ABSTRACT
Background:
It has been reported that circular RNA hsa_circ_0044907 (circ_0044907) expression is overtly elevated in acute myeloid leukemia (AML) patient-derived BMMCs. However, the effect of circ_0044907 on AML progression remains un-clarified.
Methods:
Expression of circ_0044907 in BM and AML cells were detected with real-time quantitative reverse transcription-polymerase chain reaction (RT-qPCR). Cell viability, proliferation, apoptosis, and cycle progression were determined by 3-(4,5-Dimethylthiazol-2-yl)−2,5-Diphenyltetrazolium Bromide (MTT), 5-ethynyl-2’-deoxyuridine (EDU), and flow cytometry assays. The regulatory mechanism of circ_0044907 was predicted by bioinformatics analysis and validated by dual-luciferase reporter, RNA pull-down, and RNA immunoprecipitation (RIP) assays. In vivo experiments were carried out to verify the function of circ_0044907.
Results:
Circ_0044907 was overexpressed in AML patient-derived BM and AML cells. Furthermore, circ_0044907 could distinguish AML patients from healthy controls, and high circ_0044907 expression in BM had a poor prognosis for AML patients, implying that circ_0044907 served as a diagnostic and prognostic indicator for AML. Functionally, circ_0044907 silencing reduced cell viability, restrained cell proliferation, arrested cell cycle progression, and induced cell apoptosis in AML cells in vitro. Furthermore, circ_0044907 knockdown decreased AML cell growth in xenograft mouse models. Mechanically, circ_0044907 sponged miR-186-5p to block the inhibiting effect of miR-186-5p on KIT. Silenced miR-186-5p expression weakened circ_0044907 knockdown mediated suppression on AML cell viability, proliferation, and cycle progression. Also, forced KIT expression weakened miR-186-5p upregulation mediated inhibition on AML cell viability, proliferation, and cycle progression.
Conclusion:
Circ_0044907 absorbed miR-186-5p to block the inhibiting impact of miR-186-5p on KIT, thus promoting AML progression.
KEYWORDS:
Introduction
Acute myeloid leukemia (AML), a heterogeneous hematological malignancy accounting for 7.6% of hematopoietic malignancies, is caused by the carcinogenic transformation of hematopoietic progenitor cells in the bone marrow (BM) [Citation1]. The main treatment options for AML are chemotherapy and allogeneic bone marrow transplantation [Citation2], but the 5-year survival rate is only about 25% due to chemotherapy resistance and limited bone marrow supply [Citation3–5]. Therefore, understanding the mechanisms related to its progression can help improve the treatment and prognosis of AML.
MicroRNAs (miRNAs) modulate gene expression at the post-transcriptional level [Citation6]. They are powerful regulators of various cellular activities, such as cell apoptosis and growth [Citation7]. Increasing evidence shows the significant role of miRNAs in disease processes, including AML [Citation8]. For instance, low miR-340/miR-204 expression has adverse clinical outcomes in AML patients [Citation9, Citation10]. Moreover, miR-551b is a biomarker for relapse in AML [Citation11]. Currently, the functions of miRNAs in AML have been explored a lot, but the molecular mechanism of their disorders is still unclear.
An increasing body of evidence has demonstrated that cytoplasmic circular RNAs (circRNAs) can sequester miRNAs by functioning as miRNA spongea [Citation12]. CircRNAs are covalently closed RNA biomolecules generated through back-splicing [Citation13]. They have the potential as prognostic and diagnostic biomarkers as well as therapeutic targets for personalized medicine due to their tissue/cell-specific expression patterns [Citation14]. Recent reports have demonstrated the implication of circRNAs in blood malignancies, including AML [Citation15]. For example, circ-VIMENTIN and circ-PLXNB2 act as prognostic predictors for AML prognosis [Citation16, Citation17]. For another example, circ-SPI1 contributes to AML cell malignancy through interacting with miR-1307-3p/−382-5p/−767-5p [Citation18]. At present, the function of circRNAs in AML remains largely unknown.
Hsa_circ_0044907 (circ_0044907), located at chr17: 58013575-58013902, is formed by the RPS6KB1 gene with a mature length of 141 nt. In circRNA expression profile, circ_0044907 has been reported to be overexpressed in AML patient-derived BMMCs [Citation19]. However, the function of circ_0044907 in AML is unknown. Thus, the study was to explore the influence of circ_0044907 on AML progression and its regulatory mechanism.
Materials and methods
BM collection
BM samples were obtained from 45 healthy donors and 45 AML patients who offered written informed consent at Liangjiang New Area First People's Hospital. Collection of human BM samples was performed under a protocol approved by the Ethics Committee of Liangjiang New Area First People's Hospital.
