ABSTRACT
Purpose
Cytogenetically normal acute myeloid leukemia (CN-AML) is a heterogeneous disease with variable clinical outcomes. The identification of potential biomarkers to better classify the patients with unfavorable prognoses who may require more aggressive therapies is an emergent demand. PRDM16 is a transcriptional cofactor and histone methyltransferase, playing a critical role in maintaining hematopoietic stem cells, and MLL fusion-induced leukemogenesis. However, the prognostic value of PRDM16 in CN-AML is still unclear.
Materials and Methods
We retrospectively analyzed the PRDM16 expression and its association with gene mutations in CN-AML. Then the prognostic value of PRDM16 and its comparison with WT1 were analyzed.
Results
The results showed that about 73.6% of CN-AML patients harbored higher expression of PRDM16 than the healthy controls. Furthermore, CN-AML patients with high PRDM16 expression had a lower survival rate than the low PRDM16 expression group (50.5% vs. 83.3%, p = 0.0339). Interestingly, hemopoietic stem cell transplantation significantly improved the prognosis of CN-AML with high PRDM16 expression but not those with low PRDM16 expression. In terms of molecular genetics, high PRDM16 expression was significantly associated with a lower rate of CEBPA mutation (p = 0.01) and a higher rate of FLT3-ITD and DNMT3A mutation (p = 0.032 and p = 0.004, respectively). In addition, PRDM16 expression was significantly correlated with WT1 expression in CN-AML (r = 0.7, p < 0.001). These data suggested PRDM16 expression could be used to predict the outcome of patients with CN-AML.
Conclusion
PRDM16 is significantly associated with FLT3-ITD and DNMT3A mutation and WT1 expression and serves as a potential prognostic biomarker in CN-AML.
Introduction
Acute myeloid leukemia (AML) is a highly heterogeneous disease with recurrent cytogenetic abnormalities, which are reliable biomarkers for the diagnosis and prognosis of AML [Citation1]. But there is a subgroup of AML with undetectable chromosome abnormalities by the microscope, the so-called cytogenetically normal AML (CN-AML), accounting for about 50% of all AML cases [Citation2]. It is also sharply heterogeneous [Citation3] and commonly harbors various mutations, aberrant expressed genes and microRNAs, and changes in epigenetics and metabolic signatures, which might be potential and available prognostic biomarkers. For example, mutations of FLT3-ITD, NPM1, CEBPA, and WT1 [Citation4] genes are well-established prognostic factors for patients with CN-AML. In addition, high expressions of WT1, BAALC, ERG, MN1, IDH1, TET2, and RUNX1 [Citation5–10] are related to the poor outcomes of CN-AML patients [Citation11,Citation12]. But, there was still a lack of satisfactory prognostic biomarkers. So, it is important to identify the effective prognostic factors of CN-AML to provide the best management for CN-AML patients [Citation13,Citation14].
PR domain-containing 16 (PRDM16, also known as MEL1) is highly homologous to MECOM (also called EVI1), located in human chromosome 1p36, encoding 140-kDa Zinc finger protein, and services as a transcriptional cofactor and histone methyltransferase. It is originally identified in the development of brown adipose tissue and in the translocation of MDS/AML including t(1;3)(p36;q21)/RPM1-PRDM16 and t(1;21)(p36;q22)/RUNX1-PRDM16 [Citation15,Citation16]. In addition, PRDM16 is highly expressed in AML with NUP98-NSD1, which is considered a poor prognostic factor. It is also essential for the establishment, maintenance, and update of hematopoietic stem cell (HSC) [Citation17]. Some reports showed that high expression of PRDM16 in pediatric and adult AML indicated a lower overall survival rate [Citation18–21]. But, the study on the prognostic significance of PRDM16 in adult CN-AML is still limited. Here, we retrospectively analyzed the expression of PRDM16 in 121 adult CN-AML cohorts in our clinical center to clarify its clinical significance and association with genetic aberrations, such as FLT3-ITD, NPM1, and DNMT3 mutation. Furthermore, we have compared the correlation and predictive sensitivity, and specificity between the expression of PRDM16 and WT1 in the CN-AML cohort.
