ABSTRACT
Background
Acute myeloid leukemia (AML) is one of the most common malignant myeloid diseases in adults with a dismal prognosis. We aimed to explore the effects of circNFIX on the proliferation and apoptosis of AML cells.
Methods
The expressions of circNFIX, miR-876-3p and tripartite motif (TRIM) 31 in the bone marrow specimens of AML patients and AML cell lines were detected by qRT-PCR or western blot. Cell proliferation was evaluated by 3-(4,5-Dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) and 5-ethynyl-29-deoxyuridine (EdU) assays. Cell cycle and apoptosis were analyzed by flow cytometry. Western blot was used to detect protein expression. The relationship between miR-876-3p and circNFIX or TRIM31 was identified by dual-luciferase reporter assay or RNA pull-down assay.
Results
The expression level of circNFIX was significantly increased in the bone marrow samples of AML patients and AML cells when compared with normal controls. CircNFIX silencing inhibited AML cell proliferation and promoted apoptosis. Inhibition of miR-876-3p reversed the effect of circNFIX knockdown on AML cell progression. In addition, circNFIX indirectly regulated TRIM31 through miR-876-3p. Further, TRIM31 overexpression counteracted the effect of circNFIX silencing on AML cell proliferation and apoptosis.
Conclusion
CircNFIX knockdown could suppress the proliferation and induce the apoptosis of AML cells by targeting the miR-876-3p/TRIM31 axis.
Introduction
Acute myeloid leukemia (AML) is the most usual hematological malignancy in adults with deviant myeloid hematopoietic progenitor cell proliferation, differentiation and maturation [Citation1,Citation2]. Although much progress has been achieved in chemotherapy and allogeneic stem cell transplantation, the prognosis of most patients with AML is still poor with a 5-year survival rate of 24% due to tumor relapse and drug resistance [Citation3]. Thus, developing new therapies against AML is critical to improve the prognosis.
Circular RNAs (circRNAs) belong to non-coding RNAs (ncRNAs) and are generated by back-splicing without 5′∼3′ polarity and poly adenine tails, and they act key modulatory roles in cancer progression [Citation4]. CircRNAs can act as miRNA sponges or combine with RNA-associated proteins [Citation5,Citation6]. Cheng et al. confirmed that circNFIX promoted pituitary adenoma development by mediating the miR-34a-5p/CCNB1 cascade [Citation7]. A previous study has shown that circNFIX facilitates glioma development by modulating the miR-378e/RPN2 cascade [Citation8]. Moreover, circNFIX aggravated non-small cell lung cancer (NSCLC) progression [Citation9]. Nevertheless, there is no relevant study on the effect of circNFIX on AML.
MicroRNAs are a class of short ncRNA molecules that can modulate gene expression by combining with mRNAs [Citation10]. Previous documents have indicated that miRNAs can modulate cancer cell proliferation, differentiation and apoptosis by targeting oncogenes or tumor suppressors, thus playing a significant role in cancer diagnosis and treatment [Citation11]. MiR-876-3p was downregulated in GC tissues and served as a tumor suppressor in GC [Citation12], NSCLC [Citation13], and lung cancer [Citation14]. Moreover, Mao et al. discovered that miR-876-3p was decreased in acute lymphoblastic leukemia and inhibited the growth of acute lymphocytic leukemia (ALL) cells [Citation15]. But the impact and mechanism of miR-876-3p in AML remain unclear.
Tripartite motif (TRIM) 31 belongs to the TRIM family and plays a carcinogenic role in tumor progression [Citation16–19]. Elevation of TRIM31 accelerated glioma cell proliferation, invasion and migration [Citation20]. TRIM31 was significantly upregulated in HCC tissues and could enhance HCC cell malignancy [Citation16]. Xiao et al. confirmed that TRIM31 expression was elevated in AML cells, and TRIM31 overexpression promoted AML progression [Citation21].
In this study, we evaluated the function of circNFIX in AML pathogenesis. In addition, we assembled the circNFIX/miR-876-3p/TRIM31 pathway to determine the molecular mechanism of circNFIX in this process with the hope of providing therapeutic targets for AML.
Materials and methods
Clinical assay
Bone marrow samples were acquired from 47 AML cases and 40 normal patients at Tongji Hospital, Tongji Medical College, Huazhong University of Science and Technology. This study was approved by the Tongji Hospital, Tongji Medical College, Huazhong University of Science and Technology Ethics Committee. All participants provided the informed consent.
