ABSTRACT
Background
Recently, platelet to white blood cell ratio (PWR) was reported as an independent prognostic predictor in acute promyelocytic leukemia. Acute myeloid leukemia (AML) often presents with abnormal platelet counts and white blood cell counts (WBC) at disease diagnosis. However, the clinical impact of PWR on cytogenetically normal AML (CN-AML) is still unclear. Therefore, we evaluate its prognostic impact on CN-AML patients.
Methods
We recorded the clinical information at the time of disease diagnosis, and calculated the ratio of platelet counts to WBC in 338 patients with CN-AML. To assess the prognostic value of PWR, we divided patients into low, intermediate and high group based on the values of PWR. The independent prognostic value of PWR was investigated in the context of the well-established predictors including white blood cell counts, age, and genes of NPM1, FLT3-ITD, CEBPA, and DNMT3A mutations. Receiver operating characteristic (ROC) curve was used to assess the performance of its prognostic prediction.
Results
Higher PWR have the higher levels of platelet counts, but lower levels of white blood cell counts, percentage of bone marrow blasts, FLT3-ITD and NPM1 mutations. The performance of survival prediction was comparable between PWR alone and combined molecular biomarkers. Moreover, PWR had the additional prognostic information to the molecular biomarkers. Finally, PWR was associated with favorable overall survival and event free survival in CN-AML patients independent of genetic subtypes and clinical parameters.
Conclusion
We found PWR was an independent prognostic predictor in CN-AML.
Introduction
Acute myeloid leukemia (AML) is a group of heterogeneous hematologic malignancies with poor clinical outcomes [Citation1]. One of the underlying reasons is biological and clinical heterogeneity. Hence, the lack of reliable biomarkers will hinder the improvement of prognostic prediction in clinical practice. To date, specific chromosomal aberrations like t(8;21) and inv(16) abnormalities and genes mutations including FLT3-ITD, NPM1, and CEBPA double allele mutations have been proved to be the most important predictors [Citation2]. With respect to cytogenetically normal AML (CN-AML), they did not have abnormal karyotype changes. Although there has been a significant progression in the discovery of novel predictors, a useful biomarker is still required for CN-AML prognostic stratifications.
Notably, clinical parameters are still important factors for classification in CN-AML [Citation3]. In clinical practice, patients with AML often present with leukocytosis and thrombocytopenia. Leukocytosis is usually caused by leukemia blasts, which release from bone marrow storage pool to peripheral blood. At the same time, leukemia blasts can suppress hematopoiesis including the inhibition of the generation of platelets [Citation4]. Notably, high WBC had been regarded as a reliable indicator of poor clinical outcome [Citation5]. Generally, platelet count has been used to predict bleed risk in AML. Low platelet count can contribute to bleeding complications, which is a dangerous and potentially fatal complication [Citation5]. Recently, a combinational index of platelet and WBC had been proved as an independent prognostic predictor in several diseases such as acute-on-chronic liver failure, renal malignancy, ischemic stroke, and acute promyelocytic leukemia [Citation5–8]. However, whether this ratio of platelet and WBC counts (PWR) has somewhat prognostic indication in CN-AML is still not investigated. Therefore, we analyzed the prognostic value of PWR in a large cohort of CN-AML patients.
Materials and Methods
Patients
In our cohort, we enrolled 338 patients with CN-AML in the first affiliated hospital of Zhejiang university college of medicine during 2015 and 2019. Bone marrow (BM) samples were obtained at the time of diagnosis. Patients received anthracycline and cytarabine induction chemotherapy, and consolidations chemotherapy based on the treating physician’s choice in an individualized manner. Details of the treatment protocols were reported in previous studies [Citation9,Citation10]. We excluded patients who had previous malignancy, transformed from myelodysplastic syndromes. All of the subjects were well-informed about the study and provided written informed consent to participate in the study. All procedures followed were in accordance with the ethical standards of the responsible committee on human experimentation (institutional and national) and with the Helsinki Declaration of 1975, as revised in 2000. The study was approved by the Institutional Review boards of the First Affiliated Hospital of Zhejiang University.
