ABSTRACT
Autophagy is an important cytoprotective process that mediates degradation of dysfunctional or unnecessary cellular components. In the process of autophagy, a double-membrane organelle termed the autophagosome is formed to sequestrate portions of cytoplasm and subsequently delivered into lysosome or vacuole for degradation. The accumulation of autophagic bodies in the vacuoles after treatment with concanamycin A (ConcA) is a widely used protocol for monitoring the occurrence of autophagy in plants. Here, it was found that the cytoplasmic soluble GFP was accumulated in vacuoles upon ConcA treatment. Importantly, the GFP signal showed good colocalization with the autophagic marker mCherry-ATG8f in vacuoles based on two commonly used methods, the Pearson-Spearman correlation colocalization analysis and the plot profile analysis. Further results showed that the free GFP did not interact with ATG8s. Thus, analysis of accumulation and colocalization only in vacuoles is not a trustworthy way to judge whether degradation of cytoplasmic protein is dependent on the selective autophagy pathway in plants. In this short perspective, we propose several primary steps to distinguish that the cytoplasmic proteins are degraded by selective or bulk autophagy, hoping they could contribute to identify and clarify the selective autophagic cargos and receptors in plants.
Autophagy plays an important role in the maintenance of intracellular homeostasis by degradation and recycling of cytoplasmic components, including dysfunctional organelles, unnecessary proteins, and invading pathogens. Based on their different morphological features, three types of autophagic pathways (macroautophagy, chaperone-mediated autophagy and microautophagy) have been characterized in eukaryotic cells.Citation1 During macroautophagy (hereafter autophagy), a double-membrane vesicle, called autophagosome, is formed from the expanding membrane of phagophore. The autophagosomes sequester the enclosed components and deliver them into the lysosome/vacuole for degradation. Mounting evidence suggests that autophagy is a highly regulated process by selectively recognizing the specific protein cargos.Citation2,Citation3 The molecular machinery for selective autophagy requires autophagic receptors that recruit the targeted cargos to the growing autophagosomal membranes. ATG8s, the lipid-conjugated ubiquitin-like proteins, are core components of the autophagy machinery. Most of the proteins that are specifically turned over by selective autophagy are recognized by the presence of short ATG8 interacting motifs (AIMs) that facilitate their association with the autophagy apparatus.Citation4,Citation5 Over the last decade, ground-breaking studies in yeast and mammalian models uncovered molecular details of selective cargo recognition. However, there is still a large gap in our understanding of selective autophagy process in plants. Although the precise molecular mechanisms of cargo selection by autophagy are yet to be established, an increasing number of autophagic receptors or cargos, including AtNBR1, DSK2, RPN10, Exo70E2, AtSec62 and C53 have been identified and characterized for selective autophagy degradation in plants ().Citation6–11 In the process of their function being clarified, colocalization analysis with the autophagic marker ATG8s is an important step in selective cargos identification.
Table 1. Autophagic cargos and/or receptors have been identified and characterized for selective autophagy in plants
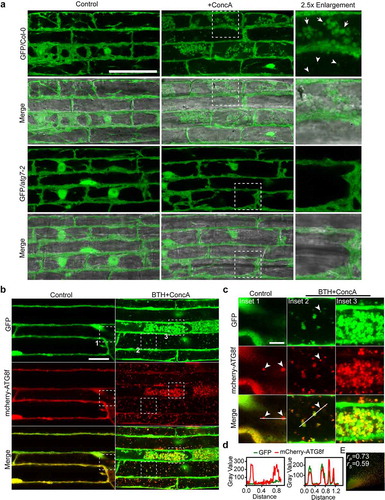
Concanamycin A (ConcA) is one of the widely used inhibitors to study autophagy in plants. ConcA interferes with vacuolar-type H+-translocating ATPase by binding to the subunit c of the proton translocating Vo complex.Citation22 Inhibition of the V-ATPases results in blocking vacuolar acidification and impeding hydrolases activity in the vacuoles.Citation23 Thus, the accumulation of autophagic bodies in vacuoles upon treatment with ConcA is applied for monitoring the occurrence of autophagy in plants.Citation24 Here, it was found that the cytoplasmic soluble GFP were accumulated in vacuoles, showing globular clusters (indicated by arrows) and punctate structures (indicated by arrowheads), after treatment with ConcA for 6 hours. These two patterns might be due to their different extents of degradation. However, the delivery of GFP protein into vacuoles was abolished in autophagic defective mutant atg7-2 ()). Upon BTH and ConcA (BTH+ConcA) treatment, a common autophagy-inducible condition,Citation25 the GFP signal were colocalized with the autophagic marker mCherry-ATG8f in vacuoles, showing punctate (Inset 2) and globular structures (Inset 3), respectively ()). These signals in vacuoles showed highly dynamic movement under time-lapse imaging (Supplemental movie 1). Remarkably, the GFP protein was not recruited to mCherry-ATG8f puncta in the cytoplasm under the control condition (Inset 1 of )). To qualitative the level of co-occurrence of these fluorescing proteins, two common colocalization methods, the Pearson-Spearman correlation analysis and the plot profile analysis were used. Both analyses showed that the GFP signals had good colocalization with the autophagic marker mCherry-ATG8f ()).