Cell culture and transfection
HS-5, HL-60 and MOLM-13 cells were acquired from the Cell Bank of Chinese Academy of Sciences (Shanghai, China). All cells were maintained in RPMI-1640 medium (Gibco, Carlsbad, CA, USA) containing 10% fetal bovine serum (FBS; Gibco), 100 U/mL penicillin (Gibco) and 100 mg/mL streptomycin (Gibco) at 37°C with 5% CO2.
Lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA) was used to introduce RNAs and plasmids into AML cells. Short hairpin RNA (shRNA) of circNFIX (sh-circNFIX), shRNA negative control (sh-NC), miR-876-3p mimics (miR-876-3p), miR-NC, miR-876-3p inhibitor (in-miR-876-3p), in-miR-NC, TRIM31 overexpression plasmid (TRIM31), and pcDNA were acquired from GenePharma (Shanghai, China). Cells were collected for further detection at the defined time points.
Quantitative real-time polymerase chain reaction (qRT-PCR)
Trizol (Invitrogen) was used to prepare RNA. Complementary DNA (cDNA) was synthesized using the RevertAid First Strand cDNA kit (Thermo Fisher Scientific, Foster City, CA, USA) and miScript reverse transcription kit (Qiagen, Hilden, Germany). Then, amplification reaction was implemented with SYBR Mix (Applied Biosystems, Foster City, CA, USA). Primer sequences are shown in . Relative gene level was evaluated by the formula of 2−ΔΔCt with glyceraldehyde-3-phosphate dehydrogenase (GAPDH) or U6 as a housekeeping gene.
Table 1. Primers used for qRT-PCR.
3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium Bromide (MTT) assay
After transfection of 0, 24, 48 or 72 h, cells were collected and inoculated in 96-well plates (1 × 105 cells per well). Then, the cells were incubated with MTT reagent (Sigma, St. Louis, MO, USA). After 4 h, the insoluble products were incubated with DMSO (Sigma). The absorbance at 490 nm was examined using a microplate reader (Nikon, Tokyo, Japan).
5-ethynyl-29-deoxyuridine (EdU) assay
Transfected HL-60 and MOLM-13 cells were selected after 48 h of transfection and inoculated in 96-well plates. AML cells were incubated with 50 μM EdU reagent (Solarbio, Beijing, China) for 2 h. DNA was labeled with 2-(4-Amidinophenyl)-6-indolecarbamidine dihydrochloride (DAPI) reagent (Sigma). Cell images were observed on a fluorescence microscope (Olympus, Tokyo, Japan).
Flow cytometry
An Annexin V-fluorescein isothiocyanate (FITC)/propidium iodide (PI) kit (BD Biosciences, Franklin Lakes, NJ, USA) was used to analyze cell apoptosis. After 48 h of transfection, AML cells were harvested and incubated with Annexin V-FITC and PI in darkness for 15 min. After centrifugation, the cells were re-suspended by PBS and analyzed using flow cytometry.
To analyze cell cycle progression, cells were cultured in 24-well plates. After 48 of transfection, the cells were collected and fixed with 70% iced ethanol at 4°C overnight. The cells were resuspended in 400 μL binding buffer and then incubated with 50 μL PI (Sigma) for 30 min under dark conditions. Cell cycle was detected by flow cytometry.
Western blot
Radio Immunoprecipitation Assay (RIPA) lysis buffer (EpiZyme, Shanghai, China) was utilized to prepare proteins. Protein quantification was carried out with BCA kit (Beyotime, Shanghai, China). Then, the proteins were separated by 12% SDS-PAGE and transferred to PVDF membranes (Millipore, Billerica, MA, USA). The membranes were blocked with TBST containing 0.05% Tween 20 and 2.5% skim milk for 1 h and then labeled with primary antibodies at 4°C overnight. The primary antibodies include anti-CyclinD1 (ab16663; 1:10000; Abcam, Cambridge, MA, USA), anti-Bcl2 (ab32124; 1:8000; Abcam), anti-cleaved-caspase 3 (C-casp3; ab32042; 1:8000; Abcam), anti-TRIM31 (ab67785; 1:5000; Abcam), and anti-β-actin (ab8226; 1:20000; Abcam). The membrane was incubated with the secondary antibody (ab288151; 1:5000; Abcam) for 1 h. Enhanced chemiluminescence (ECL) reagents (Pierce Biotechnology, Rockford, IL, USA) were used to analyze the protein signals.
Dual-luciferase reporter assay
The wild-type (WT) and mutant (MUT) reporter plasmids, including circNFIX-WT/MUT and TRIM31 3′ untranslated region (3′UTR)-WT/MUT, were purchased from Promega (Madison, WI, USA). HL-60 and MOLM-13 cells were introduced with miR-876-3p mimics, miR-NC and the luciferase reporter plasmids. After 48 h, luciferase intensity was determined by Dual-Luciferase Reporter Assay System (Promega).