Cytogenetic and gene mutation analysis
Cytogenetic and molecular studies were performed centrally at ZIH molecular laboratories. Mononuclear cells were isolated from the BM samples at primary diagnosis by Ficoll–Hypaque density-gradient centrifugation [Citation11]. DNA samples obtained from mononuclear cells were extracted using the Gentra Puregene Blood DNA kit according to the manufacturer’s instructions. Genes of NPM1, FLT3-ITD, CEBPA, and DNMT3A mutations were detected by PCR amplification of the entire coding region or a portion of it, followed by direct bidirectional DNA sanger sequencing. Samples were amplified using the following PCR conditions: 95°C for 2 min, 35 cycles at 95°C for 30 s, 60°C for 40 s, and 72°C for 40 s, the final cycle at 72°C for 30 min. The primers were listed in Table S1.
Statistical analysis
The main objective is to investigate the prognostic impact of PWR on the overall survival (OS) of CN-AML patients. OS was measured as the date from disease diagnosis to death from any cause or censoring for patients alive at their last follow-up. To characterize patients with distinct levels of PWR, we subdivided our cohort into low, intermediate and high groups based on PWR values (). Patient characteristics were summarized using descriptive statistics, which included frequency counts, median and range. To evaluate the prognostic value of PWR, the Kaplan–Meier method was used in the univariate analysis and Cox proportional hazard regression model in the multivariate analysis. Receiver operating characteristic (ROC) curve was used to assess the performance of its prognostic prediction[Citation12]. All statistical analyses were conducted with R statistic package, version 3.6.1 (www.r-project.org). P < 0.05 demonstrated statistical difference.
Results
Characteristics of patients in high PWR group
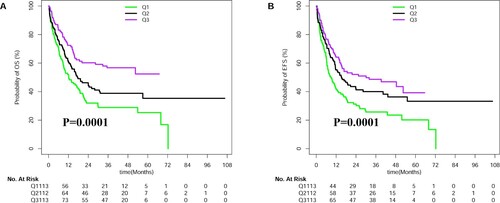
The median of PWR in this study is 3.76 with the range from 0.06 to 200. As shown in Figure S1A, PWR illustrates a positively skewed distribution. Log2 transformation of PWR was used to get closer to a normal distribution (Figure S1B). Among 338 patients, we divided patients into low, intermediate, and high groups based on the three equal quartiles (Q1: <33.3%, Q2: 33.3%–66.6%, Q3: >66.6%) of PWR values. We enrolled 100 healthy controls and found PWR ranges from 13.31 to 66.08 with the normal distribution(Figure S2). AML patients in a 3rd group are predominant in the normal value of PWR in healthy controls. Clinical features of patients are summarized in . Low PWR was associated with a significantly lower level of platelet counts, but a higher level of white blood cell counts (WBC), bone marrow blasts percentages, and was more common in genes of FLT3-ITD and NPM1 mutations. There was no statistically significant correlation between PWR levels and other variables including age, sex, hemoglobin (HB), DNMT3A, CEBPA double allele mutations, European Leukemia Net (ELN) genetic subtypes, treatment protocols and bone marrow transplantations (BMT) ().
Table 1. Clinical characteristics of CN-AML with distinct level of PWR.
