ABSTRACT
Background
Multiple myeloma (MM) is a relatively common hematologic tumor, and the study of non-coding RNAs in MM is gradually increasing. Our study aimed to reveal the regulatory mechanism of circular RNA_0003489 (circ_0003489)/microRNA-433-3p (miR-433-3p)/Pre-B-cell leukemia homeobox 3 (PBX3) axis in MM.
Methods
Circ_0003489, miR-433-3p and PBX3 contents were measured by real-time quantitative PCR or western blot. Functionally, MM cell proliferation and apoptosis were evaluated using cell counting kit-8, flow cytometry and EdU assays. Interaction between miR-433-3p and circ_0003489 or PBX3 was confirmed with the application of dual-luciferase reporter assay and RNA pull down assay.
Results
Circ_0003489 and PBX3 were upregulated, while miR-433-3p was downregulated in MM tissues. Circ_0003489 knockdown or miR-433-3p overexpression remarkably suppressed MM proliferation and increased apoptosis in vitro. In terms of mechanism, circ_0003489 could sponge miR-433-3p to regulate PBX3. Besides, miR-433-3p downregulation or PBX3 overexpression reversed the inhibition effect of circ_0003489 knockdown on MM progression.
Conclusion
Circ_0003489 facilitated MM progression by targeting miR-433-3p/PBX3 axis, suggesting that it might be a potential target for MM.
1. Introduction
Multiple myeloma (MM) is one of the common malignancies of the hematological system and is characterized by the clonal phenomenon of malignant plasma cells of the bone marrow [Citation1]. Radiation, toxins, viral exposure and chromosomal abnormalities may be associated with the development of MM, and a mutation at chromosome 14 has been found to be closely related to the growth of cancer cells in the bone marrow [Citation2]. MM is more prevalent in older age groups and has a higher incidence in males, and there are statistics that blacks have a higher incidence than whites [Citation2,Citation3]. Patients with MM usually have anemia, bone pain, kidney damage and hypercalcemia [Citation4]. Recently, research in the molecular field for MM diagnosis and treatment is gradually being carried out, which may give a huge impetus to the updating of MM treatment method.
Circular RNAs (circRNAs) are a class of endogenous regulatory non-coding RNAs with stable structures, high conservation and cell specificity [Citation5]. Currently, circRNAs have been confirmed to exert regulatory roles in various diseases such as cardiovascular disease [Citation6], osteoarthritis [Citation7], and diabetic nephropathy [Citation8]. A high amount of circRNAs have also been identified to be aberrantly expressed in MM tissues [Citation9–11]. For example, circ_0007841was found to be highly expressed in MM, which had the potential to be a biomarker for MM diagnosis [Citation9]. Circ_0000190 was dramatically downregulated in MM tissues, and it could arrest MM cell proliferation and promote apoptosis [Citation10]. Circ_0003489 (circRNA_101237) was confirmed to possess the high abundance in the circRNA microarray sequencing of 143 MM patients [Citation11]. However, the role of circ_0003489 in MM progression is still unclear.
One of the mechanisms of circRNA is to mediate the regulation of mRNA expression as a sponge of microRNA (miRNA), which is known as the circRNA/miRNA/mRNA mechanism [Citation12,Citation13]. MiRNAs do not encode proteins, but can target and regulate multiple mRNAs to down-regulate their protein levels [Citation14]. Currently miRNAs have been identified to have regulatory roles in various life activities such as cell proliferation, inflammatory response, and immune regulation [Citation15,Citation16]. MiR-433-3p had been found to function as tumor suppressor in esophageal cancer, which could inhibit cancer cell proliferation and invasive [Citation17]. However, whether miR-433-3p mediates MM progression has not been explored.
Pre-B-cell leukemia homeobox 3 (PBX3) is one of the four members of the PBX family. PBX3 is distinguished from other family members by its ability to interact stably with DNA [Citation18]. Dysregulation of PBX3 had also been observed in gastric cancer, and its overexpression facilitated cell proliferation [Citation19]. In MM, PBX3 was found to be overexpressed in MM tissues and could promote cell growth [Citation20]. Therefore, PBX3 might be an important regulator for MM progression.
