ABSTRACT
Introduction
Several studies have confirmed that mutations in the Wilms tumor 1 (WT1) gene occur in adult acute myeloid leukemia (AML). However, few data are available regarding the incidence of WT1 mutations in CEBPAmut AML and their impact.
Methods
We retrospectively analyzed the frequency and clinical impact of WT1 mutations in 220 newly diagnosed AML patients with CEBPA mutations(CEBPAmut). Chromosome karyotype analysis was performed by R or G banding method and further confirmed either by fluorescence in situ hybridization (FISH) and/or by multiple reverse transcription polymerase chain reaction (multiple RT–PCR). Mutations were detected with a panel of 112mutational genes using next-generation sequencing (NGS).
Results
Overall, 30 WT1 mutations were detected in 29 of the 220 CEBPAmut AML patients (13.18%) screened. These mutations clustered overwhelmingly in exon 7 (n=16). WT1 mutations were found to be significantly more frequent in AML patients with double-mutated CEBPA (CEBPAdm) than in AML patients with single-mutated CEBPA (17.36%vs. 8.08%, P = 0.043). Among WT1-mutated patients, the most common co-mutation was FLT3-ITD (n = 7, 24.14%), followed by NRAS (n = 5, 17.24%), CSF3R (n = 4, 13.79%), GATA2 (n = 4, 13.79%), and KIT (n = 4, 13.79%). The most frequent functional pathway was signaling pathways inas many as 62.07% of cases. Notably,the concomitant mutations in epigenetic regulatorswere inversely correlated with WT1 mutations(P = 0.003). CEBPAdm AML patients with WT1 mutations had inferior relapse-free survival, event-free survival and overall survival compared with patients CEBPAdm AML without WT1 mutations (P = 0.002, 0.004, and 0.010, respectively).
Conclusion
Our data showed that WT1 mutations are frequently identified in CEBPAmut AML, especially in CEBPAdm AML. CEBPAmut AML patients with WT1 mutations show distinct spectrum of comutations. In the context of CEBPAdm AML, WT1 mutations predict a poor prognosis.
KEYWORDS:
Introduction
CCAAT/enhancer binding protein a (CEBPA) plays a crucial role in the repression of self-renewal, cell cycle arrest, and myeloid differentiation of mature neutrophils during normal hematopoiesis[Citation1, Citation2]. Mutations in the CEBPA gene can occur across the whole coding region and have been described in approximately 6-24% of all acute myeloid leukemia (AML) patients[Citation3–5]. CEBPA-mutated (CEBPAmut) patients can be subdivided into two different subgroups: (1) AML carrying one mutation on one allele (single-mutated CEBPA, CEBPAsm) and (2) AML with two CEBPA mutations (double-mutated CEBPA, CEBPAdm), typically showing an N-terminal mutation and a basic leucine zipper gene mutation. However, only patients who harbor CEBPA double mutations are associated with favorable outcomes[Citation6, Citation7]. In the 2016 World Health Organization (WHO) classification of leukemia, AML with CEBPAdm was included as a distinctive disease entity due to its unique biological and clinical profiles[Citation8].
With the application of next-generation sequencing (NGS), >85% of CEBPAmut AML patients were observed to have other known concurrent mutations, such as WT1, CSF3R, and GATA2[Citation9, Citation10]. However, studies have shown conflicting data on the impact of CEBPA mutations on prognosis when identified with other concomitant mutations[Citation9–11]. These findings suggest that it is necessary to further explore co-mutations in the context of CEBPA mutations in AML.
Recently, mutations in the Wilms tumor 1 (WT1) gene have been observed in adult and childhood leukemia, commonly in AML[Citation12, Citation13]. In several studies investigating more than 1000 AML samples total, mutations in WT1 were found in 2%−8.3% of the patients[Citation12–15]. However, in AML patients with CEBPAdm, the frequency of WT1mutations increased to 13.6%−18.52%[Citation10, Citation14]. To the best of our knowledge, few data are available regarding the impact of WT1 mutations on CEBPAmut AML. We report here the frequency and types of WT1 mutations and their association with clinical features and outcomesin a cohort of 220 CEBPAmut AML patients.