Patients and methods
Patients
Adult CN-AML patients (n = 121) diagnosed and treated in our medical center from January 2012 to December 2017 were involved in this study. All patients met the following eligibility criteria: age (≥) 14 years old, confirmed by the R-banding technique as cytogenetically normal, receiving standard ‘3 + 7’ induction chemotherapy. In addition, the patients with specific fusion genes were also excluded even though there are cytogenetically normal. Patients with complete remission (CR) receive consolidation chemotherapy, with or without allogeneic stem cell transplant (SCT), depending on whether there is a matching relative or non-relative donor. All procedures performed in studies involving human participants were under the ethical standards of the institutional and/or national research committee and with the 1964 Helsinki declaration and its later amendments or comparable ethical standards.
Analysis of public databases
RNA-seq data (TPM) and the corresponding clinical information were obtained from TCGA_AML (https://portal.gdc.cancer.gov/), and those with any translocations or fusion-genes positive were excluded. There are 68 cases of CN-AML in the cohort who were enrolled in this study. Gene Expression profiling of De Novo AML patients with normal karyotype and clinical information was retrieved from Gene Expression Omnibus (GEO) database, including GSE71014 (n = 104) and GSE12417 (n = 162). A receiver operating characteristic (ROC) curve was used to identify the optimal cutoff levels that best discriminated patients with the outcome. Survival functions were estimated using the Kaplan–Meier method and compared using the log-rank test.
Quantitative real-time PCR
Total RNAs of BM mononuclear cells from CN-AML patients (n = 121) and healthy donors (n = 15) were extracted by TRIZOL reagent (Ambion). Real-time PCR was performed by ABI Q6 PCR systems (Application Biosystems, Waltham, MA, USA). The following primers were used for detecting PRDM16 mRNA expression: Sense, 5′- CAGCCAATCTCACCAGACACCT-3′ and antisense, 5′-GTGGCACTTGAAAGGCTTCTCC-3′ (Origene); For WT1 gene (Sense, 5′-CGAGAGCGATAACCACACAACG-3′, antisense, 5′-GTCTCAGATGCCGACCGTACAA-3′); For GAPDH: Sense, 5′-GTCTCCTCTGACTTCAACAGCG-3′, and antisense, 5′-ACCACCCTGTTGCTGTAGCCAA-3′. Relative quantification analysis was performed using the comparative ΔCt method. The relative quantitative value of one specific gene was calculated by the 2^- ΔCt formula (ΔCt = Ct value of the specific gene – Ct value of GAPDH).
Statistical analysis
Overall survival (OS) was defined as the time from treatment randomization until death from any cause. Event-free survival (EFS) was defined as the time from the start date of study treatment to the time when the primary refractory disease was confirmed (i.e. the date when the failure to achieve a response to induction therapy was determined), relapse or death. IBM SPSS Statistics 25.0 was used for statistical analysis. Mann-Whitney’s U test compared the two groups of continuous variables. The Pearson chi-square analysis compared the differences between classified variables. The Kaplan-Meier method and Cox regression model (multivariate analysis) were used to analyzing the effect of PRDM16 expression on overall survival (OS) and disease-free survival (EFS). The P-value of 0.05 is considered to be significant.
Results
The expression of PRDM16 in CN-AML
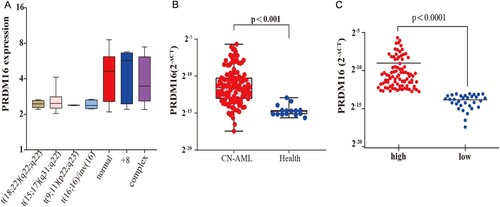
As shown in A, the expression of PRDM16 is higher in cytogenetical normal, trisomy 8, and complex cytogenetical AML patients than in other karyotypes in the TCGA AML cohort (n = 139). Subsequently, the quantitative RT–PCR analysis showed that the mean expression value of PRDM16 in CN-AML was significantly 32 times higher than that in normal bone marrow (NBM, n = 15, p < 0.001, B). About 73.6% (89/121) of CN-AML patients showed a high expression of PRDM16, which was defined as higher than the upper limit of NBM controls. The mean value of the high PRDM16 expression group was 28 times greater than that in the low PRDM16 expression group (C).