This study aimed to analyze the role and regulatory mechanism of circ_0003489 in MM progression. Through bioinformatics analysis, we found that circ_0003489 could complement with miR-433-3p, and miR-433-3p could bind to the 3’UTR of PBX3. Therefore, the hypothesis of circ_0003489/miR-433-3p/PBX3 axis is proposed and verified in our study.
2. Materials and methods
2.1. Tissue samples
All experiments for this study were approved by the Ethics committee of Lishui Second People’s Hospital. 40 MM tissues were collected from patients diagnosed with MM at Lishui Second People’s Hospital, and 10 normal bone marrow tissues were obtained from normal volunteers. The clinicopathological characteristics in 40 MM patients are shown in . All participants signed written informed consents.
Table 1. The correlation of circ_0003489 expression with the clinicopathological characteristics in 40 MM patients.
2.2. Cell culture and transfection
Normal plasma cells (nPCs) and human MM cell lines (MM.1S, U266 and H929) that purchased from China center for type culture collection (Wuhan, China) were grown in RPMI-1640 medium (Invitrogen, Carlsbad, CA, USA) containing 10% FBS (Invitrogen, Carlsbad, CA, USA)at 37 °C with 5% CO2. The circ_0003489 lentivirus short hairpin RNA (sh-circ_0003489#1 and sh-circ_0003489#2), miR-615-5p mimic, miR-433-3p mimic, miR-95-3p mimic, miR-433-3p inhibitor (anti-miR-433-3p), PBX3 overexpression vector, and negative controls were purchased from GeneChem (Shanghai, China). According to the guidelines of Lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA), cell transfection was performed.
2.3. Real-time quantitativePCR (RT-qPCR)
RNA from tissues and cells with or without actinomycin D treatment was extracted with the application of TRIzol (Invitrogen, Carlsbad, CA, USA). Subsequently, 1 µg of RNA was transformed into cDNA with the help of miScript RT Kit (TaKaRa, Dalian, China) and then SYBR Green (TaKaRa, Dalian, China) was applied to carry out RT-qPCR. Relative expression was calculated with2-ΔΔCT method and normalized by GAPDH or U6. Primer sequences were presented in . In addition, cDNA was obtained using random primers and oligo (dT)18 primers, and then the circ_0003489 expression and CDK8 mRNA expression were detected by RT-qPCR to explore whether circ_0003489 had poly-A tail. Also, the nucleus and cytoplasm RNAs from MM.1S and H929 cells were isolated by PARIS Kit (Invitrogen, Carlsbad, CA, USA). After that, circ_0003489, U6 and GAPDH expression in cell nucleus and cytoplasm was measured.
Table 2. Primer sequences used for RT-qPCR.
2.4. Cell counting kit-8 (CCK8) assay
After transfection, MM cells were seeded into 96-well plates. 10 µL of CCK8 solution (Beyotime, Shanghai, China) was incubated with the cells, and the absorbance at 450 nm was measured at selected time points.
2.5. Flow cytometry
Cell cycle and apoptosis were detected using flow cytometry. Following the guidelines of the Cell Cycle Assay Kit (Beyotime, Shanghai, China), cells were fixed with 70% ethanol, treated with RNase A and stained by Prodium Iodide (PI) solution. Then, cell cycle distribution was analyzed by a flow cytometer (BD Biosciences, San Jose, CA, USA). Apoptosis assay was executed following the guidelines of the Annexin V/PI cell apoptosis analysis kit (Beyotime, Shanghai, China). Briefly, cells were suspended with binding buffer and then stained with Annexin V-FITC solution and PI solution. Subsequently, cell apoptosis rate was detected under the flow cytometer.