RNA pull-down assay
All processes were conducted under RNase-free condition. The wild-type biotin-coupled miR-876-3p probe (Bio-miR-876-3p-WT) was mixed with C-1 magnetic beads (Life Technologies, Carlsbad, CA, USA) for 4 h to obtain probe-coated beads. Bio-miR-NC and Bio-miR-876-3p-MUT were used as the controls. Subsequently, cell extracts were mixed with the probe-coated beads at 4°C overnight. QRT-PCR was implemented to analyze circNFIX enrichment.
Statistical analysis
All results were processed by GraphPad Prism 7.0 software (GraphPad, La Jolla, CA, USA) and the data were expressed as mean ± standard deviation. The differences were evaluated by Student’s t-test or one-way analysis of variance (ANOVA). P < 0.05 was deemed as statistically significant.
Results
CircNFIX was up-regulated in AML specimens and cells
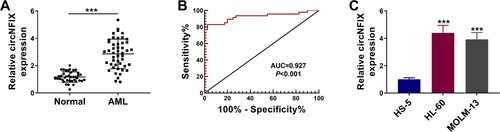
As shown in A, circNFIX was overexpressed in AML samples when compared with normal samples. To evaluate the potential diagnostic value of circNFIX, the ROC curve of serum circNFIX levels in AML patients was generated. We found that the area under ROC curve (AUC) was 0.927, the sensitivity was 83%, and the specificity was 97.5% (B). Next, the level of circNFIX in AML cell lines was monitored. As shown in C, circNFIX expression was up-regulated in HL-60 and MOLM-13 cells when compared with HS-5 cells. These results suggested that circNFIX might be involved in AML progression.
Figure 1. The expression of circNFIX was induced in AML tissues and cells. (A) The expression of circNFIX in the bone marrow samples of AML patients (N = 47) and healthy controls (N = 40) was tested by qRT-PCR. (B) ROC curve analysis of serum circNFIX level in distinguishing AML patients from normal controls. (C) The expression of circNFIX in HS-5, HL-60 and MOLM-13 cells was evaluated by qRT-PCR. ***P < 0.001.
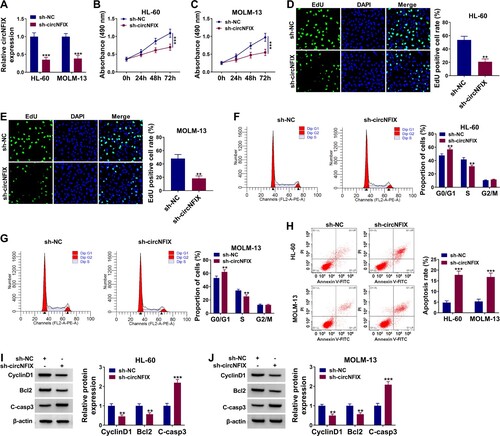
Repression of circNFIX impeded AML cell growth
The results showed that sh-circNFIX markedly reduced the abundance of circNFIX, which suggested the high transfection efficiency of sh-circNFIX (A). MTT assay revealed that circNFIX knockdown suppressed the proliferation of AML cells (B,C) but did not affect HS-5 cell proliferation (Figure S1). EdU assay showed that circNFIX silencing reduced the percentage of EdU positive cells when compared with the sh-NC group (D,E), further suggesting that circNFIX knockdown restrained AML cell proliferation. CircNFIX silencing resulted in cell cycle arrest at G0/G1 phase (F,G). In addition, circNFIX knockdown increased the apoptosis rate of HL-60 and MOLM-13 cells (H). Accordingly, in HL-60 and MOLM-13 cells transfected with sh-circNFIX, the levels of CyclinD1 and Bcl2 were decreased, while the expression of apoptosis-related protein C-casp3 was increased (I,J). These results indicated that circNFIX silencing suppressed the proliferation and induced the apoptosis of AML cells.
Figure 2. The effects of circNFIX on the proliferation and apoptosis of AML cells. (A–J) AML cells were divided into two groups: sh-NC group and sh-circNFIX group. (A) The level of circNFIX was estimated by qRT-PCR. (B-E) MTT and EdU assays were used for cell proliferation. (F–H) The cell cycle distribution and apoptosis rate were assessed by flow cytometry. (I and J) The expression levels of CyclinD1, Bcl2 and C-casp3 were monitored by western blot assay. **P < 0.01, ***P < 0.001.