Figure 1. The expression of PRDM16 in CN-AML. (A) The expression of PRDM16 (Log2 FPKM) in TCGA AML cohort (n = 139) with different karyotypes. (B) Quantitative RT-PCR showed PRDM16 expression in 121 cytogenetically normal AML (CN-AML) bone marrow samples in our clinical center and 15 normal bone marrow samples, and (C) PRDM16 expression in high and low PRDM16 expression subgroup of CN-AML. The graph showed the log2 (2-ΔCt) value against GAPDH.
The prognostic value of PRDM16 in CN-AML
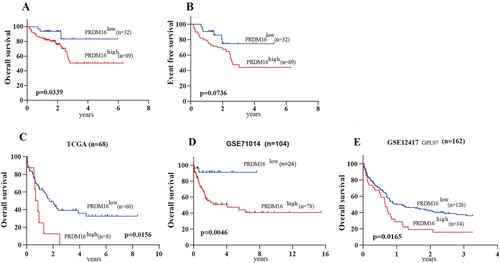
Importantly, CN-AML patients with high PRDM16 expression harbored a significantly lower rate of over survival (OS) than those with low PRDM16 expression (50.5% vs. 83.3%, p = 0.0339; A). Event-free survival rate was lower in the high RPDM16 expression group (p = 0.0736, B), although not statistically significant. Furthermore, the prognostic value of PRDM16 in OS was confirmed in 3 independent CN-AML cohorts, including TCGA (p = 0.0156, C), GSE12417 (p = 0.0165, D), and GSE71014 (p = 0.0234, E).
Figure 2. The prognostic value of PRDM16 in CN-AML. The Kaplan–Meier curves show the over survival (OS, A) and event-free survival (EFS, B) of high and low PRDM16 expression groups of the CN-AML cohort. The Kaplan–Meier curves of OS for 3 independent CN-AML cohorts, including TCGA (C), GSE71014 (D), and GSE12417 (E).
The effect of HSCT on CN-AML with different PRDM16 expression
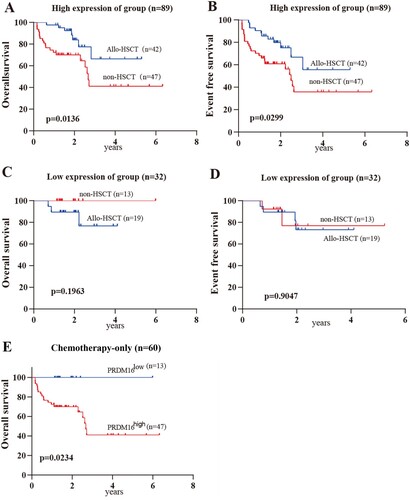
Expectedly, HSCT could significantly prolong the overall survival (p = 0.0136) and the event-free survival (p = 0.0299) of CN-AML patients with high PRDM16 expression (A,B). But unexpectedly, there was no effect on those patients with low PRDM16 expression (C,D). Notably, CN-AML patients with low PRDM16 expression could achieve long-term remission with standard intensive chemotherapy only, suggesting HSCT might not be required in this group. But, patients with high PRDM16 expression might benefit from HSCT. In addition, when excluding the effect of HSCT on the clinical outcome of this cohort, the prognostic significance of PRDM16 was more prominent in CN-AML with the chemotherapy-alone group (high vs. low PRDM16 expression group: 41.1% vs 100%, p = 0.0234; E), although there was no significance in the hematopoietic stem cell transplantation (HSCT) group (Supplementary Figures 1 and 2).
Figure 3. The effect of HSCT on CN-AML with different PRDM16 expressions. The Kaplan–Meier curves for OS (A) and EFS (B) of CN-AML cohort with High PRDM16 expressions. The Kaplan–Meier curves for OS (C) and EFS (D) of CN-AML cohort with low PRDM16 expression. E. The Kaplan–Meier curves show the over the survival of high and low PRDM16 expression groups of the CN-AML cohort with chemotherapy-only.
