ABSTRACT
Background
Resembling acute promyelocytic leukemia (APL) is a unique subtype of APL who sharing clinical, morphological, and immunophenotypic features with typical APL, but lacking evidence of PML-RARA fusion gene and usually insensitive to arsenic trioxide (ATO) and all-trans retinoic acid (ATRA). For years, RARA, RARB and RARG rearrangement were found in resembling APL continually. The confirmed partner genes of RARG rearrangement included CPSF6, NUP98, NPM1, PML, and HNRNPC. These patients were a group of resembling APL with rare molecular genetic abnormality and unfavorable prognosis. They usually were resistant to ATO and ATRA but partially sensitive to anthracycline-based chemotherapy.
Case presentation
We reported a 25-year-old female patient with a novel fusion gene RARG-HNRNPM (RARG chr12:53606869: –; HNRNPM chr19: 8527413: + based on GRCh37/hg19 Assembly) through RNA-seq as resembling APL. The patient with RARG-HNRNPM was benefited from a combined chemotherapy homoharringtonine, cytarabine, and aclacinomycin (HAA) regimen with no relapse.
Discussion and conclusions
RARG rearrangement resembling APL are various. The treatment should be switched from ATRA/ATO to AML combined chemotherapy regimen early.
1. Background
Acute promyelocytic leukemia (APL) with typically PML-RARA fusion gene caused by t (15;17) (q22; q21) was distinguished from other types of acute myeloid leukemia (AML) according to the World Health Organization (WHO) 2016 criteria [Citation1,Citation2]. The clinical features of APL include severe hemorrhagic and coagulation disorder and sensitivity to arsenic trioxide (ATO), all-trans retinoic acid (ATRA), and anthracyclines. Nowadays, APL is considered ‘curable leukemia’ [Citation1]. Resembling APL is referring to those rare cases that sharing clinical, morphological, and immunophenotypic features with APL, but lacking evidence of PML-RARA fusion gene and usually resistance to ATRA [Citation3,Citation4]. Herein, we reported a 25-year-old female case with a novel fusion gene involving the retinoic acid receptor gamma (RARG) gene and heterogeneous nuclear ribonucleoprotein M (HNRNPM) gene in a leukemic phenotype resembling APL.
2. Case presentation
A 25-year-old woman from China was admitted to Institute of Hematology and Blood Diseases Hospital with the symptoms of fever, pharyngodynia, and thoracalgia. Informed consent was obtained from the patient before bone marrow aspiration for further cytogenetic and molecular analysis.
The whole blood cell count of the female patient showed that the white cell count was 26.75 × 109/L, hemoglobin level was 67 g/L, and platelet count was 27 × 109/L. Evaluation of coagulation function was as follows: Prothrombin time (PT) 12.6 (reference 10–14 s), Activated Partial Thromboplastin Time (APTT) 24.3 (reference 23.7–36 s), Thrombin Time (TT)16.0 (reference 13.3–19.3 s), Fibrinogen 4.37 (reference 2.0–4.0 g/L), Fibrin Degradation Products(FDP) 44.9 (reference < 5 μg/mL), D-Dimer test 18.32 (reference 0.0–0.55 mg/L FEU).
Bone marrow (BM) morphology showed 80.5% atypical hypergranular promyelocytes ((A)). The positive rate of specific esterase (SE) and myeloperoxidase (MPO) for immunohistochemical staining was 100% and 98%, respectively. Immunophenotype of the patient showed that blasts strongly expressed CD13 and CD33, CD117, CD64, CD123, and MPO are positive, but CD34, HLA-DR are negative.
Figure 1. Morphology, cytogenetic and molecular analysis of the RARG-HNRNPM fusion gene. (A) BM smear morphology; (B) Chromosome analysis of the patient.