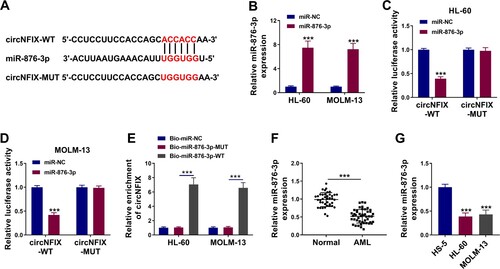
CircNFIX targeted miR-876-3p in AML cells
Circinteractome online database (https://circinteractome.irp.nia.nih.gov) was used to predict circNFIX-associated miRNAs. Five miRNAs were employed for the present work due to their inhibitory effects in cancer progression, including miR-296-5p, miR-324-5p, miR-502-5p, miR-598 and miR-876-3p. As shown in Figure S2A and B, circNFIX probe significantly enriched miR-876-3p and miR-598, especially miR-876-3p. Thus, miR-876-3p was chosen as a potential target miRNA of circNFIX. CircNFIX had binding sites for miR-876-3p (A). The content of miR-876-3p was significantly enhanced in HL-60 and MOLM-13 cells after miR-876-3p transfection (B). When compared with the miR-NC group, the luciferase activity of circNFIX-WT in HL-60 and MOLM-13 cells transfected with miR-876-3p was significantly reduced, while the luciferase activity of circNFIX-MUT was not changed (C,D). RNA pull-down assay revealed that circNFIX could be pulled down by wild-type biotin-coupled miR-876-3p probe (Bio-miR-876-3p-WT) but not by mutant probe (Bio-miR-876-3p-MUT) or Bio-miR-NC (E), further suggesting the interaction between circNFIX and miR-876-3p. In addition, experimental results confirmed that miR-876-3p level was reduced in AML tissues and cell lines when compared with normal tissues and HS-5 cell line (F,G). Taken together, miR-876-3p was a target of circNFIX in AML cells.
Figure 3. CircNFIX acted as a sponge for miR-876-3p in AML cells. (A) The targeted sequence of circNFIX with miR-876-3p was predicted by Circinteractome database. (B) The abundance of miR-876-3p in HL-60 and MOLM-13 cells transfected with miR-876-3p or miR-NC was evaluated by qRT-PCR. (C and D) The luciferase activity of HL-60 and MOLM-13 cells co-transfected with circNFIX-WT or circNFIX-MUT and miR-876-3p or miR-NC was measured by dual-luciferase reporter assay. (E) RNA pull-down assay was used to verify the relationship between circNFIX and miR-876-3p. (F and G) The expression of miR-876-3p in the bone marrow samples of AML patients (N = 47) and healthy controls (N = 40), HS-5 cells, HL-60 cells and MOLM-13 cells was examined by qRT-PCR. **P < 0.01, ***P < 0.001.
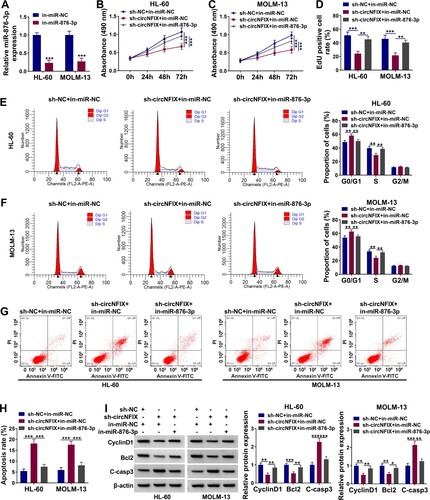
Silencing of miR-876-3p attenuated the effect of circNFIX deletion on the progression of AML cells
MiR-876-3p inhibitor down-regulated the level of miR-876-3p in HL-60 and MOLM-13 cells (A). The results of MTT and EdU assays together showed that circNFIX knockdown-induced suppressive effect on the proliferation of AML cells was reversed by silencing miR-876-3p (B–D). In addition, miR-876-3p knockdown alleviated circNFIX silencing-mediated cell cycle arrest in HL-60 and MOLM-13 cells (E,F). In-miR-876-3p could rescue the enhanced apoptosis of HL-60 and MOLM-13 cells induced by circNFIX silencing (G,H). Meanwhile, the effects of sh-circNFIX on CyclinD1, Bcl2 and C-casp3 expression were overturned after the addition of in-miR-876-3p (I). These data demonstrated that circNFIX knockdown suppressed AML progression by up-regulating miR-876-3p in vitro.