Cell culture
Two AML cell lines THP-1 (TIB-202™, ATCC, Manassas, VA, USA) and HL-60 (CCL-240™, ATCC), HS-5 cell line (CRL-11882™, ATCC), and 293 T cell line (CRL-3216™, ATCC) were used in the study. The THP-1 cell line was cultured in RPMI-1640 medium (Thermo, Waltham, MA, USA) supplemented with 10% fetal bovine serum (Biowest, Nuaillé, France) and 0.05 mM 2-mercaptoethanol [Sigma-Aldrich (SA), St. Louis, MO, USA]. The HL-60 cell line was cultured in IMDM (Thermo) supplemented with 20% fetal bovine serum (Biowest). The HS-5 cell line was cultured in DMEM (Thermo) supplemented with 10% fetal bovine serum. The 293 T cell line was cultured in DMEM supplemented with 10% fetal bovine serum and 2 mM L-glutamine [SA].
Real-time quantitative reverse transcription-polymerase chain reaction (RT-qPCR)
Total RNA isolation was performed using the TRIzol reagent (Thermo). After purification with the Direct-zolTM kit (Zymo Research, Orange, CA, USA), equal amounts (1 µg) were used for reverse transcription with the RT2 first strand kit (Qiagen, Hilden, Germany) or miScript II RT Kit (Qiagen) as per the manufacturer’s instructions. Amplification reactions were carried out using the PCR Maxima SYBR Green kit (Thermo) or miScript SYBR Green PCR Kit (Qiagen) with specific primers (). Relative quantification was calculated with the 2-ΔΔCt method with GAPDH or U6 as endogenous control.
Table 1. The primer sequences of genes.
Transient transfection
3 siRNAs against circ_0044907 [si-circ_0044907#1 (5’-TTTTCAGCAGCAACAGAGGTT-3’), si-circ_0044907#2 (5’-TTTCAGCAGCAACAGAGGTTT-3’), and si-circ_0044907#3 (5’-ATTTCTTTTCAGCAGCAACAG-3’)] were used to explore the function of circ_0044907 in vitro with si-NC (5’-AAUUCUCCGAACGUGUCACGU-3’) as a control. To knock down and overexpress miR-186-5p, miR-186-5p inhibitor (anti-miR-186-5p: 5’-AGCCCAAAAGGAGAAUUCUUUG-3’) and miR-186-5p mimic (miR-186-5p: 5’-CAAAGAAUUCUCCUUUUGGGCU-3’) were utilized with anti-NC (5’-CGGUACGAUCGCGGCGGGAUAUC-3’) and miR-NC (5’-GGUUCGUACGUACACUGUUCA-3’) as controls, respectively. The pcDNA-KIT plasmid (KIT) was constructed and used for overexpression of KIT with the empty pcDNA vector (Thermo) as a control. Transfection was carried using using TransIT-X2 (Mirus Bio., Madison, WI, USA) or FuGENE® HD Transfection Reagent (Promega, Madison, WI, USA).
3-(4,5-Dimethylthiazol-2-yl)−2,5-Diphenyltetrazolium Bromide (MTT) assay
AML cells (2 × 103) were seeded and incubated for 24-72 h. Following incubation with the MTT solution (5 mg/mL, 10 μL) (Yesen, Shanghai, China), the purple crystals were dissolved by the formazan solution (100 μL) (Yesen). Absorbance (570 nm) was measured using a plate reader (MTX Lab Systems, Bradenton, FL, USA).
5-ethynyl-2’-deoxyuridine (EDU) incorporation assay
When conducting the EDU incorporation assay (Ribobio, Guangzhou, China), AML cells were seeded with a density of 2 × 104 viable cells per well and incubated for 4 h after adding 50 μM EDU. Following fixation (4% formaldehyde, 15 min), AML cells were treated with Triton X-100 (0.2%) and Apollo mixture (100 μL). Nuclei were then labeled with blue fluorescence (DAPI), and a fluorescence microscope (Olympus, Tokyo, Japan) was utilized to photograph.
Cell apoptosis analysis
48 h after transient transfection, AML cells were stained with Annexin V-FITC/PI as per the manufacturer’s protocol of the Annexin V-FITC/PI Apoptosis Detection Kit (Yesen). Data were analyzed by FlowJo software (Tree star, San Carlos, CA, USA) following detection with the flow cytometry (BD, San Jose, CA, USA).
Cell cycle analysis
Following washing 3 times with PBS (Biowest), the cultured AML cells were adjusted to approximately 1 × 106 cells/mL. Subsequently, the cells were fixed with 4% pre-chilled 75% ethanol and then incubated with 100 μL of RNase A. 30 min later, the cells were stained with 400 μL of PI, followed by subjecting to flow cytometry (BD).
Western blotting
After washing, AML cells were lysed using RIPA buffer. Western blotting analysis was carried out as previously described [Citation20]. Primary antibodies β-actin at 1:1000 (#3700, CST, Danvers, MA, USA), PCNA at 1:2000 (#2586, CST), Cyclin D1 at 1:1000 (#55506, CST), KIT at 1:1000 (#3074, CST), and Cleaved caspase 3 at 1:1000 (#9661, CST) were used. Chemiluminescence reactions were performed as per manufacturer’s instructions (GE Healthcare).