2.6. Eduassay
Cell proliferation was assessed according to the guidelines of the EdU Cell Proliferation Assay Kit (Solarbio, Beijing, China). After cells were inoculated into 96-well plates for 24 h, cells were stained by EdU solution and DAPI solution. Then, cells were analyzed under a microscope to count the EdU positive cell rate.
2.7. Western blot
Western blot was carried out as described in He et al. [Citation21]. Proteins were extracted using RIPA buffer (Beyotime, Shanghai, China). Proteins were isolated on SDS-PAGE gel and transferred onto PVDF membrane. Then, membrane was incubated with primary antibody and secondary antibody. Protein signal was detected using enhanced chemiluminescence solution (Beyotime, Shanghai, China). All antibodies were obtained from Abcam (Cambridge, MA, USA), including anti-Ki67 (1:1,000, ab16667), anti-Bax (1:1,000, ab32503), anti-MMP2 (1:3,000, ab97779), anti-β-actin (1:1,000, ab8227), anti-PBX3 (1:1,000, ab129478), and Goat anti-rabbit IgG (1:50,000, ab205718).
2.8. Dual-luciferase reporter assay
Downstream targets of circ_0003489 and miR-433-3p were forecasted by Circbank, circInteractome and Targetscan. Next, the wild-type and mutant-type ofcirc_0003489 and PBX3 luciferase reporter vectors (circ_0003489 WT/MUT and PBX3 3’UTR WT/MUT) were constructed and then co-transfected into MM cells with miR-433-3p mimic or miR-NC. 48 h later, luciferase activity was detected using Dual-Lucy Assay Kit (Solarbio, Beijing, China).
2.9. RNA pull down assay
The biotin-labeled miR-433-3p probe (Bio-miR-433-3p), biotin-labeled mutant probe (Bio-miR-433-3p MUT), and the negative control (Bio-NC) probe were constructed by Sangon (Shanghai, China). They were transfected into cells for 48 h. Then, cell lysates were incubated with Dynabeads M-280 Streptavidin (Invitrogen, Carlsbad, CA, USA). The formed complexes were isolated to extract RNA, and the circ_0003489 and PBX3 levels were analyzed by RT-qPCR.
2.10. Data analysis
Data are expressed as mean ± SD. SPSS 24.0 (IBM Corp., Armonk, NY, USA) was applied to analyze the data. Differences between groups were analyzed using Student’s t-test or ANOVA. Pearson correlation analysis was used to analyze linear correlation. P < 0.05 was considered as statistically significant.
3. Results
3.1. Circ_0003489 in MM tissues and cells possessed higher abundance
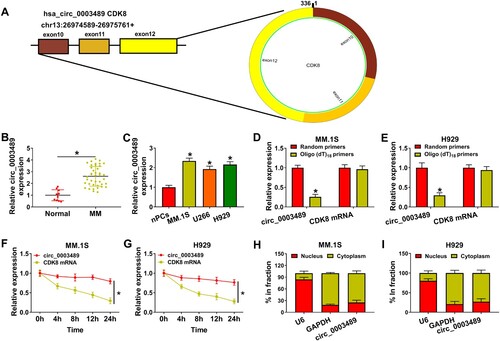
Circ_0003489 is located at chr13 and is derived from the back-splicing of exons 10–12 of CDK8 gene ((A)). A dramatic upregulation of circ_0003489 expression was observed in MM tissues and cells compared to that in normal tissues and nPCs cells ((B,C)). Through analyzing, we confirmed that high circ_0003489 expression was related to the hemoglobin level, bone injury and globulin level of patients with MM (). The inability of circ_0003489 to be amplified by Oligo (dT)18 primers was demonstrated in (D,E). Meanwhile, the expression level of circ_0003489 in actinomycin D-treated MM cells remained stable than linear CDK8 levels ((F,G)). Nucleoplasm separation assay showed that circ_0003489 was mainly expressed in the cytoplasm ((H,I)).