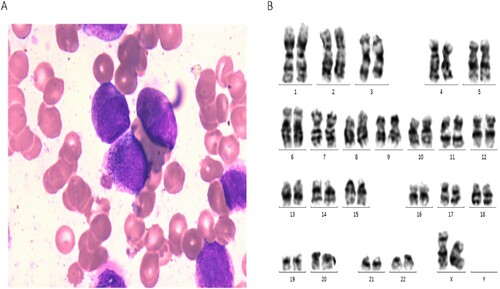
Multiplex real-time quantitative PCR of 43 fusion genes, including PLZF-RARA, PML-RARA (L), PRKAR1A-RARA, NPM-RARA, NUMA1-RARA, and STAT5b-RARA, displayed negative results (Supplementary Table 1). Also, fluorescence in situ hybridization (FISH) was negative for PML-RARA, and the karyotype was normal ((B)). Moreover, targeted-hematological disease gene mutation in the next-generation sequencing (NGS) panel also did not reveal any additional mutation.
Considering the morphology and immunophenotype were similar to APL, while we didn’t find the molecular evidence of APL, we performed RNA-seq for a definitive diagnosis. Total RNA was extracted from BM mononuclear cells. RNA-sequencing was performed with Illumina NovaSeq 6000. The RNA-seq data was available in PubMed (PRJNA719216). The STAR-Fusion software (http://star-fusion.github.io) was applied to match the UCSC Genome Browser on Human February 2009 (GRCh37/hg19) Assembly and call the fusion gene. The total fusion gene list of the patient was shown as Supplementary Table 2. Reverse transcription-polymerase chain reaction (RT-PCR) and Sanger sequencing further verified the identified genes. The following primers were used to amplify RARG-HNRNPM mRNA: 5’-AGCCAGCCCTACATGTTCCC-3’ for RARG, 5’-TGCTCTCCTGGCATGTTCACC-3’ for HNRNPM.
We identified RARG-HNRNPM fusion gene with 54 junction reads and 0 spanning flag through RNA-seq analysis. The prediction of gene split position was: RARG chr12:53606869: –; HNRNPM chr19: 8527413: + based on GRCh37/hg19 Assembly ((A)). The coding sequence (CDS) size of RARG-HNRNPM was 3087 bp. In contrast, we did not observe CDS of HNRNPM-RARG with the 1 junction reads and 0 spanning flag. The amplicon size of RARG-HNRNPM fusion was 252 bp ((B)). The chimeric fusion gene was detected in RARG exon9 and HNRNPM exon3 ((C)). Based on the above evidence, we diagnosed this patient as resembling APL with RARG-HNRNPM.
Figure 2. Detection of the RARG/HNRNPM fusion. (A) The circros visualization of the reciprocal RARG/HNRNPM fusion on chromosome. (B) Picture of the amplified fragment on agarose gel. Marker: DNA 2K marker; lane 1: PCR amplicons of RARG-HNRNPM patient; lane 2: NC negative control. (C) Sanger sequencing for RARG-HNRNPM fusion point surrounding base pairs. Verified by NCBI blast, it completely aligned with RARG exon9 and HNRNPM exon3. (D) Overview of the chimeric fusion diagram: the fusion gene structure contained RARG whole DBD (90–155 amino acids), which coincided with two zinc finger motifs (90–110 amino acids and 126–150 amino acids) and two main RNA-recognition motifs (RRMs) of HNRNPM gene.
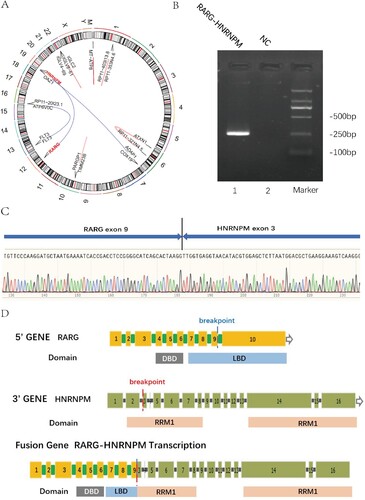
We investigated the RNA binding domain (RBD) and DNA binding domain (DBD) of RARG-HNRNPM fusion gene through UniProt (https://www.uniprot.org). The fusion gene contains RARG whole DBD (ranging from 90 to 155 amino acids) which highly coinciding with 2 zinc finger areas (90–110 amino acids, 126–150 amino acids) and 2 incomplete RRMs (RNA-recognition motifs) of HNRNPM gene ((D)).