Dial-luciferase reporter assay
The psiCHECK-2 vector (Promega) was utilized to construct recombinant luciferase reporters circ_0044907-WT, circ_0044907-MUT, KIT-3’UTR-WT, or KIT-3’UTR-MUT. For luciferase analysis, the transfection of a recombinant luciferase reporter along with miR-NC and miR-186-5p mimic into AML cells was done using TransIT-X2 (Mirus Bio.,). Luciferase activity was analyzed using the dual-luciferase assay system (Promega).
RNA pull-down assay
The AML cells transfected with Bio-miR-186-5p or Bio-NC were sonicated. The obtained supernatants were incubated with activated M-280 streptavidin beads (SA) at 4˚C overnight. RNA complexes were eluted from streptavidin beads and then subjected to RT-qPCR analysis.
RIP assay
The Magna RIP RNA-Binding Protein Immunoprecipitation Kit (Millipore, Billerica, MA, USA) with an antibody against Ago2/IgG was used for analysis of the interaction between circ_0044907 and miR-186-5p following the manufacturer’s instructions. Co-precipitated RNA complexes were purified and then analyzed by RT-qPCR.
Generation of HL-60 cells with stable circ_0044907 knockdown
To stable knock down circ_0044907, the shRNA sequence against circ_0044907 was cloned into lentiviral vector pLKO.1 with puromycin resistance. Lentiviruses were packaged using the psPAX2 and pMD2.G packaging system (Thermo). Infection of HL-60 cells was done as described previously [Citation21].
Animal experiments
All procedures and maintenance conditions were approved by the Animal Ethics Committee of Liangjiang New Area First People's Hospital. 10 BALB/c nude mice (Vital River, Beijing, China) were housed in a specific pathogen-free housing facility. After a week of adaptation, the right armpit region of each mouse was injected with HL-60 cells carrying sh-NC or sh-circ_0044907. 5 weeks after injection, mice were euthanized and tumors were excised, weighed, and photographed. Tumor volume was evaluated every 7 days (Volume = (length ×width2)/2). Tumor tissues embedded in paraffin were cut into 4-μm sections for immunohistochemistry (IHC) analysis with primary antibodies against PCNA (#2586, CST) and Ki-67 (#9449, CST).
Statistical analysis
Values from three biological replicates were analyzed with GraphPad Prism 8 software (GraphPad, La Jolla, CA, USA) and exhibited as mean ± standard deviation. Statistical analysis was performed by Student’s t-test (for two groups) or analysis of variance (ANOVA) followed by Tukey’s test (for multiple comparisons) with P < 0.05 as the significant criteria. Survival was analyzed by the log-rank test.
Results
Characterization and clinical features of circ_0044907
CircRNA expression profile showed the upregulation of circ_0044907 in BMMCs derived from AML patients [Citation19]. To address its function, changes in circ_0044907 expression in BM from AML patients and healthy controls were detected. An apparent upregulation of circ_0044907 was observed in AML patient-derived BM relative to healthy control-derived BM ((a)). We then investigated the relationship between high and low expression of circ_0044907 and FLT3-ITD, NPM1 mutation, CEBPA mutation, WT1 mutation and MLL-rearranged, and the results showed that the expression of circ_0044907 was not related to these AML genotypes (). The receiver-operating characteristic (ROC) curve showed that the AUC of BM circ_0044907 was 0.9447 (95% CI: 0.8997-0.9897), and its sensitivity and specificity for the diagnosis of AML were 77.78% and 88.89%, respectively, implying that circ_0044907 had a good diagnostic significance for AML ((b)). Also, AML patients with high expression of circ_0044907 in BM had a significantly shorter overall survival rate ((c)). Analogously, circ_0044907 was overexpressed in AML cells than that in the HS-5 cell line ((d)). The resistance of circ_0044907 to RNase R digestion was demonstrated in (e), which validated the circular structure of circ_0044907 in AML cells. These findings suggested that circ_0044907 might be involved in AML progression.
Figure 1. Change and characterization of circ_0044907 in AML. (A) Fold change of circ_0044907 in AML patient-derived BM relative to healthy control-derived BM was determined by RT-qPCR. (B) Diagnostic value (ROC) of BM circ_0044907 in 45 AML patients. (C) Correlation between BM circ_0044907 expression pattern and clinical survival rate was analyzed. (D) Relative expression changes of circ_0044907 in AML cells. (E) Analysis of circ_0044907 and linear RPS6KB1 in AML cells treated with or without RNase R. ***P < 0.001.
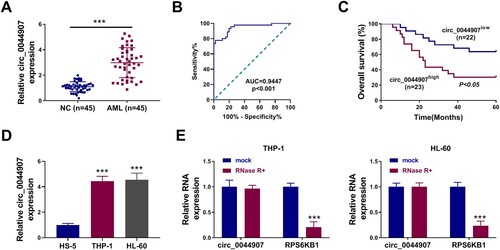
Table 2. Correlation of circ_0044907 expression with AML genotype.
