochemical localization of somatostatin-containing neurons in the rat central nervous system. Neuroscience. 1981;6:2173–2192.
- Funahashi H, Takenoya F, Guan JL, et al. Hypothalamic neuronal networks and feeding-related peptides involved in the regulation of feeding. Anat Sci Int. 2003;78:123–138.
- Luque RM, Kineman RD, Tena-Sempere M. Regulation of hypothalamic expression of KiSS-1 and GPR54 genes by metabolic factors: analyses using mouse models and a cell line. Endocrinology. 2007;148:4601–4611.
- Tolson KP, Garcia C, Yen S, et al. Impaired kisspeptin signaling decreases metabolism and promotes glucose intolerance and obesity. J Clin Invest. 2014;124:3075–3079.
- Song WJ, Mondal P, Wolfe A, et al. Glucagon regulates hepatic kisspeptin to impair insulin secretion. Cell Metab. 2014;19:667–681.
- Dudek M, Kolodziejski PA, Pruszynska-Oszmalek E, et al. Effects of high-fat diet-induced obesity and diabetes on Kiss1 and GPR54 expression in the hypothalamic-pituitary-gonadal (HPG) axis and peripheral organs (fat, pancreas and liver) in male rats. Neuropeptides. 2016;56:41–49.
- Brown RE, Imran SA, Ur E, et al. KiSS-1 mRNA in adipose tissue is regulated by sex hormones and food intake. Mol Cell Endocrinol. 2008;281:64–72.
- Beetch M, Harandi-Zadeh S, Shen K, et al. Dietary antioxidants remodel DNA methylation patterns in chronic disease. Br J Pharmacol. 2020;177:1382–1408.
- Jones PA, Ohtani H, Chakravarthy A, et al. Epigenetic therapy in immune-oncology. Nat Rev Cancer. 2019;19:151–161.
- Motti ML, Meccariello R. Minireview: the epigenetic modulation of KISS1 in reproduction and cancer. Int J Environ Res Public Health. 2019;16:2607.
- Aylwin CF, Vigh-Conrad K, Lomniczi A. The emerging role of chromatin remodeling factors in female pubertal development. Neuroendocrinology. 2019;109:208–217.
- Uenoyama Y, Tomikawa J, Inoue N, et al. Molecular and epigenetic mechanism regulating hypothalamic Kiss1 gene expression in mammals. Neuroendocrinology. 2016;103:640–649.
- Yamazaki J, Meagawa S, Jelinek J, et al. Obese status is associated with accelerated DNA methylation change in peripheral blood of senior dogs. Res Vet Sci. 2021;139:193–199.
- Navarro VM. Metabolic regulation of kisspeptin - the link between energy balance and reproduction. Nat Rev Endocrinol. 2020;16:407–420.
- Ziarniak K, Kolodziejski PA, Pruszynska-Oszmalek E, et al. High-fat diet and type 2 diabetes induced disruption of the oestrous cycle and alteration of hormonal profiles, but did not affect subpopulations of KNDy neurons in female rats. J Neuroendocrinol. 2018;30:e12651.
- Goldman JM, Murr AS, Cooper RL. The rodent estrous cycle: characterization of vaginal cytology and its utility in toxicological studies. Birth Defects Res Part B, Dev Reprod Toxicol. 2007;80:84–97.
- Paccola CC, Resende CG, Stumpp T, et al. The rat estrous cycle revisited: a quantitative and qualitative analysis. Anim Reprod. 2013;10:677–683.
- Lubecka K, Kurzava L, Flower K, et al. Stilbenoids remodel the DNA methylation patterns in breast cancer cells and inhibit oncogenic NOTCH signaling through epigenetic regulation of MAML2 transcriptional activity. Carcinogenesis. 2016;37:656–668.
- Barrett T, Wilhite SE, Ledoux P, et al. NCBI GEO: archive for functional genomics data sets–update. Nucleic Acids Res. 2013;41:D991–5.
- Skovso S. Modeling type 2 diabetes in rats using high fat diet and streptozotocin. J Diabetes Investig. 2014;5:349–358.