Figure 1. Expression characteristics of circ_0003489 in MM. (A) Circularstructure of circ_0003489, (B and C) The abundance of circ_0003489 in MM tissues and cells was assessed, (D and E) Oligo (dT)18 primer was used to explore whether circ_0003489 had poly-A tail, (F and G) Circ_0003489 and CDK8 levels were detected after actinomycin D treatment, (H and I) Subcellular localization analysis was performed to detect circ_0003489 expression in cell nucleus and cytoplasm. *P < 0.05.
3.2. MM cell progression was inhibited by circ_0003489 knockdown
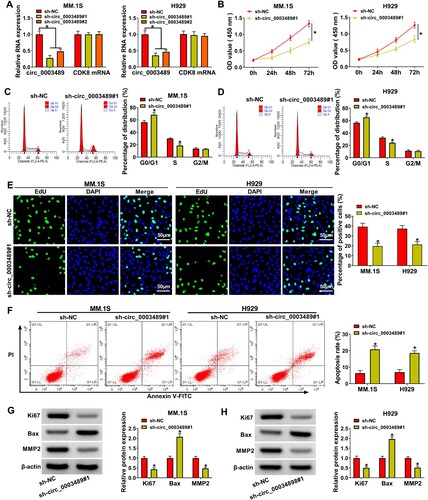
Both sh-circ_0003489#1 and sh-circ_0003489#2 notably downregulated circ_0003489 level in MM.1S and H929 cells without affecting linear RNA CDK8 mRNA expression ((A)), and sh-circ_0003489#1 was used in the follow-up assay due to its good silencing efficiency. The inhibition of MM.1S and H929 cell viability by sh-circ_0003489#1 was presented by CCK8 results ((B)). After MM.1S and H929 cells were transfected with sh-circ_0003489#1, cell cycle was dramatically blocked in G0/G1 phase ((C,D)). The results of the EdU assay demonstrated that the proliferation capacity of MM.1S and H929 cells was repressed by silencing ofcirc_0003489 ((E)). Also, the apoptosis rate of MM.1S and H929 cells in sh-circ_0003489#1 group was higher than that in sh-NC group ((F)). Besides, circ_0003489 knockdown downregulated Ki67 and MMP2 protein levels and upregulated Bax protein level ((G,H)). The above results elaborated that MM cell progression was inhibited by circ_0003489 silencing.
Figure 2. Effect of circ_0003489 silencing on MM cell function. (A) Sh-circ_0003489#1 and sh-circ_0003489#2 knockdown efficiency was analyzed. Sh-circ_0003489#1 and sh-NC were transfected into MM.1S and H929 cells, (B) cell viability, (C and D) cell cycle, (E) proliferation, (F) apoptosis, (G and H) Ki67, MMP2 and Bax protein levels were measured. *P < 0.05.
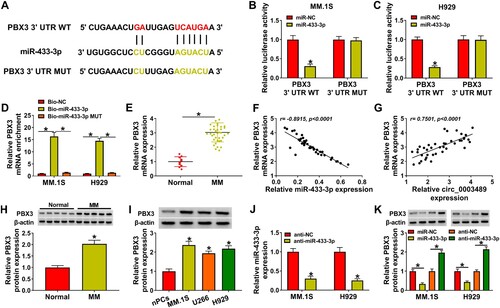
3.3. MiR-433-3p was targeted by circ_0003489
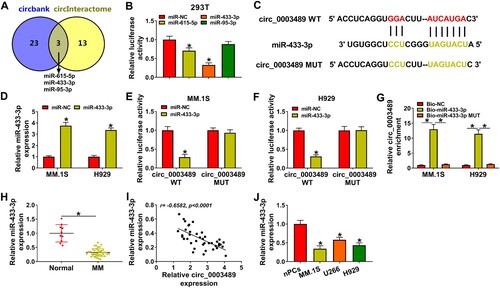
MiR-433-3p,miR-615-5p and miR-95-3p were predicted as the targets of circ_0003489 by Circbank and circInteractome software ((A)). (B) revealed that miR-433-3p mimic and miR-615-5p mimic markedly downregulated the luciferase activity of circ_0003489 WT vector, in which miR-433-3p played a stronger role. Therefore, miR-433-3p was selected as the target of circ_0003489. The binding information between miR-433-3p and circ_0003489 was displayed in (C). The overexpression effect of miR-433-3p mimic was verified by RT-qPCR ((D)). The luciferase activity of circ_0003489 WT group was greatly downregulated by miR-433-3p mimic in MM.1S and H929 cells ((E,F)). Compared to the Bio-NC probe and Bio-miR-433-3p MUT probe, the enrichment of circ_0003489 was remarkably increased in theBio-miR-433-3p probe ((G)). These data confirmed the relationship between circ_0003489 and miR-433-3p. Subsequently, miR-433-3p was found to be lowly expressed in MM tissues, and its expression was negatively correlated with circ_0003489 level ((H,I)). Meanwhile, lower level of miR-433-3p was also observed in MM cells relative to nPCs cells ((J)).