Since the coagulation disorder and morphology of this patient were similar to APL, ATRA treatment was started after admitted. But the patient could not tolerance ATRA (serious labia edema), a dosage of 60 mg/d was administered for only 12 days during the induction therapy. As PML/RARA fusion gene was negative, she received a standard DA 3 + 7 regimen (Daunorubicin 60 mg/m2/ d × 3d; cytarabine 100 mg/m2/d × 7d) as induction therapy but failed to achieve complete remission (CR). Owing to significant bleeding tendency and pulmonary infection, the patient received homoharringtonine 3 mg d1–7, cytarabine 100 mg d1–10, and aclacinomycin 20 mg d3–7 (HAA) as the salvage regimen and achieved CR. Then, 3 courses of consolidation chemotherapy regimens were given as follows: azacitidine 100 mg d1–7, aclarubicin 20 mg d4–9 and cytarabine 100 mg d4–10 (AZA + AA); azacitidine 100 mg d1–5, homoharringtonine 3 mg d4–6 and cytarabine 100 mg d4–7 (AZA + HA); cytarabine (1 g/m2/q12h d1–3). Up till now, the patient had kept in CR for 10 months.
3. Discussion and conclusions
WHO Classification of Tumors of Hematopoietic and Lymphoid Tissues (2016) only ascribes APL with PML/RARA to AML with a recurrent genetic abnormality. While 16 gene partners fused with RARA [Citation4] (such as PLZF, NPM, NUMA, BCOR, FIP1L1, STAT5b, STAT3, PRKR1a, OBFC2A, GTF2I, TNRC18, NUP98, TFG, FNDC3B, IRF2BP2, and TBLR1) and cases with TBL1XR1-RARB rearrangement [Citation5,Citation6] have been found in the presence of APL cases without classic t (15;17). Variant APL with RARA and RARB rearrangement is characterized by its vast majority resistance to ATRA treatment as lacking target of ATO and similar morphological and molecular biology characteristics with typical APL [Citation7].
To our knowledge, 12 RARG rearrangement cases in variant APL [Citation8–12] (including 10 cases of gene fusion and 2 cases of RARG abnormal expression) have been reported as follows: CPSF6-RARG (4), NUP98-RARG (3), NPM1-RARG-NPM1 (1), PML-RARG (1), and HNRNPC-RARG (1). Hitherto, we reported the first case of RARG-HNRNPM gene fusion in resembling APL. All of these patients present clinical, morphological, and immunophenotypic features similar to those of APL.
It is reported that RARA breakpoint is highly fixed in variant APL which always occurs in exon 3 [Citation7,Citation13]. While, breakpoints harboring within RARG are different, including 5’ UTR, exon1, exon 2, intron 3, exon 4, and exon 9 [Citation4,Citation13] . RARG breakpoint was also detected in exon 9 in our patient. Nevertheless, the functional domain of RARA and RARG were mostly remained in resembling APL cases regardless of different partner gene breakpoint.
RARG shares similar sequence and biological characteristics with RARA in the nucleus receptor family. It is responsible for regulating the healthy BM microenvironment and correcting hematopoiesis [Citation3]. Specifically, RARG function contains of increasing the numbers of hematopoietic progenitors and eliciting HSC self-renewal by affecting other gene expression [Citation11].
HNRNPM is an RNA binding protein (RBP) that contains the common architecture of potential RNA-binding units: two RRM structures. HNRNPM is a subtype of heterogeneous nuclear ribonucleoproteins (hnRNPs) that plays a critical role in recognizing splice site and regulating alternative splicing [Citation14].