The clinic and molecular characteristics of CN-AML with different PRDM16 expressions
In terms of clinical features, the expression of PRDM16 seemed to be not related to most factors, including sex, age, white blood cell count, hemoglobin level, platelet count, bone marrow blast ratio, or Franco-American-British (FAB) subtype (). In the molecular subsets, the ratio of FLT3-ITD and DNMT3A mutation in CN-AML with high PRDM16 expression is significantly higher than that in patients with low PRDM16 expression (35.96% vs. 15.6%, p = 0.032 and 28.9% vs. 3.1%, p = 0.004, respectively; ). In contrast, the high PRDM16 group harbors a lower ratio of CEPBA mutation than the low PRDM16 group (21.7% vs. 43.8%, p = 0.01; ). However, there was no significant difference in the mutation ratio of NPM1, RAS, or KIT (). Univariate cox analysis showed that 4 variables, allo-HSCT, induction response, age, and high PRDM16 expression, were associated with shorter OS (HR = 3.378, p = 0.046 for PRDM16 expression; Supplementary Table 1). Furthermore, multivariate cox analyses showed only induction response was significantly related to shorter OS.
Table 1. Comparison of clinical manifestations and laboratory features between CN-AML patients with low and high PRDM16 expressions.
The capacity of PRDM16 expression to predict prognosis in patients with CN-AML
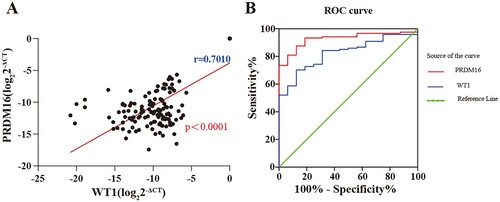
To explore the association of PRDM16 expression with another well-known adverse prognostic factor, WT1 expression in CN-AML, Spearman correlation analysis was performed. The results showed that PRDM16 expression was significantly correlated to WT1 expression in CN-AML (r = 0.7, p < 0.0001; A). Furthermore, the receiver operating characteristic (ROC) was used to evaluate the discriminative capacity of PRDM16 and WT1 expression in CN-AML. It revealed that PRDM16 expression had a similar or even higher prognostic value than WT1 expression in CN-AML (AUC = 0.928, p < 0.0001 for PRDM16 vs. AUC = 0.828, p < 0.0001 for WT1; B).
Figure 4. The comparison of the capacity of prognosis prediction between RPDM16 and WT1 expression. (A) Correlation between the expression of PRDM16 and WT in the CN-AML cohort; (B) Receiver operating characteristic (ROC) curves of PRDM16 and WT1 expression are used to determine the values for evaluating the prognosis of CN-AML.
Discussion
CN-AML is a large cytogenetic subset in AML patients and lacks sensitive prognostic biomarkers, so the identification of powerful prognostic biomarkers has important implications for these patients. Apart from gene mutations, such as FLT3, NPM1, and CEBPA, have been contributed to the risk stratification of CN-AML, abnormal gene expressions, including WT1, RUNX1, and TET2, are a common characteristic of these subsets and be served as prognostic factors. In this study, we found tPRDM16 is highly expressed in CN-AML subsets, related to some genetic alterations, such as FLT3-ITD, DNMT3A, and CEBPA mutation, and served as a potential adverse prognostic factor for survival, with a sensitivity similar to WT1 expression based on ROC analysis. Similar results have been reported in some previous studies [Citation18]. Dao et al. showed that high PRDM16 expression, associated with NPM1 mutation and FLT3-ITD, was an independent poor prognostic factor in267 adult intermediate cytogenetic risk AMLs, which included a normal cytogenetic cohort [Citation22].