Loss of circ_0044907 reduced AML cell proliferation and induced AML cell apoptosis
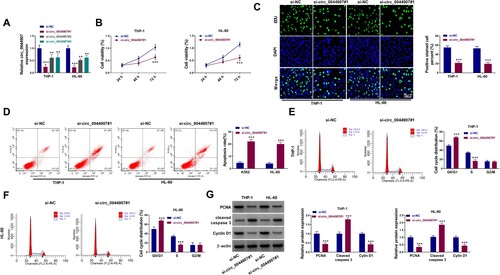
Loss-of-function experiments were then performed to explore the function of circ_0044907. Three siRNAs against circ_0044907 (si-circ_0044907#1, si-circ_0044907#2, and si-circ_0044907#3) were designed and their transfection efficiencies were validated and shown in (a), and the si-circ_0044907#1 with the highest knockdown effect was used for subsequent functional analysis. CCK-8 and EDU assays exhibited that circ_0044907 silencing decreased AML cell viability ((b)) and curbed AML cell proliferation ((c)). Apoptosis analysis exhibited that circ_0044907 knockdown resulted in an elevation in the number of apoptotic AML cells ((d)). Cycle analysis showed that circ_0044907 silencing increased the number of AML cells in the G0/G1 stage and lowered the number of AML cells in the S stage when compared with the control cells ((e,f)). WB showed that circ_0044907 inhibition decreased PCNA and Cyclin D1 protein levels and elevated Cleaved caspase 3 protein levels in AML cells ((g)). These results manifested that circ_0044907 silencing restrained cell proliferation and promoted cell apoptosis in AML cells.
Figure 2. Circ_0044907 silencing decreased AML cell proliferation and contributed to AML cell apoptosis. (A) Analysis of the interference efficiency of 3 siRNAs targeting circ_0044907 (si-circ_0044907#1, si-circ_0044907#2, and si-circ_0044907#3) was done. (B-F) MTT, EdU, and flow cytometry assays were utilized for analysis of cell viability (B), proliferation (C), apoptosis (D), and cycle progression (E and F) in AML cells with si-NC or si-circ_0044907#1. (G) Relative protein levels of PCNA, Cyclin D1, and Cleaved caspase 3 in the above AML cells. **P < 0.01 and ***P < 0.001.
Circ_0044907 interacted with miR-186-5p
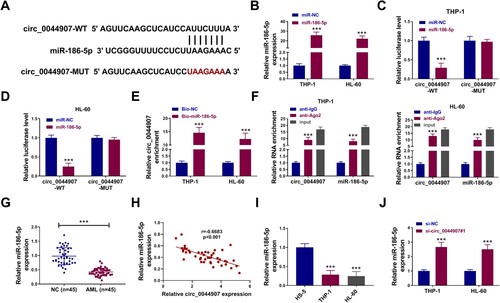
Based on the theory of competing for endogenous RNA (ceRNA), we further explored whether circ_0044907 could function as a miRNA sponge. Bioinformatics analysis (circinteractome: https://circinteractome.nia.nih.gov/mirna_target_sites.html) predicted that circ_0044907 may interact with miR-186-5p ((a)). MiR-186-5p mimic was used to overexpress miR-186-5p in AML cells, as demonstrated by RT-qPCR ((b)). We then explored the binding capability of miR-186-5p to circ_0044907 through luciferase assays, and the luciferase activity of the circ_0044907-WT reporter was overtly decreased after miR-186-5p overexpression ((c,d)). RNA pull-down assays with Bio-miR-186-5p probe showed that circ_0044907 were efficiently pulled down by the Bio-miR-186-5p probe ((e)). Anti-Ago2 RIP assays exhibited that circ_0044907 and miR-186-5p were highly enriched in the anti-Ago2 group compared with those in the anti-IgG group ((f)). We also observed that miR-186-5p was underexpressed in AML patient-derived BM ((g)) and it had a negative correlation with circ_0044907 ((h)). Consistently, miR-186-5p had lower levels in AML cells ((i)). As expected, circ_0044907 silencing led to an overt elevation in miR-186-5p expression in AML cells ((j)). In sum, these results indicated that circ_0044907 interacted with miR-186-5p.
Figure 3. MiR-186-5p was sponged by circ_0044907. (A) The putative binding sites of miR-186-5p in the circ_0044907 sequence. (B) The transfection efficiency of miR-186-5p mimic. (C and D) Luciferase activity of circ_0044907-WT/circ_0044907-MUT reporter in AML cells co-transfected with miR-186-5p mimic or miR-NC. (E and F) The interaction between circ_0044907 and miR-186-5p was demonstrated by RNA pull-down and RIP assays. (G) Relative expression changes of miR-186-5p in AML patient-derived BM. (H) Pearson’s correlation coefficient analysis of the correlation between miR-186-5p and circ_0044907 in AML patient-derived BM. (I) Relative expression levels of miR-186-5p in AML cells. (J) Influence of circ_0044907 silencing on miR-186-5p in AML cells. ***P < 0.001.