Figure 1. Cytoplasmic soluble GFP protein is nonselectively engulfed by autophagosomes and delivered into vacuole upon autophagy induction condition
GFP has a capacity to bind aggregated proteins like amyloid fibrils in mammal.Citation26 To test whether GFP could be degraded by the selective autophagy pathway, an in vivo recruitment assay was adopted. In this system, the transmembrane domain of calnexin (CNX) is fused to N-terminal of fluorescent tag RFP and then connected with ATG8e (CNX-RFP-ATG8e). When co-expression of the interest cargos, they would be recruited to endoplasmic reticulum (ER)-localized CNX-RFP-ATG8e, showing the linear ER patterns.Citation7 Compared with the positive control AtNBR1-GFP, the free GFP cannot be recruited to CNX-RFP-ATG8e ()), indicating that GFP was not recognized by ATG8e. In general, the macroautophagy and microautophagy confers two mainly pathways for vacuolar targeted degradation.1,Citation27–29 The soluble GFP protein distributed in the cytoplasm might be nonselectively engulfed by autophagosome or directly invaginated at the tonoplast and then delivered into vacuole ()). Thus, the use of vacuole-targeted accumulation and colocalization analysis as a reporter in autophagy study should be carefully considered. However, looking at the autophagic cargos or receptors identified over the last decade in the field of plant research, we found that nearly half of the findings were based on co-localization analysis in vacuoles after treatment with ConcA ().
Figure 2. Proposed model for the nonselective cytoplasmic soluble protein and the selective cargos engulfed by autophagosomes
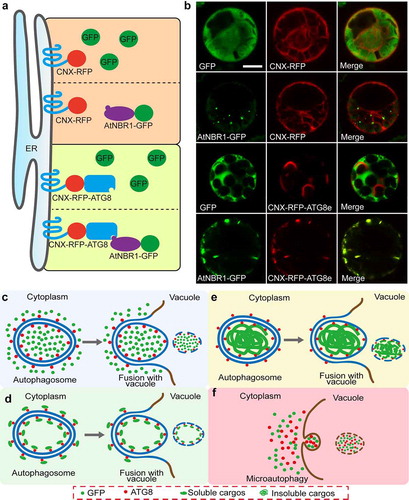
How to identify one cargo, especially the cytoplasmic soluble protein, was degraded by selective autophagy or bulk autophagy or microautophagy? To better distinguish these pathways, it is easier to get some hints from the autophagic process shown in )). For the nonselective proteins, the autophagosomes are formed to sequestrate portions of the cytoplasm that contains cytosolic materials like GFP protein, and subsequently deliver the inclusions into vacuole for degradation. The nonselective cytosolic materials are shown by colocalization with the autophagic marker ATG8s after the autophagic bodies accumulated in vacuoles ()). For the soluble cargos/receptors, they are specially recognized and recruited by ATG8s to the double membrane of autophagosomes. In this case, these proteins should be colocalized with ATG8s and sometimes show ring-like structures in cytoplasm ()), which have been already verified in some receptors like C53 and AtNBR1.Citation7,Citation9 For the insoluble cargos, like protein aggregates, they are recognized by ATG8s and engulfed by autophagosome, showing colocalization in cytoplasm ()). Additionally, microautophagy might directly invaginate the cytoplasmic components at the level of the tonoplast or of the membranes of the MVB (multivesicular body) and then deliver into the vacuolar lumen ()). Conspicuously, both the nonselective and the selective cytosolic materials are shown to colocalization after the autophagic bodies accumulated in vacuoles. Thus, colocalization analysis of interest protein with the autophagic marker ATG8s in cytoplasm, but not in vacuoles in the presence of ConcA, is an important and simple method to distinguish whether the degradation is dependent upon the selective autophagy pathway in plants.