Patients and methods
Patients
A total of 220 selected de novo AML patients with CEBPA mutations from five medical institutions of hematology (Affiliated Changzhou Second Hospital of Nanjing Medical University, Affiliated Hospital of Jiangnan University, Wuxi No. 2 People's Hospital, The Third Affiliated Hospital of Soochow University, and The First Affiliated Hospital of Soochow University) between August 2014 and November 2020 were analyzed. Of the CEBPAmut AML patients, 114 were male and 106 were female. The median age was 39 years (range: 18-88years), with 190 patients being <60 years and 30 patients being ≥60 years. According to the French–American–British (FAB) classification, 14, 96, 33, 62, 5, and 10 patients were diagnosed with M1, M2, M4, M5, M6 and undetermined types, respectively. All CEBPAmut AML patients gave their written informed consent for genetic analysis and for the use of the laboratory results for scientific studies. The study was approved by the research ethics board of each participating hospital and adhered to the tenets of the Declaration of Helsinki.
Cytomorphology, cytogenetics and immunophenotyping
Cytomorphologic assessment was performed by May-Grünwald-Giemsa staining, myeloperoxidase (MPO) reaction, and nonspecific esterase (NSE) staining using alpha-naphthyl-acetate following the FAB and WHO classifications. Karyotyping by the G or R–banding method was performed following standard methods on bone marrow (BM) cells after short culture. When possible, at least 20 metaphases were analyzed for each case. Immunophenotyping was performed as previously described[Citation16].
Mutation screening
BM smear or peripheral blood (PB) samples at the time of initial diagnosis were collected. A sensitive next-generation amplicon deep-sequencing assay was used with an Illumina next-generation sequencer. A high depth of coverage (1000×) was obtained for 112 genes, including the whole coding regions that are known to be frequently mutated in hematologic malignancies. Altered DNA sequences were deemed mutations or variants in IGV software analysis and were identified by the COSMIC database and needed to exclude dbSNPs. Mutations in NPM1, FLT3, and CEBPAwere identified by polymerase chain reaction (PCR) followed by direct Sanger sequencing, as previously described[Citation17–19]. The mutations of the 112 genes analyzed in this study are listed in supplemental (Table S1).
Table 1. Pretreatment clinical characteristics according to CEBPA mutation status.
Fusion gene and WT1 mRNA expression
Forty-one fusion genes were detected by multiple reverse transcription PCR amplification, which was performed as 8 parallel multiplex reactions on a 7500 Real-time PCR System (Applied Biosystems). We evaluated WT1 mRNA expression in BM or PB using a WT1 mRNA assay kit (Otsuka Pharmaceutical, Tokyo, Japan), as previously reported[Citation20].
Treatment and outcome
Of the 220 AML patients with CEBPA mutations, 161 patients received a standard chemotherapy regimen (DA/IA, daunorubicin/idarubicin 60/10 mg/m2 d1-3, cytarabine 100 mg/m2 d1-7) for induction therapy, 31 patients were prescribed a reduced intensity DA/IA (daunorubicin/idarubicin 45/6-8 mg/m2 d1-3, cytarabine 100 mg/m2 d1-7) regimen, 13 patients received an HAD regimen (homoharringtonine 2 mg/m2 d1-7, cytarabine 100 mg/m2 d1-7, daunorubicin 40 mg/m2 d1-3), 14 patients received decitabine-based induction due to older age or underlying comorbidity, and 1 patient discontinued chemotherapy because of severe infection and respiratory failure.
Complete remission (CR) was observed in patients after one course of induction chemotherapy. Relapse-free survival (RFS) was defined as the time from diagnosis to relapse or the last follow-up in CR. Event-free survival (EFS) was calculated as the time interval from the date of diagnosis to the date of first evidence of an event, which included relapse, progression, recurrence, change, and death. Overall survival (OS) was defined as the time from diagnosis to death or the last follow-up.
Statistical analysis
Student's t-test was used to analyze continuous factors with a normal distribution, and the Mann–Whitney U test was used for comparisons of data that failed the normality test between different groups. Analysis of categorical variables between different cohorts was performed by the chi-square test and Fisher's exact test. Survival curves were calculated for RFS, OS and EFS based on the Kaplan–Meier method and compared using the two-sided log rank test. Patients who had undergone allogeneic bone marrow transplantation were censored at the time of transplantation. SPSS software version 20.0 (SPSS version 20.0; SPSS Institute, Chicago, IL, USA) was used for statistical analysis, and the results were considered significant at P < 0.05.