Circ_0044907 mediated AML cell proliferation and apoptosis through sequestering miR-186-5p
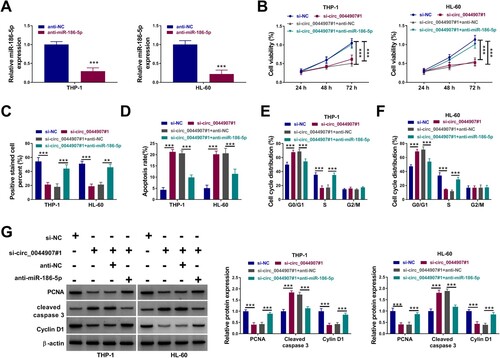
Rescue experiments were then performed to validate whether circ_0044907 exerts its promoting effect via repressing miR-186-5p. We used miR-186-5p inhibitor to knock down miR-186-5p in AML cells ((a)). Furthermore, the decreased viability ((b)), inhibited proliferation ((c)), boosted apoptosis ((d)), and arrested cycle ((e,f)) of AML cells prompted by circ_0044907 silencing were partially overturned after miR-186-5p knockdown. In addition, silenced miR-186-5p expression weakened the decreased protein levels of PCNA and Cyclin D1 and the elevated protein levels of Cleaved caspase 3 in AML cells provoked by circ_0044907 knockdown ((g)). Together, these results suggested that circ_0044907 mediated cell proliferation and apoptosis in AML cells through interacting with miR-186-5p.
Figure 4. Circ_0044907 mediated AML cell proliferation and apoptosis through interacting with miR-186-5p. (A) The transfection efficiency of miR-186-5p inhibitor. (B-F) Analysis of cell viability (B), proliferation (C), apoptosis (D), and cycle progression (E and F) in AML cells with si-NC, si-circ_0044907#1, si-circ_0044907#1 + anti-NC, or si-circ_0044907#1 + anti-miR-186-5p. (G) Detection of PCNA, Cyclin D1, and Cleaved caspase 3 protein levels in the above AML cells. **P < 0.01 and ***P < 0.001.
KIT served as a target of miR-186-5p
Subsequently, we further investigated the downstream targets of miR-186-5p. Bioinformatics prediction (trgetscan) showed that miR-186-5p contained the putative binding sites for the 3’UTR of KIT ((a)). Moreover, the luciferase activity of the KIT-3’UTR-WT reporter was restrained by miR-186-5p overexpression but not the KIT-3’UTR-MUT reporter ((b)). We also observed that miR-186-5p overexpression repressed KIT protein levels, but miR-186-5p silencing exerted an opposing effect ((c,d)). In addition, circ_0044907 silencing caused a decrease in KIT protein levels, but this decrease was impaired after miR-186-5p inhibition ((e)). Changes in KIT expression were then detected in AML patient-derived BM. Analogously, KIT mRNA levels were overexpressed in AML patient-derived BM compared with the control samples ((f)), and it had a negative correction with miR-186-5p and a positive correction with circ_0044907 ((g,h)). As expected, KIT protein levels were upregulated in AML patient-derived BM and AML cells ((i,j)). Collectively, circ_0044907 mediated KIT expression through interacting with miR-186-5p.
Figure 5. MiR-186-5p directly targeted KIT. (A) The putative binding sites of miR-186-5p in the 3’UTR of KIT. (B) Luciferase activity of KIT-3’UTR-WT/KIT-3’UTR-MUT reporter in AML cells co-transfected with miR-186-5p mimic or miR-NC. (C and D) Effects of miR-186-5p overexpression and knockdown on KIT protein levels in AML cells. (E) KIT protein levels in AML cells with si-NC, si-circ_0044907#1, si-circ_0044907#1 + anti-NC, or si-circ_0044907#1 + anti-miR-186-5p. (F) Relative expression changes of KIT mRNA in AML patient-derived BM. (G and H) Pearson’s correlation coefficient analysis of the correlation between KIT mRNA and miR-186-5p or circ_0044907 in AML patient-derived BM. (I and J) Detection of KIT protein levels in AML patient-derived BM and AML cells. *P < 0.05 and ***P < 0.001.
Circ_0044907 mediated AML cell proliferation and apoptosis via the miR-186-5p/KIT axis
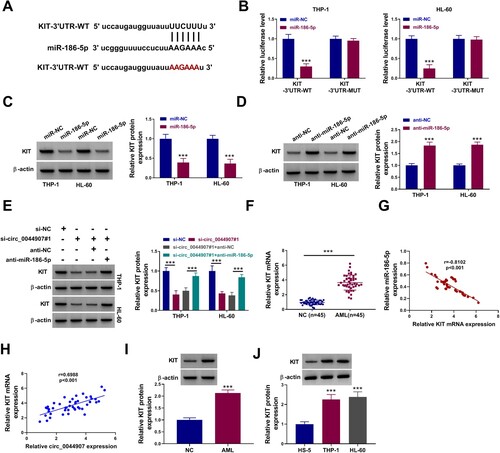
Whether circ_0044907 mediates AML cell proliferation and apoptosis via the miR-186-5p/KIT axis was investigated. We observed that inhibition of circ_0044907 reduced KIT protein levels in AML cells, but co-introduction of the KIT overexpression vector weakened this reduction mediated by circ_0044907 knockdown (Supplementary Figure 1A). Also, KIT overexpression partially reversed circ_0044907 knockdown-mediated repression of AML cell viability and proliferation, as well as induction of AML cell apoptosis and cycle arrest (Supplementary Figure 1B-G). To validate whether miR-186-5p exerts an inhibiting effect via targeting KIT, rescue experiments were then carried out through co-transfection of miR-186-5p mimic and KIT overexpression plasmids, and the transfection efficiency of the KIT overexpression plasmid alone was shown in (a). KIT overexpression weakened the inhibitory effect of miR-186-5p mimic on the KIT protein level (Supplementary Figure 2). Moreover, miR-186-5p upregulation reduced cell viability ((b)), restrained cell proliferation ((c)), facilitated cell apoptosis ((d)), and arrested cell cycle progression ((e,f)) in AML cells, while these impacts were weakened after KIT overexpression. Additionally, forced KIT expression partly overturned the reduced protein levels of PCNA and Cyclin D1 and the increased protein levels of Cleaved caspase 3 in miR-186-5p-overexpressed AML cells ((g)). These results manifested that circ_0044907 regulated the miR-186-5p/KIT axis to mediate cell proliferation and apoptosis in AML cells.