- Kleinert M, Clemmensen C, Hofmann SM, et al. Animal models of obesity and diabetes mellitus. Nat Rev Endocrinol. 2018;14:140–162.
- Leitner DR, Fruhbeck G, Yumuk V, et al. Obesity and Type 2 diabetes: two diseases with a need for combined treatment strategies - EASO can lead the way. Obes Facts. 2017;10:483–492.
- Shen Y, Zhao H, Zhang L, et al. The roles of DNA methylation and hydroxymethylation at short interspersed nuclear elements in the hypothalamic arcuate nucleus during puberty. Mol Ther Nucleic Acids. 2021;26:242–252.
- Wyatt AK, Zavodna M, Viljoen JL, et al. Changes in methylation patterns of kiss1 and kiss1r gene promoters across puberty. Genet Epigenet. 2013;5:51–62.
- Kang HS, Baba T, Mandai M, et al. GPR54 is a target for suppression of metastasis in endometrial cancer. Mol Cancer Ther. 2011;10:580–590.
- Rodriguez-Rodero S, Fernandez AF, Fernandez-Morera JL, et al. DNA methylation signatures identify biologically distinct thyroid cancer subtypes. J Clin Endocrinol Metab. 2013;98:2811–2821.
- Taiwo O, Wilson GA, Emmett W, et al. DNA methylation analysis of murine hematopoietic side population cells during aging. Epigenetics. 2013;8:1114–1122.
- Vazquez MJ, Toro CA, Castellano JM, et al. SIRT1 mediates obesity- and nutrient-dependent perturbation of pubertal timing by epigenetically controlling Kiss1 expression. Nat Commun. 2018;9:4194.
- Velasco I, Leon S, Barroso A, et al. Gonadal hormone-dependent vs. -independent effects of kisspeptin signaling in the control of body weight and metabolic homeostasis. Metabolism. 2019;98:84–94.
- Jones PA, Takai D. The role of DNA methylation in mammalian epigenetics. Science. 2001;293:1068–1070.
- Semaan SJ, Dhamija S, Kim J, et al. Assessment of epigenetic contributions to sexually-dimorphic Kiss1 expression in the anteroventral periventricular nucleus of mice. Endocrinology. 2012;153:1875–1886.
- Gebauer F, Hentze MW. Molecular mechanisms of translational control. Nat Rev Mol Cell Biol. 2004;5:827–835.
- Song P, Yang F, Jin H, et al. The regulation of protein translation and its implications for cancer. Signal Transduct Target Ther. 2021;6:68.
- Subramaniam D, Ramalingam S, Linehan DC, et al. RNA binding protein CUGBP2/CELF2 mediates curcumin-induced mitotic catastrophe of pancreatic cancer cells. PloS One. 2011;6:e16958.
- Link JC, Reue K. Genetic basis for sex differences in obesity and lipid metabolism. Annu Rev Nutr. 2017;37:225–245.
- Jeffery E, Wing A, Holtrup B, et al. The adipose tissue microenvironment regulates depot-specific adipogenesis in obesity. Cell Metab. 2016;24:142–150.
- De Bond JP, Tolson KP, Nasamran C, et al. Unaltered hypothalamic metabolic gene expression in Kiss1r knockout mice despite obesity and reduced energy expenditure. J Neuroendocrinol. 2016;28(10).
- Dudek M, Kolodziejski PA, Pruszynska-Oszmalek E, et al. Effects of orchidectomy and testosterone replacement on numbers of kisspeptin-, neurokinin B-, and dynorphin A-immunoreactive neurons in the arcuate nucleus of the hypothalamus in obese and diabetic rats. J Neuroendocrinol. 2017;29. DOI:10.1111/jne.12453
- Ziarniak K, Kolodziejski PA, Pruszynska-Oszmalek E, et al. Effects of ovariectomy and sex hormone replacement on numbers of kisspeptin-, neurokinin B- and dynorphin A-immunoreactive neurons in the arcuate nucleus of the hypothalamus in obese and diabetic rats. Neuroscience. 2020;451:184–196.
- Cockwell H, Wilkinson DA, Bouzayen R, et al. KISS1 expression in human female adipose tissue. Arch Gynecol Obstet. 2013;287:143–147.