Previously, a basic set of guidelines for the selection and interpretation of assays for studying autophagy in different organisms has been well documented.Citation30 Here, we propose several primary steps to distinguish that the cytoplasmic proteins are degraded by selective or nonselective autophagy, hoping these suggested points could contribute to standardize research of autophagy in plants.
Step 1. Screening ATG8s-interacted cargos/receptors
Identifying the interacted proteins of ATG8s is the first and most important step. To elucidate and identify the interacting cargo/receptor of ATG8s, some commonly methods including the yeast two-hybrid (Y2H) screen and immunoprecipitation followed by mass spectrometry (IP-MS) analysis have been widely used. Recently, new methods like proximity labeling have been developed for capturing protein–protein interactions in a vast array of biological contexts.Citation31 The effectiveness of proximity labeling in mapping ATG8s-interacted networks will open up a new line of research in the autophagy field in plants.
Step 2. Performing colocalization analysis
Colocalization with the fluorescent tag fused ATG8s in images is an important way to verify special autophagic cargos or receptors. At this step, to avoid random separation of cytoplasmic components by nonselective autophagy, we strongly suggest to do colocalization analysis in cytoplasm but not in vacuoles (). For the soluble proteins, the ring-like structures sometimes could be observed in the cytoplasm. Since autophagy is activated under various abiotic and biotic stresses, it is necessary to consider induction of autophagy with a proper condition. Starvation by deprivation of carbon or nitrogen source is one of the most used methods. To monitor distinct organelle-specific autophagy, specific treatment conditions need to be carried out, such as induction of endoplasmic reticulum (ER) stress by tunicamycin (TM) and dithiothreitol (DTT),Citation8 as well as induction of mitophagy by some uncouplers like 2,4-dinitrophenol (DNP) and carbonyl cyanide ptrifluoro-methoxyphenyl hydrazone (FCCP).Citation32 Additionally, data obtained from overexpression assays should always be considered with caution, because overexpressing proteins may affect its distribution causing aggregate formation and autophagy.
Step 3. Interaction analysis and AIM sites identification
Physical interaction between the cargos/receptors and ATG8-family proteins is required for subjecting them to autophagic degradation. It is important to verify protein–protein interactions both in vitro and in vivo through methods such as yeast two-hybrid assays (Y2H), bimolecular fluorescence complementation (BiFC), Förster (Fluorescence) resonance energy transfer (FRET), co-immunoprecipitation (Co-IP), and pull-down assay. Recently, we have developed an ER-targeted in vivo recruitment assay to screen the cargos recruited by autophagic receptor AtNBR1.Citation7 The in vivo recruitment assay is a powerful tool to analyze the interact partners of ATG8s, especially for the cytoplasmic soluble cargos. By relocating the targeted proteins at the subcellular level, this system could effectively avoid background interference caused by the dispersive distribution of soluble proteins.
Most of ATG8-family interacting proteins contain a core consensus sequence W/F/Y-x-x-L/I/V (x = any amino acid) called LIR (LC3-interacting region) motif, also named ATG8 interacting motif (AIM).Citation4 AIM binds to the hydrophobic pocket of ATG8s that comprise a docking surface known as LIR docking site (LDS). Mutational analysis of the AIM sites is important to validate the interest proteins as an autophagy substrate. Recently, different from the canonical AIM, new ATG8 binding interfaces like the ubiquitin-interacting motif (UIM) and the shuffled ATG8 interacting motifs (sAIM) have been found.Citation9,Citation11,Citation33 These findings extend the range of autophagic receptors and cargos, and reveal a higher complexity in autophagic cargos selection.