Figure 3. The relationship between circ_0003489 and miR-433-3p was verified. (A) Downstream targets of circ_0003489 were predicted, (B) Dual-luciferase reporterassay was analyzed to screen the targeted miRNA for circ_0003489, (C) Complementary binding sequence between circ_0003489 and miR-433-3p, (D) The overexpression efficiency of miR-433-3p mimic was determined, (E and F) Dual-luciferase reporter assay was employed to analyze the relationship between circ_0003489 and miR-433-3p, (G) RNA pull down assay was employed to analyze the relationship between circ_0003489 and miR-433-3p, (H) The abundance of miR-433-3p in MM tissues was measured and (I) Correlation analysis of circ_0003489 and miR-433-3pabundance in MM tissues, (J) Determination of miR-433-3p abundance in MM cells. *P < 0.05.
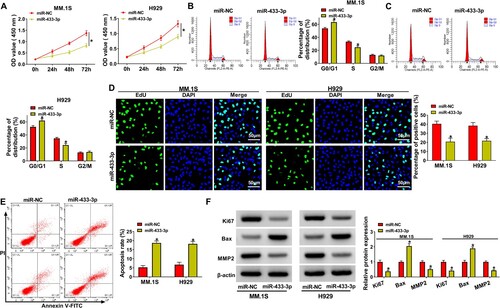
3.4. miR-433-3p upregulation suppressed MM progression in vitro
MM.1S and H929 cell viability and cell cycle process were repressed after miR-433-3p upregulation ((A–C)). Also, the proliferation of MM.1S and H929 cells was retarded after the upregulation of miR-433-3p ((D)). We also observed that the apoptosis rate of MM.1S and H929 cells was boosted in the miR-433-3p overexpression group ((E)). Finally, the inhibitory effect of miR-433-3p upregulation on Ki67 and MMP2 protein levels and the promotive effect on Bax were demonstrated by western blot results ((F)). The above results exhibited the inhibitory effect of miR-433-3p on MM progression in vitro.
Figure 4. MiR-433-3p upregulation regulated MM progression. MiR-433-3p mimic and miR-NC were transfected into MM.1S and H929 cells, respectively. (A) CCK8 assay was employed to analyze cell viability, (B and C) Cell cycle was analyzed using flow cytometry assay, (D) EdU assay was used to assess MM cell proliferation, (E) MM cell apoptosis was analyzed by flow cytometry assay and (F) Ki67, MMP2 and Bax protein levels were determined using western blot. *P < 0.05.