Results
Patients’ characteristics
Among the 220 patients with CEBPAmut AML, a total of 99 patients (45.0%) harboring single CEBPA mutations and 121 patients (55.0%) withbiallelic double mutations were identified. Compared with CEBPAsm AML patients, CEBPAdmAML patients had lower platelet counts (median: 29×109/L vs. 43×109/L, P = 0.002), higher hemoglobin levels (median: 93 g/Lvs. 82 g/L, P = 0.0162), and lower WT1 expression levels (median: 12.385% vs. 42.35%, P = 0.000). A total of 52.07% (63/121) of CEBPAdm AML patients presented with the FAB-M2 subtype, which occurred at a distinctively higher rate than that in CEBPAsm patients (52.07% vs. 33.33%, P = 0.0053). AML patients with CEBPAsm were more likely to show the M5 subtype than those with CEBPAdm (41.4% vs. 17.35%, P = 0.0001). No significant differences were identified regarding sex, age, white blood cell (WBC) count or cytogenetickaryotypebetween the CEBPAsm and CEBPAdm AML groups. lists different categories of clinical information of the enrolled patients.
Frequency and types of WT1 mutations
Overall, 30 WT1mutations were detected in 29 of the 220 CEBPAmut AML patients (13.18%) screened. WT1 mutations occurred significantly more frequently in CEBPAdm AML (21/121, 17.36%) than in CEBPAsm (8/99, 8.08%) AML patients (P = 0.043). Mutations clustered overwhelmingly in exon 7 (16 mutations in 15 patients), but they were also detected in exon 8 (n = 6) and exon 9 (n = 8). One patient presented with two kinds of mutations (no. 29). The amino acid changes and mutation information of the WT1gene are shown in and .
Figure 1. Schematic showing the locations of WT1 mutations found in CEBPA mutated AML. One patient had both the R380G and S381fs mutations.
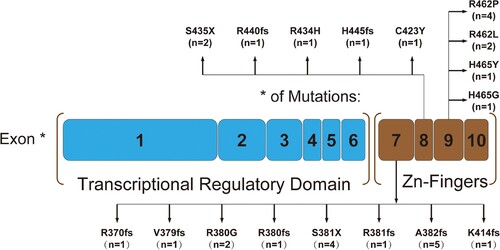
Table 2. WT1. mutations information in 29 patients with CEBPAmut AML.
Companion gene mutations
Based on NGS, we detected 59 mutated genes by screening the 112-gene panel in 220 CEBPAmut AML patients. In addition to WT1, 33 of the 59 genes could be categorized as signaling pathways, epigenetic regulators, transcription factors, spliceosomes, tumor suppressors, chromatin modifiers, and cohesin. A total of 94.54% (208/220) of CEBPA mutations were accompanied by additional mutations and frequently co-occurred with FLT3-ITD (n = 39, 17.73%), NRAS (n = 34, 15.45%), GATA2 (n = 34, 15.45%), RUNX1 (n = 30, 13.64%), WT1 (n = 29, 13.18%), FAT1 (n = 29, 13.18%), TET2 (n = 23, 10.45%), CSF3R (n = 20, 9.09%), KIT (n = 21, 9.54%) and NOTCH2 (n = 18, 8.18%). Other genes had a mutation prevalence of <8%. Functionally, mutations involved in signaling pathways(n = 123, 55.91%) and transcription factors (n = 47, 21.36%) were found frequently in CEBPAmut AML patients.
Among WT1-mutated AML patients with CEBPAmut, the most common co-mutation was FLT3-ITD (n = 7, 24.14%), followed by NRAS (n = 5, 17.24%), CSF3R (n = 4, 13.79%), GATA2 (n = 4, 13.79%), KIT (n = 4, 13.79%), and FAT1 (n = 3, 10.34%). Fewer common co-mutations (2%–10.25%) were found in NPM1, SF3B1, NOTCH1/2, PTPN11, JAK2 and others. The most frequent functional pathway was signaling pathways inas many as 62.07% of cases. Notably, no mutations in TET2, IDH1/2, or DNMT3A were identified in WTmut patients. The concomitant mutations in epigenetic regulatorswere inversely correlated with WT1 mutations(P = 0.003). There were no differences in the incidences of FLT3-ITD, NRAS, CSF3R, GATA2, PTPN11, SF3B1 and NOTCH1/2 between WTmut and WTwt patients. The difference in themutational spectrum between WT-mutated and WT1 wild-type CEBPAmut AML is shown in . The frequencies of the detected gene mutations and gene functional groups are shown in .