Figure 6. MiR-186-5p mediated AML cell proliferation and apoptosis via negatively regulating KIT expression. (A) Western blotting analysis of the transfection efficiency of the KIT overexpression plasmid. (B-F) Determination of cell viability (B), proliferation (C), apoptosis (D), and cycle progression (E and F) in AML cells with miR-NC, miR-186-5p, miR-186-5p + vector, or miR-186-5p + KIT. (G) Detection of PCNA, Cyclin D1, and Cleaved caspase 3 protein levels in the above AML cells. **P < 0.01 and ***P < 0.001.
Circ_0044907 silencing decreased AML cell growth in mouse models
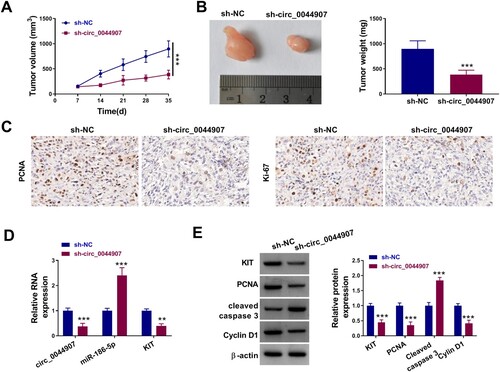
Next, xenograft assays were conducted to verify the effect of circ_0044907 on AML cell growth in vivo. Injection of HL-60 cells with circ_0044907 knockdown showed smaller tumor volume and lighter tumor weight compared to the control group ((a,b)). IHC assays exhibited that circ_0044907 knockdown decreased the levels of PCNA and Ki-67 in xenograft samples ((c)). Concordantly low levels of circ_0044907 and KIT were found in xenograft samples from the circ_0044907-knockdown group, but miR-186-5p had higher levels ((d)). Moreover, in addition to the upregulation of Cleaved caspase-3 protein levels in xenograft samples in the circ_0044907-knockdown group, KIT, PCNA and Cyclin D1 protein levels were downregulated ((e)). Altogether, these findings suggested that circ_0044907 promoted AML cell in vivo.
Figure 7. Circ_0044907 facilitated AML cell in vivo. (A) The tumor volume of sh-circ_0044907 and sh-NC groups. (B) Representative images of xenograft tumors in the above two groups (Left). The average weight of xenograft tumors in the above two groups (Right). (C) Representative images of IHC staining for PCNA and Ki-67 in xenograft tumors in the above two groups. (D) Expression levels of circ_0044907, miR-186-5p, and KIT mRNA in xenograft tumors in the above two groups. (E) Protein levels of Cleaved caspase-3, KIT, PCNA, and Cyclin D1 in xenograft tumors in the above two groups. **P < 0.01 and ***P < 0.001.
Discussion
Dysregulation of circRNAs is gaining recognition for its vital function in AML. Here, we identified a novel mechanism by which circ_0044907 mediates AML growth through regulating KIT expression via interacting with miR-186-5p, indicating the promoting role of circ_0044907 in AML.
Our data exhibited an overt elevation in circ_0044907 expression in AML patient-derived BM and AML cells. Moreover, BM circ_0044907 could distinguish patients with AML from healthy controls, implying that circ_0044907 had good diagnostic significance for AML. Also, high circ_0044907 expression in BM had a poor prognosis for AML patients, suggesting that circ_0044907 served as a prognostic indicator for AML. Loss-of-function experiments manifested that circ_0044907 silencing reduced AML cell viability, restrained AML cell proliferation, arrested AML cell cycle progression, and induced AML cell apoptosis in vitro. Furthermore, circ_0044907 knockdown decreased AML cell growth in xenograft mouse models, and PCNA, Ki67, and Cyclin D1 levels were lower in the xenograft tumors from the circ_0044907-knockdown group, but Cleaved caspase 3 levels were higher. These results highlighted the oncogenic role of circ_0044907 in AML.