Based on the cases reported above, patients with RARG rearrangement were expected to be resistant to ATRA/ATO but partially sensitive to anthracycline-based chemotherapy. Six out of eleven patients (including our patient) with RARG arrangement achieved CR with chemotherapy, while four out of eleven patients died during induction therapy. Although our patient showed intolerance to treatment with ATRA and no benefit from the standard DA regimen, she achieved CR after HAA regimen chemotherapy and kept in CR for 10 months. Therefore, RARG rearrangement APL should early switch from ATRA/ATO to AML combined chemotherapy regimen after distinguishing from typical APL. In fact, the proper treatment and prognosis for variant APL still needs to be accumulated.
Taken together, we reported a novel RARG-HNRNPM fusion gene in a patient resembling APL. It broadens the spectrum of variant APL genetics type.
Ethics approval and consent to participate
This study was approved by the National Clinical Research Center for Blood Diseases (Ethics number KT2020004-EC-2).
Supplemental Material
Download MS Word (13.5 KB)Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
RNA-seq data were available in NCBI with accession number PRJNA719216 (https://submit.ncbi.nlm.nih.gov/subs/bioproject/SUB9406085/overview). All data will be shared with qualified scientific and medical researchers, upon the researcher’s request, as necessary for conducting legitimate research.
Additional information
Funding
References
- Grimwade D, Lo Coco F. Acute promyelocytic leukemia: a model for the role of molecular diagnosis and residual disease monitoring in directing treatment approach in acute myeloid leukemia. Leukemia. 2002;16(10):1959–1973.
- Dohner H, Weisdorf DJ, Bloomfield CD. Acute myeloid leukemia. N Engl J Med. 2015;373(12):1136–1152.
- Conserva MR, Redavid I, Anelli L, et al. RARG gene dysregulation in acute myeloid leukemia. Front Mol Biosci. 2019;6:114.
- Zhang X, Sun J, Yu W, et al. Current views on the genetic landscape and management of variant acute promyelocytic leukemia. Biomark Res. 2021;9(1):33.
- Shiba N, Yoshida K, Hara Y, et al. Transcriptome analysis offers a comprehensive illustration of the genetic background of pediatric acute myeloid leukemia. Blood Adv. 2019;3(20):3157–3169.
- Osumi T, Tsujimoto SI, Tamura M, et al. Recurrent RARB translocations in acute promyelocytic leukemia lacking RARA translocation. Cancer Res. 2018;78(16):4452–4458.
- Geoffroy MC, de The H. Classic and variants APLs, as viewed from a therapy response. Cancers. 2020;12(4):967.
- Qin YZ, Huang XJ, Zhu HH. Identification of a novel CPSF6-RARG fusion transcript in acute myeloid leukemia resembling acute promyelocytic leukemia. Leukemia. 2018;32(10):2285–2287.
- Such E, Cervera J, Valencia A, et al. A novel NUP98/RARG gene fusion in acute myeloid leukemia resembling acute promyelocytic leukemia. Blood. 2011;117(1):242–245.
- Zhang X, Li F, Wang J, et al. RARgamma-rearrangements resemble acute promyelocytic leukemia and benefit from 3 + 7 regimen. Leuk Lymphoma. 2019;60(7):1831–1834.
- Su Z, Liu X, Xu Y, et al. Novel reciprocal fusion genes involving HNRNPC and RARG in acute promyelocytic leukemia lacking RARA rearrangement. Haematologica. 2020;105(7):e376-e8.
- Ha JS, Do YR, Ki CS, et al. Identification of a novel PML-RARG fusion in acute promyelocytic leukemia. Leukemia. 2017;31(9):1992–1995.
- Wen L, Xu Y, Yao L, et al. Clinical and molecular features of acute promyelocytic leukemia with variant retinoid acid receptor fusions. Haematologica. 2019;104(5):e195–e1e9.
- Han SP, Tang YH, Smith R. Functional diversity of the hnRNPs: past, present and perspectives. Biochem J. 2010;430(3):379–392.