3.5. Circ_0003489 sponged miR-433-3p to regulate PBX3
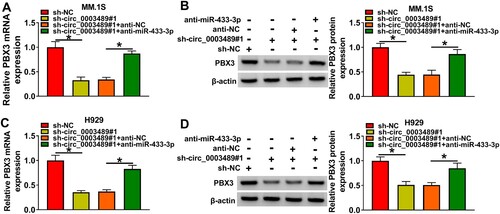
The complementary sequences of miR-433-3p and PBX3 3’UTRwere indicated in (A). The luciferase reporter system indicated that only the luciferase activity of the PBX3 3’UTR WT group was repressed by miR-433-3p mimic ((B,C)). The enrichment of PBX3 was markedly increased in the Bio-miR-433-3p probe rather than in the Bio-NC probe and Bio-miR-433-3p MUT probe ((D)). These data substantiated the association between miR-433-3p and PBX3. PBX3 expressions was higher in MM tissue ((E)), which was negatively correlated with miR-433-3p expression ((F)) and positively correlated with circ_0003489 expression ((G)). We observed that the PBX3 protein level in MM tissues and cells was also dramatically increased than normal tissues and nPCs cells ((H,I)). In addition, anti-miR-433-3p was constructed and we confirmed that it had a good silencing effect on miR-433-3p expression ((J)). As displayed in (K), we found that PBX3 protein level could be inhibited by miR-433-3p upregulation, but promoted by miR-433-3p silencing. These results confirmed that PBX3 was targeted by miR-433-3p. Additionally, the repression effect of circ_0003489 downregulation on PBX3 levels in MM.1S and H929 cells was reverted by anti-miR-433-3p ((A–D)). To sum up, circ_0003489 positively regulated PBX3 expression by sponging miR-433-3p.
Figure 5. PBX3 and miR-433-3p targeting relationship was validated. (A) The complementary sequences of miR-433-3p and PBX3, (B and C) PBX3 and miR-433-3p targeting relationship was validated using dual-luciferase reporter assay, (D) PBX3 and miR-433-3p targeting relationship was validated using RNA pull down assay, (E) The abundance of PBX3 in MM tissue was analyzed, (F and G) Correlation analysis of PBX3 level with miR-433-3p and circ_0003489 levels in MM tissues, (H and I) PBX3 protein level in MM tissues and cells were measured, (J) The silencing effect of anti-miR-433-3p was analyzed and (K) The protein level of PBX3 was determined in MM.1S and H929 cells transfected with miR-433-3p mimic or inhibitor. *P < 0.05.
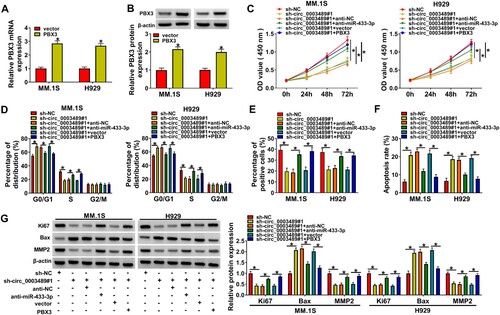
3.6. The suppressive effect of sh-circ_0003489 knockdown on MM progression was attenuated by miR-433-3p downregulation or PBX3 overexpression
The mRNA and protein levels of PBX3 were distinctly boosted by the PBX3 overexpression vector ((A,B)). The suppressive effect of circ_0003489 knockdown on cell viability could be rescued by anti-miR-433-3p or PBX3 overexpression ((C)). Meanwhile, circ_0003489 knockdown induced cell cycle arrest and impeded cell proliferation in MM.1S and H929 cells, while silencing miR-433-3p or PBX3 upregulation abolished these impacts ((D,E)). Moreover, miR-433-3p downregulation or PBX3 overexpression relieved the effects of circ_0003489 knockdown on apoptosis and the protein levels of Ki67, MMP2 and Bax in MM.1S and H929 cells ((F,G)). These data demonstrated that circ_0003489 regulated miR-433-3p/PBX3 axisto promote MM cell function.
Figure 7. Circ_0003489/miR-433-3p /PBX3 axis regulated MM cell function. (A and B) The efficiency of PBX3 overexpression vector was determined. The sh-NC, sh-circ_0003489, sh-circ_0003489 + anti-NC, sh-circ_0003489 + anti-miR-433-3p, sh-circ_0003489 + vector, sh-circ_0003489 + PBX3 were transfected into MM.1S and H929 cells, (C) Cell viability, (D) cell cycle, (E) proliferation, (F) apoptosis and (G) Ki67, MMP2 and Bax protein levels were measured. *P < 0.05.