Considering that circRNAs act as miRNA sponges [Citation22], we further explored the function of circ_0044907 as a miRNA sponge. Bioinformatics analysis with validation experiments identified the function of circ_0044907 as a miR-186-5p sponge. Reports uncovered that miR-186-5p exerts a tumor-inhibiting function in a set of tumors, including ovarian cancer [Citation23], gastric cancer [Citation24], osteosarcoma [Citation25], and glioblastoma [Citation26]. A previous report uncovered that low miR-186-5p level showed overtly shorter overall survival in AML [Citation27]. Also, miR-186-5p played an inhibiting impact on chronic myeloid leukemia progression [Citation28, Citation29]. Here, miR-186-5p was apparently underexpressed in AML patient-derived BM and AML cells. Moreover, miR-186-5p silencing weakened circ_0044907 knockdown-mediated effects on AML cell viability, proliferation, cycle progression, and apoptosis. These results suggested that circ_0044907 mediated AML progression via interacting with miR-186-5p.
KIT is a trans-membrane receptor tyrosine kinase that binds the SCF ligand to activate MAPK, JAK/STAT, and PI3K pathways to facilitate cell survival [Citation30]. It is implicated in cell transformation through overexpression or oncogenic mutation [Citation31, Citation32]. A report from Hu et al. manifested that miR-137 decreased AML progression by targeting KIT [Citation33]. Furthermore, TUG1 silencing induced AML cell apoptosis and curbed AML cell viability via targeting KIT [Citation34]. Gao et al. showed that miR-193b restoration curbed AML cell growth through decreasing KIT expression [Citation35]. Moreover, KIT upregulation frequently occurs in AML, resulting in promoting abnormal cell proliferation and poor outcomes [Citation35, Citation36]. Here, KIT as a miR-186-5p target was identified. Moreover, KIT was overexpressed in AML patient-derived BM and AML cells, and KIT upregulation impaired miR-186-5p mimic mediated suppression on AML cell viability, proliferation, and cycle progression. Importantly, circ_0044907 could mediate KIT expression via interacting with miR-186-5p. Therefore, we concluded that circ_0044907 promoted AML progression via sequestering miR-186-5p and upregulating KIT.
In conclusion, our findings demonstrated that circ_0044907 promotes AML progression, at least in part, via the miR-186-5p/KIT axis, highlighting the critical role of circ_0044907 in AML. Further studies are needed to explore the role of circ_0044907 in AML.
Competing interests
The authors declare that they have no competing interest.
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request
Authors’ contributions
Ling Liu designed the study, analyzed the data and wrote the manuscript. Xing Qiang performed the experiments and analyzed the data.
Ethics Approval
The research related to human use has been complied with all the relevant national regulations, institutional policies and has been approved by the Ethics Committee of Liangjiang New Area First People's Hospital.
Acknowledgment
None.
Disclosure statement
No potential conflict of interest was reported by the author(s).
References
- Pasquer H, Tostain M, Kaci N, et al. Descriptive and functional genomics in acute myeloid leukemia (AML): paving the road for a cure. Cancers (Basel). 2021;13(4):748.
- Almond LM, Charalampakis M, Ford SJ, et al. Myeloid sarcoma: presentation, diagnosis, and treatment. Clin Lymphoma Myeloma Leuk. 2017;17(5):263–267.
- Westhus J, Noppeney R, Dührsen U, et al. FLAG salvage therapy combined with idarubicin in relapsed/refractory acute myeloid leukemia. Leuk Lymphoma. 2019;60(4):1014–1022.
- Aldoss I, Marcucci G. More options for older patients with acute myeloid leukemia: venetoclax in combination with low dose cytarabine. Chin Clin Oncol. 2019;8(S1):S25.
- Xu S, Zhang M, Fang X, et al. A novel CD123-targeted therapeutic peptide loaded by micellar delivery system combats refractory acute myeloid leukemia. J Hematol Oncol. 2021;14(1):193.
- Bushati N, Cohen SM. microRNA functions. Annu Rev Cell Dev Biol. 2007;23:175–205.
- Saliminejad K, Khorram Khorshid HR, Soleymani Fard S, et al. An overview of microRNAs: biology, functions, therapeutics, and analysis methods. J Cell Physiol. 2019;234(5):5451–5465.
- Bhatnagar B, Garzon R. Clinical applications of MicroRNAs in acute myeloid leukemia: A mini-review. Front Oncol. 2021;11:679022.
- Wang Q, Feng T, Xu J, et al. Mitochondrial inner membrane protein, Mic60/mitofilin in mammalian organ protection. J Cell Physiol. 2019;234(4):3383–3393.
- Butrym A, Rybka J, Baczyńska D, et al. Low expression of microRNA-204 (miR-204) is associated with poor clinical outcome of acute myeloid leukemia (AML) patients. J Exp Clin Cancer Res. 2015;34(1):68.
- de Leeuw DC, Verhagen HJ, Denkers F, et al. MicroRNA-551b is highly expressed in hematopoietic stem cells and a biomarker for relapse and poor prognosis in acute myeloid leukemia. Leukemia. 2016;30(3):742–746.