Step 4. Degradation pathway analysis
The proteasome and the autophagosome constitute two major cytoplasmic protein degradation pathways in eukaryotic cells.Citation34 Their activities are carefully orchestrated to control various protein abundance and maintain cellular homeostasis. The GFP tagged cargo protein is often used to monitor autophagic flux due to their different sensibility to degradation in vacuole (GFP is relatively resistant to hydrolysis). A concentration-curve and time-course analysis with or without autophagic flux inhibitors could help to evaluate the extent of vacuolar delivery and proteolysis of interest cargos. Moreover, genetic approaches to block autophagy with several core autophagy-related gene mutants are required for validating the autophagic protein degradation pathway. Importantly, studies should also compare the effects of proteasomal degradation inhibitors like MG132 to determine the contribution from the proteasome. Recent findings strongly suggest that there is crosstalk and cooperation between the proteasome and the autophagosome pathways. Actually, these two degradation pathways have been found coordinately regulating BES1 (BRI1-EMS SUPRESSOR 1) degradation.Citation10 The proteasome is itself degraded by autophagy upon nitrogen starvation and proteasome inhibition.Citation11 Thus, comparative analyses of the proteasomal and the autophagy pathways are helpful to clarify their substrate selectivity and compensatory regulation of protein degradation.
Conclusions
The selective autophagy has emerged as a new and potent modulator of various abiotic and biotic stress responses in plants. Even though a growing number of ATG8s-interacting proteins have been identified, there remain challenges to our understanding of how specific cargos are targeted for degradation. Colocalization analysis with autophagic marker ATG8s has always been considered as an important step to determine whether the degradation of a protein is dependent on autophagy. Here, the cytoplasmic soluble GFP protein was found accumulating in the vacuoles and showing good colocalization with the autophagic marker mCherry-ATG8f after treatment with ConcA. Therefore, when reporting special cytosol proteins degraded by selective autophagy, we need to do colocalization analysis in cytoplasm rather than in vacuoles to avoid the cytoplasmic components that are nonselectively delivered into vacuoles by bulk autophagy or microautophagy. Additionally, colocalization analysis is just a beginning, and multiple parallel verifiable experiments, especially protein–protein interaction between ATG8 and the cargo/receptor, need to be carried out to reach a proper conclusion.
Supplemental Material
Download Zip (15.1 MB)Disclosure Statement
No potential conflicts of interest are disclosed.
Supplementary material
Supplemental data for this article can be accessed on the publisher’s website.
Additional information
Funding
References
- Mizushima N, Komatsu M. Autophagy: renovation of cells and tissues. Cell. 2011. 147(4):1–7. doi:10.1016/j.cell.2011.10.026.
- Johansen T, Lamark T. Selective autophagy mediated by autophagic adapter proteins. Autophagy. 2011. 7(3):279–296. doi:10.4161/auto.7.3.14487.
- Gatica D, Lahiri V, Klionsky DJ. Cargo recognition and degradation by selective autophagy. Nat Cell Biol. 2018. 20(3):233–242. doi:10.1038/s41556-018-0037-z.
- Noda NN, Ohsumi Y, Inagaki F. Atg8-family interacting motif crucial for selective autophagy. FEBS Lett. 2010. 584(7):1379–1385. doi:10.1016/j.febslet.2010.01.018.
- Birgisdottir ÅB, Lamark T, Johansen T. The LIR motif-crucial for selective autophagy. J Cell Sci. 2013. 126(15):3237–3247. doi:10.1242/jcs.126128.
- Svenning S, Lamark T, Krause K, Johansen T. 2011. Plant NBR1 is a selective autophagy substrate and a functional hybrid of the mammalian autophagic adapters NBR1 and p62/SQSTM1. Autophagy. 7(9):993–1010. doi:10.4161/auto.7.9.16389.
- Ji C, Zhou J, Guo R, Lin Y, Kung C-H, Hu S, Ng WY, Zhuang X, Jiang L. AtNBR1 is a selective autophagic receptor for AtExo70E2 in Arabidopsis. Plant Physiol. 2020. 184(2):777–791. doi:10.1104/pp.20.00470.
- Hu S, Ye H, Cui Y, Jiang L. AtSec62 is critical for plant development and is involved in ER-phagy in Arabidopsis thaliana. J Integr Plant Biol. 2020. 62(2):181–200. doi:10.1111/jipb.12872.