PWR added the prognostic value to molecular predictors
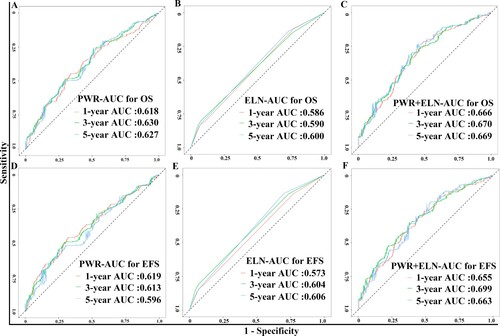
The median follow-up for our CN-AML patients was 570 days. Considering PWR as a continuous variable(Figure S3), we found PWR illustrates a negative linear relationship with overall survival. To determine whether PWR added some prognostic value to the well-established molecular predictors like ELN genetic subtypes [mutated NPM1 without FLT3-ITD and CEBPA double allele mutations], we first selected ELN genetic subtypes and DNMT3A mutations to construct the Cox regression Model 1. Secondly, we entered PWR (log2 transformed) to Model 1 as Model 2. The Akaike’s information criterion (AIC) of Model 1 decreases from 2021 to 2004 of Model 2, implying the better fitting of model 2 is contributed from PWR. For respectively predicting 1-, 3-, 5-year overall survival, the area under the survivalROC curve for the prediction of PWR alone was 0.618, 0.630, and 0.627, that for Model 1 was 0.586, 0.590, 0.600, and that for Model 2 was 0.666, 0.670, and 0.669 (A-C). Similar results were observed in the survivalROC curves of EFS(D–F). These results of greater values of AUC from PWR than those in Model 1 implied that PWR alone had the more accurately predictive value than the well-established ELN genetic subtypes. What’s more, AUC values of Model 2 are higher than Model 1, implying PWR added additional prognostic information to the molecular models of survival predictions. There were significant relapse rates among three groups in the univariate survival analysis (Figure S4). However, the statistical significance did not remain in the multivariate analysis (Figure S5). Concerning treatment-related toxicity, there were no differences among the three groups, when we compared WBC, Neutrocyte and platelet counts at day 15 after induction therapy (see Table S3).
Independent prognostic values of PWR
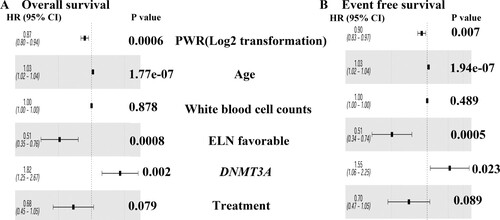
When we treated PWR as a third categorical variable or a continuous variable, higher PWR had more favorable overall survival (OS) and event-free survival (EFS) ( and Table S2). Even if we take the potential predictors as confounders, PWR was still as an independent prognostic factor in multivariate analyses after adjusting for age, WBC, and ELN genetic subtypes and DNMT3A mutations [for OS HR (95%CI), 0.87 (0.80,0.94); P = 0.0006 (); for EFS HR (95%CI), 0.90 (0.83,0.97); P = 0.007 ()]. Additionally, the impacts of PWR on OS and EFS were the same when we excluded the cases with FLT3-ITD (see Table S4).
Discussion
Acute myeloid leukemia is a group of clinically and molecularly heterogeneous disease. To data, molecular markers derived from gene expression profiling and next-generation sequencing are important to refine AML classification, and have strongly contributed to a better prognostication. However, several questions remain before these novel biomarkers can be implemented in clinical practice. First, the detection of these molecular markers is required for high-technical approaches, which are often expensive and limited the adoption in some developing countries. Second, even these equipments can be affordable in every routine laboratory, robustness is still needed to be considered because different sequencing pipelines and statistical methods might affect the ability to resolve disease subtypes. Clinical parameters are critical to initially establish a diagnosis of leukemia in clinical practice. What’s more, some of clinical parameters are still useful for disease classification and outcome prediction. Considering cost-effective services, utilities of the composition of two or more clinical available parameters to construct the novel biomarker would be a better method to search for robust and practical predictors. For instance, a complete blood count test measures several components of blood cells, at least partially including white blood cells (WBC), red blood cells and platelets. Leukemia patients often present with abnormal counts of WBC as well as platelets. High WBC is associated with poor clinical outcome due to increased early mortality and relapse [Citation13]. Low platelet indicates the propensity of bleeding. Additionally, the ratio of WBC to platelet can be used to predict early death in APL [Citation5]. Thus the combination of WBC and platelet counts(PWR) may have additional clinical significance for AML patients.