- Shang Q, Yang Z, Jia R, et al. The novel roles of circRNAs in human cancer. Mol Cancer. 2019;18(1):6.
- Kristensen LS, Hansen TB, Venø MT, et al. Circular RNAs in cancer: opportunities and challenges in the field. Oncogene. 2018;37(5):555–565.
- Li X, Yang L, Chen LL. The biogenesis, functions, and challenges of circular RNAs. Mol Cell. 2018;71(3):428–442.
- de Acha O P, Rossi M, Gorospe M. Circular RNAs in blood malignancies. Front Mol Biosci. 2020;7:109.
- Yi YY, Yi J, Zhu X, et al. Circular RNA of vimentin expression as a valuable predictor for acute myeloid leukemia development and prognosis. J Cell Physiol. 2019;234(4):3711–3719.
- Lin L, Wang Y, Bian S, et al. A circular RNA derived from PLXNB2 as a valuable predictor of the prognosis of patients with acute myeloid leukaemia. J Transl Med. 2021;19(1):123.
- Wang X, Jin P. CircSPI1 acts as an oncogene in acute myeloid leukemia through antagonizing SPI1 and interacting with microRNAs. Cell Death Dis. 2021;12(4):297.
- Ding Y, Dong Y, Lu H, et al. Circular RNA profile of acute myeloid leukaemia indicates circular RNA annexin A2 as a potential biomarker and therapeutic target for acute myeloid leukaemia. Am J Transl Res. 2020;12(5):1683–1699.
- Alanazi B, Munje CR, Rastogi N, et al. Integrated nuclear proteomics and transcriptomics identifies S100A4 as a therapeutic target in acute myeloid leukemia. Leukemia. 2020;34(2):427–440.
- Johnson DC, Taabazuing CY, Okondo MC, et al. DPP8/DPP9 inhibitor-induced pyroptosis for treatment of acute myeloid leukemia. Nat Med. 2018;24(8):1151–1156.
- Hall IF, Climent M, Quintavalle M, et al. Circ_Lrp6, a circular RNA enriched in vascular smooth muscle cells, acts as a sponge regulating miRNA-145 function. Circ Res. 2019;124(4):498–510.
- Dong S, Wang R, Wang H, et al. HOXD-AS1 promotes the epithelial to mesenchymal transition of ovarian cancer cells by regulating miR-186-5p and PIK3R3. J Exp Clin Cancer Res. 2019;38(1):110.
- Ouyang Y, Li Y, Huang Y, et al. CircRNA circPDSS1 promotes the gastric cancer progression by sponging miR-186-5p and modulating NEK2. J Cell Physiol. 2019;234(7):10458–10469.
- Zhang Z, Zhang W, Mao J, et al. miR-186-5p functions as a tumor suppressor in human osteosarcoma by targeting FOXK1. Cell Physiol Biochem. 2019;52(3):553–564.
- Bi Y, Mao Y, Su Z, et al. HOXB-AS1 accelerates the tumorigenesis of glioblastoma via modulation of HOBX2 and HOBX3 at transcriptional and posttranscriptional levels. J Cell Physiol. 2021;236(1):93–106.
- Zhang TJ, Wang YX, Yang DQ, et al. Down-Regulation of miR-186 correlates with poor survival in de novo acute myeloid leukemia. Clin Lab. 2016;62(1-2):113–120.
- Lin J, Ma JC, Yang J, et al. Arresting of miR-186 and releasing of H19 by DDX43 facilitate tumorigenesis and CML progression. Oncogene. 2018;37(18):2432–2443.
- Wen XM, Zhang TJ, Ma JC, et al. Establishment and molecular characterization of decitabine-resistant K562 cells. J Cell Mol Med. 2019;23(5):3317–3324.
- Roskoski R., The role of small molecule Kit protein-tyrosine kinase inhibitors in the treatment of neoplastic disorders. Pharmacol Res. 2018;133:35-52.
- Bougherara H, Subra F, Crépin R, et al. The aberrant localization of oncogenic kit tyrosine kinase receptor mutants is reversed on specific inhibitory treatment. Mol Cancer Res. 2009;7(9):1525–1533.
- Ashman LK, Griffith R. Therapeutic targeting of c-KIT in cancer. Expert Opin Investig Drugs. 2013;22(1):103–115.
- Hu Y, Dong X, Chu G, et al. miR-137 downregulates c-kit expression in acute myeloid leukemia. Leuk Res. 2017;57:72–77.
- Zhang X, Yang L, Xu G. Silencing of long noncoding RNA TUG1 inhibits viability and promotes apoptosis of acute myeloid leukemia cells by targeting microRNA-221-3p/KIT axis. Clin Hemorheol Microcirc. 2020;76(3):425-437.
- Gao XN, Lin J, Gao L, et al. MicroRNA-193b regulates c-Kit proto-oncogene and represses cell proliferation in acute myeloid leukemia. Leuk Res. 2011;35(9):1226–1232.
- Malani D, Yadav B, Kumar A, et al. KIT pathway upregulation predicts dasatinib efficacy in acute myeloid leukemia. Leukemia. 2020;34(10):2780–2784.