Figure 2. Spectrum of acquired co-mutations in 29WT1 mutated patients with CEBPA mutated AML divided into different mutational categories. Each bar represents a distinct driver gene.
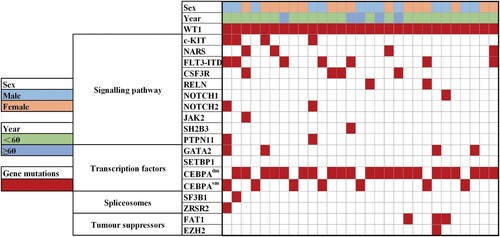
Table 3. Concomitant gene abnormalities according to WT1 mutation status.
Prognostic impact of WT1mutations in the context of CEBPA double mutations
Of 121 patients with CEBPA double mutations, 111 patients were evaluable for complete remission (CR) after the first induction therapy. No differences were observedin the CR rate between patients with or without WT1mutations in CEBPAdm AML (89.5% vs. 91.3%, P = 1.000). The median follow up was 30.5 (range 0.5-60.6) months. We next investigated the impact of WT1mutations on survival in CEBPAdm AML. WT1-mutatedAML patients with CEBPAdm had significantly inferior RFS, EFS, and OS than WT1 wild-type patients (3-year RFS rate, 40% vs. 70%, median, 8.9 months vs. NR, P = 0.002; 3-year EFS rate, 36% vs. 65%, median, 9.9 months vs. NR, P = 0.004; 3-year OS rate, 48% vs. 72%, 20.4 months vs. NR, P = 0.010) ().(). Influence of mutations in WT1on survival has been shown in a/b/c.
Figure 3. Influence of mutations in WT1on survival. a. Kaplan–Meier estimates of Relapse-free survival (RFS), b.event-free survival (EFS) and c. overall survival (OS) in newly diagnosed patients with CEBPAdouble mutated AML, according to the presence or absence of mutations in WT1.
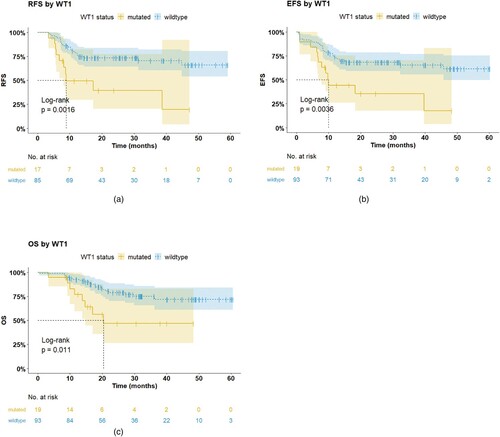
Table 4. Univariable outcome analyses according to WT1 mutation status.
Discussion
The WT1 gene, located on chromosome 11p13, encodes a zinc-finger protein that exists in multiple isoforms and functions as a transcription factor involved in cellular growth, proliferation and differentiation and may therefore act as an oncogene[Citation21–23]. The WT1 gene was initially identified as a tumor suppressor gene linked with nephroblastoma[Citation24, Citation25]. Over the past years, numerous studies have highlighted that WT1 is aberrantly expressed or mutated in hematopoietic malignancies, including acute leukemia (AL), myelodysplastic syndrome (MDS) and chronic myelogenous leukemia (CML)[Citation26–30]. However, few data are available regarding the frequency of WT1 mutations and their impact on CEBPAmut AML.
Recently, WT1mutations were detected in 13.6%−18.52% of biallelic CEBPAmut AML patients[Citation3, Citation10, Citation14]. We revealed that WT1 mutations occurred in 17.36% of CEBPAdm AML patients and 8.08% of CEBPAdm AML patients. This percentage of CEBPAdm AML cases is slightly higher than that reported by Krauth MT et al.[Citation14]and Fasan Aet al.[Citation3], but similar to report fromSu L et al.[Citation10]. The difference could be explained by the fact that in the present study, we analyzed WT1 mutations in all exons, while in the study by Krauth MT et al., only mutations in exons 7 and 9 were analyzed. Similar to the findings of Fasan A et al.[Citation3]. We also found that WT1 was significantly more frequently mutated in CEBPAdm patients than in CEBPAsm patients, suggesting that the implication of the WT1 mutation may differ in CEBPAdm and CEBPAsm AML.