It has been reported that EVI1 (PRDM3) and PRDM16 have similar functions in normal and malignant hematopoiesis [Citation23]. Similar to EVI1, PRDM16, as a proto-oncogene, can be activated and upregulated by retrovirus infection and induce leukemogenesis. In normal hematopoiesis, PRDM16 is highly and selectively expressed in the normal HSPC compartment and plays an essential role in their maintenance. Notably, in myeloid leukemia, PRDM16 is actually selectively overexpressed in some AML subsets, including AML with NUP98-NSD1, PRDM16 translocation, trisomy 8, complex karyotype, and cytogenetic normal, but low expressed in AML with t(8;21), t(15;17), inv(16) and 11q23 (MLL) translocation based on the TCGA cohort (A and Ref. [Citation23]). Matsuo et al. found that high EVI1 expression was mainly detected in MLL-rearranged AML and megakaryocytic-lineage AML, while high PRDM16 expression was detected in myelocytic-lineage AML and myelomonocytic lineage AML without MLL-rearrangements. So, the mechanisms for the upregulation of PRDM16 might be different from EVI1 overexpression. Further studies were needed to investigate how theses factors are upregulated and activated and elucidate their roles in CN-AML pathology and development.Importantly, the study showed that the patients with higher PRDM16 expression have significantly lower overall survival rates (A, p = 0.0339). It suggested that PRDM16 expression is a valuable predictor and could be used for risk stratification of CN-AML patients. Furthermore, its predictive impact has been validated in 3 other CN-AML cohorts, including TCGA, GSE71014, and GSE12417. Interestingly, in patients with chemotherapy-treated only, it seemed that the predicted value of PRDM16 is more potential and significant. (C, p = 0.0234) It might be very important to the risk stratification and management of those patients who are unfit and non-essential for HSCT. Just as univariate analyses of OS in CN-AML, allo-HSCT could significantly improve the survival of the entire cohort. Unexpectedly, allo-HSCT has significantly increased the overall and event-free survival rate (A,B, p = 0.0136 and p = 0.0299) in patients with high PRDM16 expression, but it appeared not to improve in patients with low PRDM16 expression. It suggested that HSCT might not be necessary for the CN-AML patients, at least in part, with low PRDM16 expression, avoided some side effects such as GVHD for a better life.
Yoon et al. have firstly reported that WT1 expression associated with NPM1 and FLT3-ITD mutation is defined as a high-risk subgroup in CN-AML [Citation4]. Here, we found that the expressions of WT1 and PRDM16 were significantly relevant, which suggested high PRDM16 expression was also an adverse prognostic factor in CN-AML, although it was not an independent factor in this study, similar to other factors, such as FLT3-ITD, CEBPA, and NPM1 mutation. It might be related to the high transplantation ratio (50%) in our CN-AML cohort. Of course, it required more investigation. However, it seemed that PRDM16 could be as sensitive as WT1 in predicting outcomes of CN-AML. But it might require more cases for validation.
Overall, we revealed the prognostic impact of PRDM16 expression in adult CN-AML, which might contribute to better risk stratification and management of these patients.
Supplemental Material
Download TIFF Image (330.4 KB)Supplemental Material
Download TIFF Image (266.2 KB)Supplemental Material
Download MS Word (15.6 KB)Supplemental Material
Download MS Word (17.1 KB)Acknowledgements
All the samples were from Hematological Biobank, Jiangsu Biobank of Clinical Resources.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Additional information
Funding
References
- Cancer Genome Atlas Research N, Ley TJ, Miller C, et al. Genomic and epigenomic landscapes of adult de novo acute myeloid leukemia. N Engl J Med. 2013;368:2059–2074.
- Mrózek K, Heerema NA, Bloomfield CD. Cytogenetics in acute leukemia. Blood Rev. 2004;18:115–136.
- Dohner H, Estey E, Grimwade D, et al. Diagnosis and management of AML in adults: 2017 ELN recommendations from an international expert panel. Blood. 2017;129:424–447.
- Yoon JH, Kim HJ, Jeon YW, et al. Outcome of allogeneic hematopoietic stem cell transplantation for cytogenetically normal AML and identification of high-risk subgroup using WT1 expression in association with NPM1 and FLT3-ITD mutations. Genes Chromosomes Cancer. 2015;54:489–499.
- Zhou JD, Yang L, Zhang YY, et al. Overexpression of BAALC: clinical significance in Chinese de novo acute myeloid leukemia. Med Oncol. 2015;32:386.
- Metzeler KH, Dufour A, Benthaus T, et al. ERG expression is an independent prognostic factor and allows refined risk stratification in cytogenetically normal acute myeloid leukemia: a comprehensive analysis of ERG, MN1, and BAALC transcript levels using oligonucleotide microarrays. J Clin Oncol. 2009;27:5031–5038.