Figure 4. MiR-876-3p knockdown reversed the effect of circNFIX silencing on AML progression. (A) The expression of miR-876-3p in HL-60 and MOLM-13 cells transfected with in-miR-NC or in-miR-876-3p was assessed by qRT-PCR. (B-I) AML cells were divided into three groups: sh-NC + in-miR-NC group, sh-circNFIX + in-miR-NC group, and sh-circNFIX + in-miR-876-3p group. (B–D) The cell proliferation of HL-60 and MOLM-13 cells after transfection was explored by MTT and EdU assays. (E–H) Flow cytometry was utilized to monitor cell cycle and cell apoptosis of HL-60 and MOLM-13 cells. (I) The expression levels of CyclinD1, Bcl2 and C-casp3 in HL-60 and MOLM-13 cells were detected by western blot assay. *P < 0.05, **P < 0.01, ***P < 0.001.
TRIM31 was a target of miR-876-3p
TargetScan (https://www.targetscan.org) database predicted mRNA with miR-876-3p-binding sites. As a result of prediction, TRIM31, RHOBTB2, INPP4A, CDK9, RCC2 and MSI2 were employed because of their abnormal expression in AML. Subsequent qRT-PCR analysis showed that miR-876-3p introduction dramatically downregulated TRIM31 and MSI2 expression, especially TRIM31 expression (Figure S2C and D). Therefore, TRIM31 was chosen as a target gene of miR-876-3p. The specific binding sites of miR-876-3p for TRIM31 were shown in A. After transfection with miR-876-3p, luciferase activity of TRIM31 3′UTR-WT was decreased in both HL-60 and MOLM-13 cells (B,C). However, the luciferase activity of mutant reporter plasmid (TRIM31 3′UTR-MUT) was not affected after the transfection of miR-876-3p or miR-NC (B,C). The mRNA and protein levels of TRIM31 were up-regulated in AML samples when compared with normal samples (D,E). Meanwhile, we found that the mRNA and protein levels of TRIM31 were elevated in AML cell lines in comparison with HS-5 cell line (F,G). MiR-876-3p overexpression down-regulated TRIM31 expression, while miR-876-3p inhibitor up-regulated TRIM31 expression (H–K). These results indicated that miR-876-3p interacted with TRIM31. Further, sh-circNFIX significantly reduced TRIM31 expression level in HL-60 and MOLM-13 cells, while in-miR-876-3p reversed this effect (A,B). Taken together, circNFIX could sponge miR-876-3p to up-regulate TRIM31 expression in AML cells.
Figure 5. TRIM31 was targeted by miR-876-3p. (A) The targeting sequence of TRIM31 and miR-876-3p was predicted by TargetScan database. (B and C) The luciferase activity of TRIM31 3′UTR-WT and TRIM31 3′UTR-MUT in HL-60 and MOLM-13 cells transfected with miR-876-3p or miR-NC was examined by dual-luciferase reporter assay. (D and E) TRIM31 expression in the bone marrow samples of AML patients and healthy controls was evaluated by qRT-PCR and western blot assays. (F and G) TRIM31 expression in HS-5, HL-60 and MOLM-13 cells was detected by qRT-PCR and western blot assays. (H–K) QRT-PCR and western blot assays were used to detect the mRNA and protein levels of TRIM31 in HL-60 and MOLM-13 cells transfected with miR-876-3p, in-miR-876-3p or matched controls (miR-NC or in-miR-NC). ***P < 0.001.
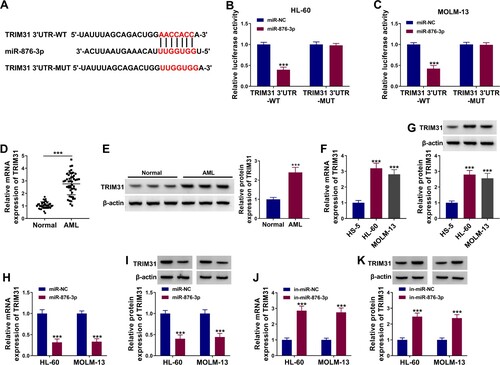
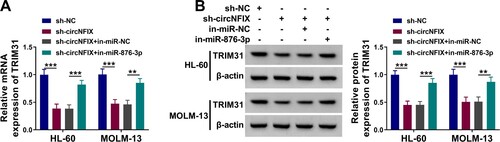
Figure 6. CircNFIX regulated TRIM31 expression by miR-876-3p. (A and B) The expression of TRIM31 in HL-60 and MOLM-13 cells transfected with sh-circNFIX, sh-circNFIX + in-miR-876-3p or corresponding controls (sh-NC or sh-circNFIX + in-miR-NC) was detected by qRT-PCR and western blot assays. **P < 0.01, ***P < 0.001.