Becker H et al. found that in WT1-mutated patients, FLT3-ITD more frequently occurred inolder patients with primary normal karyotype AML (NK-AML)[Citation31]. Krauth MT and colleagues found that WT1 mutations were rarely concomitant with DNMT3A (4.4%, P = 0.014), ASXL1 (1.7%, P = 0.001), IDH2R140 (1.7%, P = 0.001) and IDH1R132 (0.9%,P = 0.001) mutations[Citation14]. In this study, concomitant mutations in epigenetic regulatorswere inversely correlated with WT1 mutations in CEBPAmut AML patients(P = 0.003). This observation suggests that WT1 mutations are anticorrelated with epigenetic regulatormutationsunder the background of CEBPAmut AML. Rampal R et al. observed that WT1-mutant AML patients had reduced 5-hydroxymethylcytosine (5hmC) levels, similar to TET2/IDH1/IDH2 mutant AML patients[Citation32]. Wang Y et al. showed that WT1 physically interacts with and recruits TET2 to its target genes to activate their expression. The interaction between WT1 and TET2 is disrupted by multiple AML-derived TET2 mutations[Citation33]. These findings, including our results, suggest that mutations in WT1 and TET2 may have interactive effects. Further studies are needed to reveal the interaction mechanism between WT1 and epigenetic regulatorsin the context of CEBPA mutations.
There are still some controversies regarding the prognostic significance of WT1 mutations in patients with AML. Studies from Hou HA and Virappane P demonstrated that the WT1 mutation was associated with poor prognosis in NK-AML and nonselective AML patients[Citation34, Citation35]. Tien Feng-Ming and colleagues found that WT1mut patients with CEBPAdm AML tended to have a lower CR rate, a higher relapse rate, and significantly shorter OS and disease-free survival (DFS) than those with wild-typeWT1[Citation36]. Krauth MT et al. indicated thatWT1mut was associated with shorter EFS in NK-AML patients, but it had no impact on prognosis in subgroups with high WT1mut incidences (CEBPAdm, PML-RARA)[Citation14]. An analysis based on the TCGA database showed that the WT1-mutated group had lower rates of CR but higher rates of minimal residual disease (MRD) after one and two courses of induction chemotherapy. In NK-AML pediatric patients, those with WT1 mutations had significantly worse OS and EFS in both univariate and multivariate survival analyses[Citation37]. In our study, CEBPAmut AMLpatients with WT1 mutations had significantly inferior 3-year RFS, EFS and OS rates than WT1wt patientsin the context of CEBPA double mutations. This result is consistent with that reported by Tien Feng-Ming et al. but different from that reported by Krauth MT et al. This inconsistency may be attributed to differences in the characteristics of the populations investigated, such as sex, karyotype, age, combination with other mutations such as FLT3-ITD, or different ethnicities.
In conclusion, this study confirms that WT1mutations are frequently identified in CEBPAmutAMLpatients, especially in CEBPAdmAMLpatients. CEBPAmut AML patients with WT1mutations show distinct spectrum of comutations. In the context of CEBPAdm AML, WT1mutations predict a poor prognosiscompared to WT1wild-type patients.
Authors’ contributions
TW, HYH, BW, WQ, PW and HYC had contributions in collecting clinical information of selected patients. ZW, BW, HYC and PW completed the entire experimental processes. TW and HYC completed all statistical analyses and constructed figures. TW wrote the manuscript. HYC and XZL were the main contributors in revising the manuscript. Other authors provided some advice on the manuscript. All authors read and approved the final manuscript.
Availability of data and materials
The datasets used and/or analysed during the current study available from the corresponding author on reasonable request.
Competing interests
The authors declare no competing financial interests.
Consent for publication
Written informed consent for publication was obtained from all participants.
Ethics approval and consent to participate
All patients gave their written informed consent for genetic analysis and for the use of the laboratory results for scientific studies. The study was performed in accordance with the World Medical Association Declaration of Helsinki and Ethics Committee approval of Changzhou No.2 People’s Hospital was obtained ([2015]KY004-1).
Acknowledgements
The authors thank all the physicians who provided leukemia samples and clinical data.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Additional information
Funding
References
- Koschmieder S, Halmos B, Levantini E, et al. Dysregulation of the C/EBPα differentiation pathway in human cancer. J Clin Oncol. 2009;27(4):619–628.