To investigate the prognostic impact of PWR in this study, we first explore the distribution of PWR and found the non-normal distribution of PWR. Log2 transformation of PWR was used to get a closely normal distribution. Using the log2 transformed PWR as a predictor, we found it was negatively correlated to OS in terms of a linear shape. To illustrate the survival curves, we also divided patients into three equal groups. Consistently, their survival trends of high-, intermediate-, and low group decreased (). Among the distinct levels of PWR, we found high PWR was negatively associated with percentages of bone marrow blasts, WBC, FLT3-ITD, and NPM1 mutations. These results indicate these well-known predictors might interact with the predictive ability of PWR. In this study, we found the predictive performance of PWR alone was better than the combined four genes of FLT3-ITD, NPM1, CEBPA, and DNMT3A mutations (Model 1) due to the greater AUC values of PWR( A–B, D–E). What’s more, PWR can also add some prognostic information to the molecular prediction models( C, F). Additionally, we conducted multivariable models in the context the well-established clinical and molecular factors. As a result, we found PWR can be used as an independent predictor in the context of WBC, age, genes of FLT3-ITD, NPM1, CEBPA, and DNMT3A mutations. Thus we believe PWR can be used as a practical predictor for CN-AML.
There are still some limitations in this study. First, CN-AML patients were retrospectively enrolled from a single center; therefore, selection bias may exist. Second, we only examine genes of FLT3-ITD, NPM1, CEBPA, and DNMT3A mutations, thus we could not exclude other mutated genes like IDH1, IDH2, ASXL1, and TET2 mutations, which will also confound the prognostic value of PWR in AML patients. Third, further large-scale and prospective research is still required to validate the prognostic value PWR. Therefore, caution in the application of our findings is still warranted.
In conclusion, we found the ratio of platelet to white blood cell counts (PWR) can be used as an independent predictor for overall survival and event-free survival in patients with CN-AML.
Supplemental Material
Download MS Word (565.9 KB)Acknowledgement
We thank for patients who participated in this study.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Additional information
Funding
References
- Bain BJ, Bene MC. Morphological and immunophenotypic clues to the WHO categories of acute myeloid leukaemia. Acta Haematol. 2019;141(4):232–244.
- Cai SF, Levine RL. Genetic and epigenetic determinants of AML pathogenesis. Semin Hematol. 2019;56(2):84–89.
- de Jonge HJ, Huls G, de Bont ES. Gene expression profiling in acute myeloid leukaemia. Neth J Med. 2011;69(4):167–176.
- Boyiadzis M, Whiteside TL. Exosomes in acute myeloid leukemia inhibit hematopoiesis. Curr Opin Hematol. 2018;25(4):279–284.
- Lou Y, Ma Y, Sun J, et al. Effectivity of a modified Sanz risk model for early death prediction in patients with newly diagnosed acute promyelocytic leukemia. Ann Hematol. 2017;96(11):1793–1800.
- Chen Z, Huang Y, Li S, et al. Platelet-to-white blood cell ratio: A prognostic predictor for 90-day outcomes in ischemic stroke patients with intravenous thrombolysis. J Stroke Cerebrovasc Dis Official J National Stroke Assoc. 2016;25(10):2430–2438.
- Jie Y, Gong J, Xiao C, et al. Low platelet to white blood cell ratio indicates poor prognosis for acute-on-chronic liver failure. BioMed Res Int. 2018;2018:7394904.
- Garbens A, Wallis CJD, Bjarnason G, et al. Platelet to white blood cell ratio predicts 30-day postoperative infectious complications in patients undergoing radical nephrectomy for renal malignancy. Canadian Urol Assoc J. 2017;11(11):E414–E420.
- Wang JH, Chen WL, Li JM, et al. Prognostic significance of 2-hydroxyglutarate levels in acute myeloid leukemia in China. Proc Natl Acad Sci U S A. 2013;110(42):17017–17022.
- Wang J, Li F, Ma Z, et al. High expression of TET1 predicts poor survival in cytogenetically normal acute myeloid leukemia from Two cohorts. EBioMedicine. 2018;28:90–96.
- Ma QL, Wang JH, Wang YG, et al. High IDH1 expression is associated with a poor prognosis in cytogenetically normal acute myeloid leukemia. Int J Cancer. 2015;137(5):1058–1065.
- Heagerty PJ, Lumley T, Pepe MS. Time-dependent ROC curves for censored survival data and a diagnostic marker. Biometrics. 2000;56(2):337–344.
- Kelaidi C, Ades L, Fenaux P. Treatment of acute promyelocytic leukemia with high white cell blood counts. Mediterr J Hematol Infect Dis. 2011;3(1):e2011038.