4. Discussion
As the research further progressed, circRNAs are considered as more promising cancer markers due to their high abundance and stability in the cytoplasm [Citation22]. Our study revealed a higher abundance of circ_0003489 in MM tissues and cells, and its downregulation restrained MM cell proliferation and promoted apoptosis. These data suggested that circ_0003489 had the potential to be a molecular target for future MM therapy. Upregulation of circ_0003489 was also monitored in lung cancer and its knockdown restrained cancer cell proliferation and invasion through miRNA-490-3p/MAPK1 axis [Citation23]. In our research, we pointed out that circ_0003489 might promote MM progression by regulating miR-433-3p/PBX3 axis.
Hou et al. observed that miR-433-3p upregulation repressed osteosarcoma cell proliferation and metastasis, showing that miR-433-3p acted as a tumor suppressor in osteosarcoma [Citation24]. MiR-433-3p upregulation strikingly arrested cancer cell proliferation and metastasis, as well as improved temozolomide chemotherapy sensitivity in glioma cells [Citation25]. These results demonstrated the anti-tumor role of miR-433-3p. Similarly, lower abundance of miR-433-3p was observed in MM tissues and cells, and its downregulation attenuated MM cell progression. Importantly, miR-433-3p downregulation also reversed the repressive effects of circ_0003489 knockdown on MM progression, confirming that circ_0003489 enhanced MM progression by sponging miR-433-3p.
PBX3 was identified to be notably upregulated in papillary thyroid cancer tissues, and high level of PBX3 was dramatically associated with tumor volume and lymph node metastasis and poor prognosis [Citation26]. Xu et al. identified that PBX3 also acted as an oncogene to enhance glioblastoma cell metastasis [Citation27]. Consistent with previous study [Citation20], high expression of PBX3 in MM tissues was observed in present study. PBX3 was confirmed to be targeted by miR-433-3p, and its expression was positively regulated by circ_0003489. In addition, PBX3 upregulation reverted the effects of circ_0003489 downregulation on MM cell proliferation and apoptosis, which not only confirmed the carcinogenic effect of PBX3, but also confirmed that circ_0003489 mediated MM process by regulating PBX3.
5 Conclusion
In summary, our data showed that circ_0003489 facilitated MM progression by targeting miR-433-3p/PBX3 axis. The uncovering of circ_0003489/miR-433-3p/PBX3 axis in MM progression laid the foundation for searching novel marker for the treatment of MM.
Ethics statement
All experiments for this study were approved by the Ethics committee of Lishui Second People’s Hospital, and all participants signed written informed consent.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The datasets used and analyzed during the current study are available from the corresponding author on reasonable request.
References
- Kazandjian D. Multiple myeloma epidemiology and survival: A unique malignancy. Semin Oncol. 2016;43(6):676–681.
- Firth J. Haematology: multiple myeloma. Clin Med (Lond). 2019;19(1):58–60.
- Landgren O, Graubard BI, Katzmann JA, et al. Racial disparities in the prevalence of monoclonal gammopathies: a population-based study of 12,482 persons from the National Health and Nutritional Examination Survey. Leukemia. 2014;28(7):1537–1542.
- Eslick R, Talaulikar D. Multiple myeloma: from diagnosis to treatment. Aust Fam Physician. 2013;42(10):684–688.
- Liu B, Zhao N, Zhou Y, et al. Circular RNA circ_ABCB10 in cancer. Clin Chim Acta. 2021;518:93–100.
- Zhang J, Gao C, Zhang J, et al. Circ_0010729 knockdown protects cardiomyocytes against hypoxic dysfunction via miR-370-3p/TRAF6 axis. EXCLI J. 2020;19:1520–1532.
- Zhang J, Cheng F, Rong G, et al. Hsa_circ_0005567 activates autophagy and suppresses IL-1β-induced chondrocyte apoptosis by regulating miR-495. Front Mol Biosci. 2020;7:216.