- Quintana-Bustamante O, Smith S L-L, Griessinger E, et al. Overexpression of wild-type or mutants forms of CEBPA alter normal human hematopoiesis. Leukemia. 2012;26(7):1537–1546.
- Fasan A, Haferlach C, Alpermann T, et al. The role of different genetic subtypes of CEBPA mutated AML. Leukemia. 2014;28(4):794–803.
- Ahmad F, Rajput S, Mandava S, et al. Molecular Evaluation of CEBPA GeneMutation in Normal Karyotype Acute Myeloid Leukemia: A Comparison of Two Methods and Report of Novel CEBPA Mutations from Indian Acute Myeloid Leukemia Patients. Genet Test Mol Biomarkers. 2012;16(7):707–715.
- Ahn JY, Seo K, Weinberg O, et al. A comparison of two methods for screening CEBPA mutations in patients with acute myeloid leukemia. J Mol Diagn. 2009;11(4):319–323.
- Pabst T, Eyholzer M, Fos J, et al. Heterogeneity within AML with CEBPA mutations; only CEBPA double mutations, but not single CEBPA mutations are associated with favourable prognosis. Br J Cancer. 2009;100(8):1343–1346.
- Taskesen E, Bullinger L, Corbacioglu A, et al. Prognostic impact, concurrent genetic mutations, and gene expression features of AML with CEBPA mutations in a cohort of 1182 cytogenetically normal AML patients: further evidence for CEBPA double mutant AML as a distinctive disease entity. Blood. 2011;117(8):2469–2475.
- Arber DA, Orazi A, Hasserjian R, et al. The 2016 revision to the world health organization classification of myeloid neoplasms and acute leukemia. Blood. 2016;127(20):2391–2405.
- Zhang Y, Wang F, Chen X, et al. Companion gene mutations and their clinical significance in AML with double mutant CEBPA. Cancer Gene Ther 2020;27(7-8):599–606.
- Su L, Tan Y, Lin H, et al. Mutational spectrum of acute myeloid leukemia patients with doubleCEBPA mutations based on next-generation sequencing and its prognostic significance. Oncotarget. 2018;9(38):24970–24979.
- Grossmann V, Haferlach C, Nadarajah N, et al. CEBPA double-mutated acute myeloid leukaemia harbours concomitant molecular mutations in 76.8% of cases with TET2 and GATA2 alterations impacting prognosis. Br J Haematol. 2013;161(5):649–658.
- Ho PA, Zeng R, Alonzo TA, et al. Prevalence and prognostic implications of WT1 mutations in pediatric acute myeloid leukemia (AML): a report from the children's oncology group. Blood. 2010;116(5):702–710.
- Haider I, Kumar C, Jain G, et al. Hotspots mutational analysis of wilms tumor 1 gene in acute myeloid leukaemia; prevalence and clinical correlation in north Indian population. Am J Blood Res. 2020;10(5):179–189.
- Krauth MT, Alpermann T, Bacher U, et al. WT1 mutations are secondary events in AML, show varying frequencies and impact on prognosis between genetic subgroups. Leukemia. 2015;29(3):660–667.
- Luo S, Yu K, Yan QX, et al. Analysis of WT1 mutations, expression levels and single nucleotide polymorphism rs16754 in de novo non-M3 acute myeloid leukemia. Leuk Lymphoma. 2014;55(2):349–357.
- Kern W, Bacher U, Haferlach C, et al. The role of multiparameter flow cytometry for disease monitoring in AML. Best Pract Res Clin Haematol. 2010;23(3):379–390.
- Pabst T, Mueller BU, Zhang P, et al. Dominant-negative mutations of CEBPA, encoding CCAAT/enhancer binding protein-α (C/EBPα), in acute myeloid leukemia. Nat Genet. 2001;27(3):263–270.
- Kumar D, Mehta A, Panigrahi MK, et al. NPM1 mutation analysis in acute myeloid leukemia: comparison of three techniques - sanger sequencing, pyrosequencing, and real-time polymerase chain reaction. Turk J Haematol. 2018;35(1):49–53.
- Thiede C, Steudel C, Mohr B, et al. Analysis of FLT3-activating mutations in 979 patients with acute myelogenous leukemia: association with FAB subtypes and identification of subgroups with poor prognosis. Blood. 2002;99(12):4326–4335.