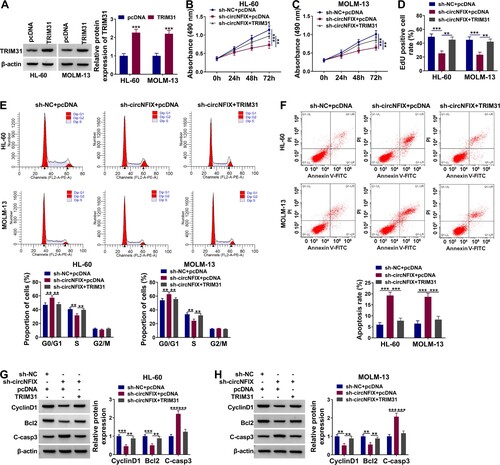
Ectopic TRIM31 expression rescued the effect of circNFIX silencing on AML cell progression
TRIM31 expression was dramatically increased in HL-60 and MOLM-13 cells after transfection with TRIM31 (A). MTT and EdU assays displayed that the proliferation of HL-60 and MOLM-13 cells was reduced after transfection with sh-circNFIX, while this effect was reversed in the sh-circNFIX + TRIM31 group (B–D). Sh-circNFIX induced cell cycle arrest in G0/G1 phase, but overexpression of TRIM31 eliminated this effect in HL-60 and MOLM-13 cells (E). In addition, TRIM31 addition abolished the increase in HL-60 and MOLM-13 cell apoptosis induced by sh-circNFIX transfection (F). Meanwhile, the effects of sh-circNFIX on CyclinD1, Bcl2, and C-casp3 expression were significantly reversed by TRIM31 overexpression (G,H). Overall, these data suggested that circNFIX knockdown restrained AML progression largely by down-regulating TRIM31 in vitro.
Figure 7. TRIM31 overexpression weakened the inhibitory effects of sh-circNFIX on AML cell proliferation and apoptosis. (A) The protein expression of TRIM31 in HL-60 and MOLM-13 cells transfected with pcDNA or TRIM31 was evaluated by western blot assay. (B–H) AML cells were divided into three groups: sh-NC + pcDNA group, sh-circNFIX + pcDNA group, and sh-circNFIX + TRIM31 group. (B–D) Cell proliferation was analyzed by MTT and EdU assays. (E and F) Cell cycle and cell apoptosis rate of HL-60 and MOLM-13 cells were explored by flow cytometry. (G and H) The protein expression levels of CyclinD1, Bcl2 and C-casp3 in HL-60 and MOLM-13 cells were examined by Western blot assay. **P < 0.01, ***P < 0.001.
Discussion
For the past few years, molecular targeted therapy, immunotherapy and hematopoietic stem cell transplantation for AML have made great progress, but many patients still die from disease progression every year due to recurrence and drug resistance [Citation22]. Therefore, it is particularly urgent to find novel molecular targets and biomarkers for AML patients.
CircRNA is a special endogenous ncRNA that can administrate gene expression [Citation23]. It is highly conserved in evolution and has better stability when compared with linear mRNA [Citation24]. CircRNA is involved in multiple stages of life activities and can be used not only as a biomarker for diagnosing human diseases, but also as a proxy for disease activity or severity [Citation25,Citation26]. CircRNA can regulate many biological processes by working as a molecular sponge of miRNA [Citation27,Citation28]. For example, circ_0000526 repressed the development of BC by sponging miR-492 [Citation29]. Hsa_circ_0000515 could absorb miR-326 to expedite cervical cancer progression by increasing ELK1 abundance [Citation30]. We observed a significant increase in circNFIX in AML. Further analysis exhibited that circNFIX knockdown hampered AML cell proliferation and facilitated cell apoptosis. Moreover, circNFIX silencing decreased CyclinD1 and Bcl2 protein expression and increased cleaved caspase 3 production. The above evidence suggested that circNFIX might be an oncogene in AML progression.
We also analyzed the possible mechanism by which circNFIX silencing inhibited AML cell growth. MiRNA is an endogenous small ncRNA that is widely involved in the regulation of growth and apoptosis of organisms and is associated with the pathogenesis of a variety of disorders. Accumulating evidence has suggested that circRNAs can regulate cell biological behaviors by sponging miRNAs [Citation31,Citation32]. In this study, bioinformatics screening showed that circNFIX targeted miR-876-3p. Dual-luciferase and RNA pull-down tests identified the interaction between circNFIX and miR-876-3p. In addition, miR-876-3p expression was downregulated in AML tissues and AML cells. MiR-876-3p silencing attenuated circNFIX knockdown-mediated effects in AML cells, which suggested that circNFIX knockdown suppressed AML cell malignant potential largely by enhancing miR-876-3p.