- Yao T, Zha D, Hu C, et al. Circ_0000285 promotes podocyte injury through sponging miR-654-3p and activating MAPK6 in diabetic nephropathy. Gene. 2020;747:144661.
- Gao M, Li C, Xiao H, et al. Hsa_circ_0007841: A novel potential biomarker and drug resistance for multiple myeloma. Front Oncol. 2019;9:1261.
- Feng Y, Zhang L, Wu J, et al. CircRNA circ_0000190 inhibits the progression of multiple myeloma through modulating miR-767-5p/MAPK4 pathway. J Exp Clin Cancer Res. 2019;38(1):54.
- Liu X, Tang H, Liu J, et al. Hsa_circRNA_101237: A novel diagnostic and prognostic biomarker and potential therapeutic target for multiple myeloma. Cancer Manag Res. 2020;12:2109–2118.
- Xiong DD, Dang YW, Lin P, et al. A circRNA-miRNA-mRNA network identification for exploring underlying pathogenesis and therapy strategy of hepatocellular carcinoma. J Transl Med. 2018;16(1):220.
- Yang G, Zhang Y, Yang J. Identification of potentially functional CircRNA-miRNA-mRNA regulatory network in gastric carcinoma using bioinformatics analysis. Med Sci Monit. 2019;25:8777–8796.
- Lu TX, Rothenberg ME. MicroRNA. J Allergy Clin Immunol. 2018;141(4):1202–1207.
- Krol J, Loedige I, Filipowicz W. The widespread regulation of microRNA biogenesis, function and decay. Nat Rev Genet. 2010;11(9):597–610.
- Bayraktar R, Van Roosbroeck K. miR-155 in cancer drug resistance and as target for miRNA-based therapeutics. Cancer Metastasis Rev. 2018;37(1):33–44.
- Shi Q, Wang Y, Mu Y, et al. MiR-433-3p inhibits proliferation and invasion of esophageal squamous cell carcinoma by targeting GRB2. Cell Physiol Biochem. 2018;46(5):2187–2196.
- Morgan R, Pandha HS. PBX3 in cancer. Cancers (Basel). 2020;12(2):431.
- Li Y, Sun Z, Zhu Z, et al. PBX3 is overexpressed in gastric cancer and regulates cell proliferation. Tumour Biol. 2014;35(5):4363–4368.
- Lu Y, Wu D, Wang J, et al. miR-320a regulates cell proliferation and apoptosis in multiple myeloma by targeting pre-B-cell leukemia transcription factor 3. Biochem Biophys Res Commun. 2016;473(4):1315–1320.
- He S, Yang J, Jiang S, et al. Circular RNA circ_0000517 regulates hepatocellular carcinoma development via miR-326/IGF1R axis. Cancer Cell Int. 2020;20:404.
- Jeck WR, Sorrentino JA, Wang K, et al. Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA. 2013;19(2):141–157.
- Zhang ZY, Gao XH, Ma MY, et al. CircRNA_101237 promotes NSCLC progression via the miRNA-490-3p/MAPK1 axis. Sci Rep. 2020;10(1):9024.
- Hou XK, Mao JS. Long noncoding RNA SNHG14 promotes osteosarcoma progression via miR-433-3p/FBXO22 axis. Biochem Biophys Res Commun. 2020;523(3):766–772.
- Sun S, Wang X, Xu X, et al. MiR-433-3p suppresses cell growth and enhances chemosensitivity by targeting CREB in human glioma. Oncotarget. 2017;8(3):5057–5068.
- Chen Q, Yu WY, Zhang HH, et al. PBX3 promotes tumor growth and angiogenesis via activation of AT1R/VEGFR2 pathway in papillary thyroid carcinoma. Biomed Res Int. 2020;2020:8954513.
- Xu X, Bao Z, Liu Y, et al. PBX3/MEK/ERK1/2/LIN28/let-7b positive feedback loop enhances mesenchymal phenotype to promote glioblastoma migration and invasion. J Exp Clin Cancer Res. 2018;37(1):158.