- Stephani M, Picchianti L, Gajic A, Beveridge R, Skarwan E, Sanchez De Medina Hernandez V, Mohseni A, Clavel M, Zeng Y, Naumann C, et al. 2020. A cross-kingdom conserved ER-phagy receptor maintains endoplasmic reticulum homeostasis during stress. Elife. 9:e58396. doi:10.7554/eLife.58396.
- Nolan TM, Brennan B, Yang M, Chen J, Zhang M, Li Z, Wang X, Bassham DC, Walley J, Yin Y, et al. 2017. Selective autophagy of BES1 mediated by DSK2 balances plant growth and survival. Dev Cell. 41(1):33–46. doi:10.1016/j.devcel.2017.03.013.
- Marshall RS, Li F, Gemperline DC, Book A, Vierstra R. Autophagic degradation of the 26S proteasome is mediated by the dual ATG8/ubiquitin receptor RPN10 in Arabidopsis. Mol Cell. 2015. 58(6):1053–1066. doi:10.1016/j.molcel.2015.04.023.
- Michaeli S, Honig A, Levanony H, Peled-Zehavi H, Galili G. Arabidopsis ATG8-INTERACTING PROTEIN1 Is Involved in Autophagy-Dependent Vesicular Trafficking of Plastid Proteins to the Vacuole. Plant Cell. 2014. 26(10):4084–4101. doi:10.1105/tpc.114.129999.
- Sjøgaard IMZ, Bressendorff S, Prestel A, Kausika S, Oksbjerg E, Kragelund BB, Brodersen P. 2019. The transmembrane autophagy cargo receptors ATI1 and ATI2 interact with ATG8 through intrinsically disordered regions with distinct biophysical properties. Biochem J. 476(3):449–465. doi:10.1042/BCJ20180748.
- Wu J, Michaeli S, Picchianti L, Dagdas Y, Galili G, Peled-Zehavi H. 2021. ATI1 (ATG8-interacting protein 1) and ATI2 define a plant starvation-induced reticulophagy pathway and serve as MSBP1/MAPR5 cargo receptors. Autophagy. doi:10.1080/15548627.2021.1872886.
- Brillada C, Teh K, Ditengo FA, Lee CW, Klecker T, Saeed B, Furlan G, Zietz M, Hause G, Eschen-Lippold L, et al. 2020. Exocyst subunit Exo70B2 is linked to immune signalling and autophagy. Plant Cell. 33(2):404–419. doi: 10.1093/plcell/koaa022.
- Acheampong AK, Shanks C, Cheng CY, Schaller GE, Dagdas Y, Kieber JJ. 2020. EXO70D isoforms mediate selective autophagic degradation of type-A ARR proteins to regulate cytokinin sensitivity. P Natl Acad Sci. 117(43):27034–27043. doi:10.1073/pnas.2013161117.
- Yang F, Kimberlin AN, Elowsky CG, Liu Y, Gonzalez-Solis A, Cahoon EB, Alfano JR. 2019. A plant immune receptor degraded by selective autophagy. Mol Plant. 12(1):113–123. doi:10.1016/j.molp.2018.11.011.
- Zhan N, Wang C, Chen L, Yang H, Feng J, Gong X, Ren B, Wu R, Mu J, Li Y, et al. 2018. S-nitrosylation targets GSNO reductase for selective autophagy during hypoxia responses in plants. Mol Cell. 71(1):142–154. doi:10.1016/j.molcel.2018.05.024.
- Vanhee C, Zapotoczny G, Masquelier D, Ghislain M, Batoko H. 2011. The Arabidopsis Multistress Regulator TSPO Is a Heme Binding Membrane Protein and a Potential Scavenger of Porphyrins via an Autophagy-Dependent Degradation Mechanism. Plant Cell. 23(2):785–805. doi:10.1105/tpc.110.081570.
- Thirumalaikumar VP, Gorka M, Schulz K, Masclaux-Daubresse C, Sampathkumar A, Skirycz A, Vierstra RD, Balazadeh S. 2020. Selective autophagy regulates heat stress memory in Arabidopsis by NBR1-mediated targeting of HSP90 and ROF1. Autophagy. 1–16. doi:10.1080/15548627.2020.1820778..