TRIM31 is the E3 ubiquitin protein ligase [Citation33]. Research data have demonstrated that TRIM31 acts an important part in the occurrence and progression of various cancers [Citation19,Citation33–36]. For example, deletion of TRIM31 promoted breast cancer progression by regulating K48- and K63-linked ubiquitination of p53 [Citation34]. TRIM31 was declined in NSCLC and worked as a potential tumor suppressor [Citation35]. TRIM31 regulated chronic inflammation and expedited colorectal cancer cell invasion and metastasis through the NF-κB pathway [Citation36]. Our data showed that miR-876-3p targeted TRIM31. MiR-876-3p negatively regulated TRIM31 expression. TRIM31 abundance was prominently increased in AML tissues and cells. The data of rescue experiments revealed that circNFIX knockdown blocked AML progression largely by down-regulating TRIM31 in vitro. Moreover, circNFIX could indirectly regulate TRIM31 expression through miR-876-3p. Thus, circNFIX regulated AML cell malignancy by inducing TRIM31 expression through miR-876-3p.
Our study was the first one to discover that circNFIX was augmented in the bone marrow samples of AML patients and AML cells. CircNFIX promoted AML progression through the miR-876-3p/TRIM31 axis. CircNFIX may be an effective target for future AML therapy.
Supplemental Material
Download Zip (308.4 KB)Disclosure statement
No potential conflict of interest was reported by the author(s).
References
- Cheng Z, Dai Y, Huang W, et al. Prognostic value of MicroRNA-20b in acute myeloid leukemia. Front Oncol. 2020;10:553344.
- Döhner H, Weisdorf DJ, Bloomfield CD. Acute myeloid leukemia. N Engl J Med. 2015;373:1136–1152.
- Shallis RM, Wang R, Davidoff A, et al. Epidemiology of acute myeloid leukemia: recent progress and enduring challenges. Blood Rev. 2019;36:70–87.
- Yu J, Xu QG, Wang ZG, et al. Circular RNA cSMARCA5 inhibits growth and metastasis in hepatocellular carcinoma. J Hepatol. 2018;68:1214–1227.
- Han B, Chao J, Yao H. Circular RNA and its mechanisms in disease: from the bench to the clinic. Pharmacol Ther. 2018;187:31–44.
- Zhang M, Zhao K, Xu X, et al. A peptide encoded by circular form of LINC-PINT suppresses oncogenic transcriptional elongation in glioblastoma. Nat Commun. 2018;9:4475.
- Cheng J, Nie D, Li B, et al. CircNFIX promotes progression of pituitary adenoma via CCNB1 by sponging miR-34a -5p. Mol Cell Endocrinol. 2021;525:111140.
- Ding C, Wu Z, You H, et al. CircNFIX promotes progression of glioma through regulating miR-378e/RPN2 axis. J Exp Clin Cancer Res. 2019;38:506.
- Lu J, Zhu Y, Qin Y, et al. CircNFIX acts as a miR-212-3p sponge to enhance the malignant progression of non-small cell lung cancer by up-regulating ADAM10. Cancer Manag Res. 2020;12:9577–9587.
- Guo T, Zhang J, Yao W, et al. CircINHA resists granulosa cell apoptosis by upregulating CTGF as a ceRNA of miR-10a-5p in pig ovarian follicles. Biochim Biophys Acta Gene Regul Mech. 2019;1862:194420.
- Marcucci G, Mrozek K, Radmacher MD, et al. The prognostic and functional role of microRNAs in acute myeloid leukemia. Blood. 2011;117:1121–1129.
- Peng C, Huang K, Liu G, et al. MiR-876-3p regulates cisplatin resistance and stem cell-like properties of gastric cancer cells by targeting TMED3. J Gastroenterol Hepatol. 2019;34:1711–1719.
- Li Q, Wang Y, Hu R, et al. Dysregulation of SPRR3/miR-876-3p axis contributes to tumorigenesis in non-small-cell lung cancer. Onco Targets Ther. 2020;13:2411–2419.
- Zhang Z. Silencing LINC00504 inhibits cell proliferation, invasion as well as migration and promotes cell apoptosis in lung cancer cells via upregulating miR-876-3p. Cytotechnology. 2020;72(6):807–817.