- Kitamura K, Nishiyama T, Ishiyama K, et al. Clinical usefulness of WT1 mRNA expression in bone marrow detected by a new WT1 mRNA assay kit for monitoring acute myeloid leukemia: a comparison with expression of WT1 mRNA in peripheral blood. Int J Hematol. 2016;103(1):53–62.
- Call KM, Glaser T, Ito CY, et al. Isolation and characterization of a zinc finger polypeptide gene at the human chromosome 11 wilms' tumor locus. Cell. 1990;60(3):509–520.
- Yang L, Han Y, Suarez SF, et al. A tumor suppressor and oncogene: the WT1 story. Leukemia. 2007;21(5):868–876.
- Yamagami T, Sugiyama H, Inoue K, et al. Growth inhibition of human leukemic cells by WT1 (wilms tumor gene) antisense oligodeoxynucleotides: implications for the involvement of WT1 in leukemogenesis. Blood. 1996;87(7):2878–2884.
- Sugiyama H. Wilms’ tumor GeneWT1: Its oncogenic function and clinical application. Int J Hematol. 2001;73(2):177–187.
- Huff V. Wilms' tumours: about tumour suppressor genes, an oncogene and a chameleon gene. Nat Rev Cancer. 2011;11(2):111–121.
- Lyu X, Xin Y, Mi R, et al. Overexpression of wilms tumor 1 gene as a negative prognostic indicator in acute myeloid leukemia. PLoS One. 2014;9(3):e92470.
- Šálek C, Vydra J, Cerovská E, et al. WT1 expression in peripheral blood at diagnosis and during the course of early consolidation treatment correlates With survival in patients With intermediate and poor-risk acute myeloid leukemia. Clin Lymphoma Myeloma Leuk. 2020;20(12):e998–e1009.
- Ahmad F, D’Souza W, Mandava S, et al. Molecular analysis of WT1 and KIT mutations in patients from an Indian population with de novo acute myeloid leukemia: determination of incidence, distribution patterns, and report of a novelKITmutation. Leuk Lymphoma. 2011;52(5):865–876.
- Luo P, Jing W, Yi K, et al. Wilms’ tumor 1 gene in hematopoietic malignancies: clinical implications and future directions. Leuk Lymphoma. 2020;61(9):2059–2067.
- Zhu H, Yang B, Liu J, et al. Case report of acute myeloid leukemia with “WT1, ATRX, CEBPA, CSMD1, IKZF1, and LRP1B mutation and translocation between chromosome 1 and 19” developing from Philadelphia-negative chronic myeloid leukemia after TKI therapy. Medicine (Baltimore). 2020;99(3):e18888.
- Becker H, Marcucci G, Maharry K, et al. Mutations of the wilms tumor 1 gene (WT1) in older patients with primary cytogenetically normal acute myeloid leukemia: a cancer and leukemia group B study. Blood. 2010;116(5):788–792.
- Rampal R, Alkalin A, Madzo J, et al. DNA hydroxymethylation profiling reveals that WT1 mutations result in loss of TET2 function in acute myeloid leukemia. Cell Rep. 2014;9(5):1841–1855.
- Wang Y, Xiao M, Chen X, et al. WT1 recruits TET2 to regulate Its target gene expression and suppress leukemia cell proliferation. Mol Cell. 2015;57(4):662–673.
- Hou HA, Huang TC, Lin LI, et al. WT1 mutation in 470 adult patients with acute myeloid leukemia: stability during disease evolution and implication of its incorporation into a survival scoring system. Blood. 2010;115(25):5222–5231.
- Virappane P, Gale R, Hills R, et al. Mutation of the wilms’ tumor 1 gene is a poor prognostic factor associated with chemotherapy resistance in normal karyotype acute myeloid leukemia: the United Kingdom medical research council adult leukaemia working party. J Clin Oncol. 2008;26(33):5429–5435.
- Tien FM, Hou HA, Tang JL, et al. Concomitant WT1 mutations predict poor prognosis in acute myeloid leukemia patients with double mutant CEBPA. Haematologica. 2018;103(11):e510–e513.
- Xu J, Zhang Y, Hu J, et al. Clinical features and prognosis of normal karyotype acute myeloid leukemia pediatric patients with WT1 mutations: an analysis based on TCGA database. Hematology. 2020;25(1):79–84.