- Aref S, Ibrahim L, Morkes H, et al. Meningioma 1 (MN1) expression: refined risk stratification in acute myeloid leukemia with normal cytogenetics (CN-AML). Hematology. 2013;18:277–283.
- Ma QL, Wang JH, Wang YG, et al. High IDH1 expression is associated with a poor prognosis in cytogenetically normal acute myeloid leukemia. Int J Cancer. 2015;137:1058–1065.
- Zhang TJ, Zhou JD, Yang DQ, et al. TET2 expression is a potential prognostic and predictive biomarker in cytogenetically normal acute myeloid leukemia. J Cell Physiol. 2018;233:5838–5846.
- Fu HF L, Tian L, Xu K, et al. High expression of RUNX1 is associated with poorer outcomes in cytogenetically normal acute myeloid leukemia. Oncotarget. 2016;7(13):15828–15839.
- Mrozek K, Marcucci G, Paschka P, et al. Clinical relevance of mutations and gene-expression changes in adult acute myeloid leukemia with normal cytogenetics: are we ready for a prognostically prioritized molecular classification? Blood. 2007;109:431–448.
- Marcucci G, Mrozek K, Radmacher MD, et al. The prognostic and functional role of microRNAs in acute myeloid leukemia. Blood. 2011;117:1121–1129.
- Weber S, Haferlach T, Alpermann T, et al. Feasibility of BAALC gene expression for detection of minimal residual disease and risk stratification in normal karyotype acute myeloid leukaemia. Br J Haematol. 2016;175:904–916.
- Marjanovic I, Karan-Djurasevic T, Ugrin M, et al. Use of wilms tumor 1 gene expression as a reliable marker for prognosis and minimal residual disease monitoring in acute myeloid leukemia with normal karyotype patients. Clin Lymphoma Myeloma Leuk. 2017;17:312–319.
- Sakai I, Tamura T, Narumi H, et al. Novel RUNX1-PRDM16 fusion transcripts in a patient with acute myeloid leukemia showing t(1;21)(p36;q22). Genes Chromosomes Cancer. 2005;44:265–270.
- Roche-Lestienne C, Deluche L, Corm S, et al. RUNX1 DNA-binding mutations and RUNX1-PRDM16 cryptic fusions in BCR-ABL+ leukemias are frequently associated with secondary trisomy 21 and may contribute to clonal evolution and imatinib resistance. Blood. 2008;111:3735–3741.
- Aguilo F, Avagyan S, Labar A, et al. Prdm16 is a physiologic regulator of hematopoietic stem cells. Blood. 2011;117:5057–5066.
- Yamato G, Yamaguchi H, Handa H, et al. Clinical features and prognostic impact of PRDM16 expression in adult acute myeloid leukemia. Genes Chromosomes Cancer. 2017;56:800–809.
- Shiba N, Ohki K, Kobayashi T, et al. High PRDM16 expression identifies a prognostic subgroup of pediatric acute myeloid leukaemia correlated to FLT3-ITD, KMT2A-PTD, and NUP98-NSD1: the results of the Japanese Paediatric Leukaemia/Lymphoma Study Group AML-05 trial. Br J Haematol. 2016;172:581–591.
- Jo A, Mitani S, Shiba N, et al. High expression of EVI1 and MEL1 is a compelling poor prognostic marker of pediatric AML. Leukemia. 2015;29:1076–1083.
- Miyamura T, Moritake H, Nakayama H, et al. Clinical and biological features of paediatric acute myeloid leukaemia (AML) with primary induction failure in the Japanese Paediatric Leukaemia/Lymphoma Study Group AML-05 study. Br J Haematol. 2019;185:284–288.
- Dao F, Chen W, Long L, et al. High PRDM16 expression predicts poor outcomes in adult acute myeloid leukemia patients with intermediate cytogenetic risk: a comprehensive cohort study from a single Chinese center. Leuk Lymphoma. 2020;62(1):185–193.
- Matsuo H, Goyama S, Kamikubo Y, et al. The subtype-specific features of EVI1 and PRDM16 in acute myeloid leukemia. Haematologica. 2015;100:e116–e117.