- Mao J, Gao W, Xue L, et al. The lncRNA SLCO4A1-AS1/miR-876-3p/RBBP6 axis regulates cell proliferation and apoptosis in acute lymphocytic leukemia via the JNK signaling pathway. Int J Lab Hematol. 2021;43:1050–1061.
- Guo P, Ma X, Zhao W, et al. TRIM31 is upregulated in hepatocellular carcinoma and promotes disease progression by inducing ubiquitination of TSC1-TSC2 complex. Oncogene. 2018;37:478–488.
- Li H, Zhang Y, Hai J, et al. Knockdown of TRIM31 suppresses proliferation and invasion of gallbladder cancer cells by down-regulating MMP2/9 through the PI3K/Akt signaling pathway. Biomedic Pharmacother Biomed Pharmacotherapie. 2018;103:1272–1278.
- Xiao Y, Deng T, Ming X, et al. TRIM31 promotes acute myeloid leukemia progression and sensitivity to daunorubicin through the Wnt/β-catenin signaling. Biosci Rep. 2020;40(4):BSR20194334.
- Yu C, Chen S, Guo Y, et al. Oncogenic TRIM31 confers gemcitabine resistance in pancreatic cancer via activating the NF-κB signaling pathway. Theranostics. 2018;8:3224–3236.
- Shi G, Lv C, Yang Z, et al. TRIM31 promotes proliferation, invasion and migration of glioma cells through Akt signaling pathway. Neoplasma. 2019;66:727–735.
- Xiao Y, Deng T, Ming X, et al. TRIM31 promotes acute myeloid leukemia progression and sensitivity to daunorubicin through the Wnt/beta-catenin signaling. Biosci Rep. 2020;40(4):BSR20194334.
- Tallman MS, Wang ES, Altman JK, et al. Acute myeloid leukemia, version 3.2019, NCCN clinical practice guidelines in oncology. J Natl Compr Canc Netw. 2019;17:721–749.
- Harper KL, McDonnell E, Whitehouse A. CircRNAs: from anonymity to novel regulators of gene expression in cancer (review). Int J Oncol. 2019;55:1183–1193.
- Rybak-Wolf A, Stottmeister C, Glažar P, et al. Circular RNAs in the mammalian brain are highly abundant, conserved, and dynamically expressed. Mol Cell. 2015;58:870–885.
- Zhang HD, Jiang LH, Sun DW, et al. CircRNA: a novel type of biomarker for cancer. Breast Cancer. 2018;25:1–7.
- Zhang Z, Yang T, Xiao J. Circular RNAs: promising biomarkers for human diseases. EBioMedicine. 2018;34:267–274.
- Zhou Z, Sun B, Huang S, et al. Roles of circular RNAs in immune regulation and autoimmune diseases. Cell Death Dis. 2019;10:503.
- Barrett SP, Salzman J. Circular RNAs: analysis, expression and potential functions. Development. 2016;143:1838–1847.
- Wang WB, Ren P, Ren FH, et al. Circ_0000526 blocks the progression of breast cancer by sponging miR-492. Cancer Biother Radiopharm. 2021;36:467–476.
- Tang Q, Chen Z, Zhao L, et al. Circular RNA hsa_circ_0000515 acts as a miR-326 sponge to promote cervical cancer progression through up-regulation of ELK1. Aging (Albany NY). 2019;11:9982–9999.
- Hansen TB, Jensen TI, Clausen BH, et al. Natural RNA circles function as efficient microRNA sponges. Nature. 2013;495:384–388.
- Panda AC. Circular RNAs Act as miRNA sponges. Adv Exp Med Biol. 2018;1087:67–79.
- Zhang H, Deng Y, Liang L, et al. Knockdown of TRIM31 enhances colorectal cancer radiosensitivity by inducing DNA damage and activating apoptosis. Onco Targets Ther. 2019;12:8179–8188.
- Guo Y, Li Q, Zhao G, et al. Loss of TRIM31 promotes breast cancer progression through regulating K48- and K63-linked ubiquitination of p53. Cell Death Dis. 2021;12:945.
- Li H, Zhang Y, Zhang Y, et al. TRIM31 is downregulated in non-small cell lung cancer and serves as a potential tumor suppressor. Tumour Biol J Int Soc Oncodevelop Biol Medic. 2014;35:5747–5752.
- Wang H, Yao L, Gong Y, et al. TRIM31 regulates chronic inflammation via NF-kappaB signal pathway to promote invasion and metastasis in colorectal cancer. Am J Transl Res. 2018;10:1247–1259.