- Zeng Y, Li B, Ji C, Feng L, Niu F, Deng C, Chen S, Lin Y, Cheung KCP, Shen J, et al. 2021. A unique AtSar1D-AtRabD2a nexus modulates autophagosome biogenesis in Arabidopsis thaliana. P Natl Acad Sci. 118(17):e2021293118. doi:10.1073/pnas.2021293118..
- Droese S, Bindseil KU, Bowman EJ, Siebers A, Zeeck A, Altendorf K. 1993. Inhibitory effect of modified bafilomycins and concanamycins on P-and V-type adenosinetriphosphatases. Biochemistry. 32(15):3902–3906. doi:10.1021/bi00066a008.
- Huss M, Ingenhorst G, König S, Gaßel M, Dröse S, Zeeck A, Altendorf K, Wieczorek H. 2002. Concanamycin A, the specific inhibitor of V-ATPases, binds to the Vo subunit c. J Biol Chem. 277(43):40544–40548. doi:10.1074/jbc.M207345200.
- Yoshimoto K, Hanaoka H, Sato S, Kato T, Tabata S, Noda T, Ohsumi Y. 2004. Processing of ATG8s, ubiquitin-like proteins, and their deconjugation by ATG4s are essential for plant autophagy. Plant Cell. 16(11):2967–2983. doi:10.1105/tpc.104.025395.
- Yoshimoto K, Jikumaru Y, Kamiya Y, Kusano M, Consonni C, Panstruga R, Ohsumi Y, Shirasu K. 2009. Autophagy negatively regulates cell death by controlling NPR1-Dependent salicylic acid signaling during senescence and the innate immune response in Arabidopsis. Plant Cell. 21(9):2914–2927. doi:10.1105/tpc.109.068635.
- Xu SC, LoRicco JG, Bishop AC, James NA, Huynh WH, McCallum SA, Roan NR, Makhatadze GI. 2020. Sequence-independent recognition of the amyloid structural motif by GFP protein family. P Natl Acad Sci. 117(36):22122–22127. doi:10.1073/pnas.2001457117.
- Chanoca A, Kovinich N, Burkel B, Stecha S, Bohorquez-Restrepo A, Ueda T, Eliceiri KW, Grotewold E, Otegui MS. 2015. Anthocyanin vacuolar inclusions form by a microautophagy mechanism. Plant Cell. 27(9):2545–2559. doi:10.1105/tpc.15.00589.
- Nakamura S, Hidema J, Sakamoto W, Ishida H, Izumi M. 2018. Selective elimination of membrane-damaged chloroplasts via microautophagy. Plant Physiol. 177(3):1007–1026. doi:10.1104/pp.18.00444.
- Sieńko K, Poormassalehgoo A, Yamada K, Goto-Yamada S. 2020. Microautophagy in plants: consideration of its molecular mechanism. Cells. 9(4):887. doi:10.3390/cells9040887.
- Klionsky DJ, Abdel-Aziz AK, Abdelfatah S, Abdellatif M, Abdoli A, Abel S, Abeliovich H, Abildgaard MH, Abudu YP, Acevedo-Arozena A, et al. 2021. Guidelines for the use and interpretation of assays for monitoring autophagy (4th edition). Autophagy. 17(1):1–382. doi: 10.1080/15548627.2020.1797280
- Branon TC, Bosch JA, Sanchez D, Udeshi ND, Svinkina T, Carr SA, Feldman JL, Perrimon N, Ting AY. 2018. Efficient proximity labeling in living cells and organisms with TurboID. Nat Biotech. 36(9):880–887. doi:10.1038/nbt.4201.
- Ma J, Liang Z, Zhao J, Wang P, Ma W, Mai KK, Fernandez Andrade JA, Zeng Y, Grujic N, Jiang L, et al. 2021. Friendly mediates membrane depolarization-induced mitophagy in Arabidopsis. Curr Biol. 31(9):1931–1944.e4. doi:10.1016/j.cub.2021.02.034..
- Marshall RS, Hua Z, Mali S, McLoughlin F, Vierstra RD. 2019. ATG8-binding UIM proteins define a new class of autophagy adaptors and receptors. Cell. 177(3):766–781. doi:10.1016/j.cell.2019.02.009.
- Pohl C, Dikic I. 2019. Cellular quality control by the ubiquitin-proteasome system and autophagy. Science. 366(6467):818–822. doi:10.1126/science.aax3769.
