ABSTRACT
Objectives
Since the publication of the third edition, the WHO classification of tumors of hematopoietic and lymphoid disorders has introduced the disease entity of ‘myeloid/lymphoid neoplasms with eosinophilia and PDGFRB rearrangement’, in which the most common chromosomal abnormality is t(5;12) (q32;p13.2), and this abnormality generates the ETV6::PDGFRB fusion gene. However, there have been patients with hematologic features and chromosomal abnormalities that are extremely similar to those carrying ETV6::PDGFRB fusion. These rare disorders harbor ETV6::ACSL6 fusion, and only sporadic cases have been reported at present.
Methods
We report a patient with chronic eosinophilic leukemia (CEL) carrying chromosome translocation t(5;12)(q32;p13.2), and we present the clinical features. In addition, we conducted a literature review to collect all reported cases and summarized the genetic and clinical profiling as well as the treatments and outcomes.
Result
In addition to our patient, a total of 19 cases have been previously reported, including 6 variants of ETV6::ACSL6 and 3 reciprocals. We identified a novel variant of the ETV6::ACSL6 transcript in our patient, and the breakpoint was flanked by exon 2 of ETV6 and exon 2 of ACSL6. The cellular morphology features consisted of myeloproliferative neoplasm (MPN); myelodysplastic/myeloproliferative neoplasm (MDS/MPN), specifically CEL; and acute myelocytic leukemia (AML). The treatments and outcomes varied greatly depending on the type of disease, although tyrosine kinase inhibitors (TKIs) were not effective.
Conclusion
In contrast to neoplasms with ETV6::PDGFRB fusion, myeloid neoplasms with ETV6::ACSL6 fusion have unique characteristics.
KEYWORDS:
Introduction
In the WHO classification of tumors of hematopoietic and lymphoid tissues, there is a distinct type of myeloid neoplasm with a t(5;12)(q32;p13.2) karyotype, and the translocation forms an ETV6::PDGFRB fusion gene, which characterizes the disease entity of myeloid/lymphoid neoplasms with PDGFRB rearrangement. The hematological features of this entity are most often those of MDS/MPN or MPN, typically with eosinophilia, including chronic myelomonocytic leukemia (CMML, usually with eosinophilia), atypical chronic myeloid leukemia (aCML), BCR::ABL1-negative (usually with eosinophilia), and CEL, and some cases are classified into myeloproliferative neoplasms with eosinophilia (MPN-eos). TKIs exert therapeutic effects on MPNs with PDGFRB rearrangement [Citation1]. However, another entity harboring the same t(5;12)(q32;p13.2) karyotype has been sporadically reported, in which the produced fusion gene is ETV6::ACSL6 rather than ETV6::PDGFRB. The patients carrying ETV6::ACSL6 share similar hematological features with those carrying ETV6::PDGFRB, although the former show no response to TKIs. Several variants of ETV6::ACSL6 have been found previously. Here, we report a male patient with chronic eosinophilic leukemia who experienced disease progression with poor curative effects. Moreover, a novel variant of ETV6::ACSL6 was identified. In addition, we reviewed the literature, collected a case series harboring the ETV6::ACSL6 fusion, and summarized the landscape of the disease, including genetic and clinical profiling, treatments and outcomes.
Patients and methods
Case report
The patient was admitted for the first time due to fatigue lasting for 2 months. All four limbs were scattered with papules and nodules, of which the largest diameter was approximately 3-4 cm, with a clear boundary, dark red color, hardness, and no tenderness. The liver was 3 cm below the costal margin, and the spleen was 5 cm below the costal margin. Routine blood examination showed a white blood cell (WBC) count of 133.63×109/L, hemoglobin level of 80 g/L, and platelet count of 44×109/L. A blood smear revealed 46% eosinophils and 20% basophils. Bone marrow smears showed active bone marrow hyperplasia, with myeloblasts accounting for 5%. Markedly increased numbers of eosinophils and eosinophil precursors were observed at all stages, accounting for 40.5% of bone marrow, and basophils accounted for 5.5% of bone marrow. Bone marrow biopsy showed extremely active bone marrow hyperplasia, with mainly myelocytes and cells at a more mature stage and a significantly increased eosinophil proportion (A). The increase in reticular fibers was moderate. Chromosome analysis revealed 46,XY,t(5;12)(q33;p13)[7]/46,XY[13] (B). Flow cytometry revealed a group of abnormal cells, accounting for 31.4% of nuclear cells, and these cells were positive for CD45, CD33 (77.6%), CD11b (98.6%), CD13 (97.7%), CD123 (91.4%), and CD11c (94.7%) but negative for MPO, cyCD79a, cyCD3, CD7, CD19, CD34, CD15, CD117, CD14, HLA-DR, CD4, CD22, and CD64. The detection results using a screening panel for 40 fusion genes in leukemia were negative, including PML::RARA and BCR::ABL. A diagnosis of chronic eosinophilic leukemia was confirmed. He received initial treatment with hydroxyurea and leukapheresis, and cytarabine (0.2 g days 1-3) was given for the first course of chemotherapy thereafter. The patient had repeated fever during myelosuppression, which improved after antibiotic treatment. The patient then underwent five courses of chemotherapy as follows: TA (theprubicin and cytarabine), IA (doxorubicin and cytarabine), TA, IA and TA. Blood cells were reduced after each course of chemotherapy, whereas leukocytosis reappeared when myelosuppression occurred (Supplemental Figure 1). Therefore, the 7th course of treatment was altered to the COPEA regimen (cyclophosphamide, vindesine, prednisone, etoposide, and cytarabine). During this period of myelosuppression, the patient developed perianal abscess and fever with a maximum body temperature of 40.5 °C, which was accompanied by loss of spirit, blurred consciousness, frequent urination, urination pain, and dysuria. Atrial fibrillation and ineffective platelet transfusion occurred consecutively. The body temperature returned to normal after antibiotic treatment, and amiodarone restored sinus rhythm. Nevertheless, the platelet count remained low. The patient was discharged automatically and died a short time later.
Figure 1. Laboratory examinations and molecular biological characteristics of the present case. (A) Marked eosinophilia in the patient's bone marrow smear. (B) G-banded karyotyping detected t(5;12)(q33;p13) abnormalities. (C) Agarose gel electrophoresis of PCR products. (D) Sanger sequencing of the PCR amplicon. Sequences flanking the breakpoint are shown. (E) Schematic of ETV6 exon 2 fusing with ACSL6 exon 2 in the present case. Red arrows represent the breakpoint site, and the horizontal dotted line indicates that the display of partial sequences is omitted. CDS, coding sequence; UTR, untranslated region.
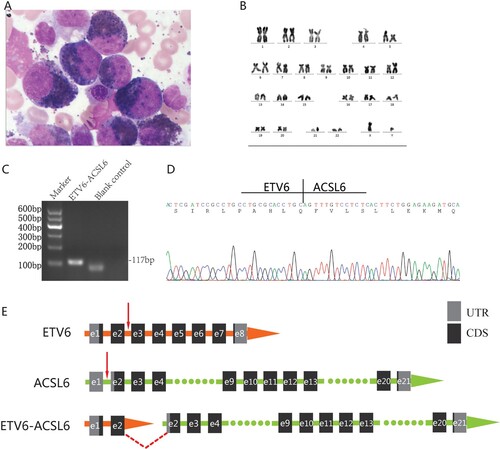
We employed a series of molecular biological methods to clarify the diagnosis. None of the ETV6::PDGFRB or FIP1L1::PDGFRA fusion genes were detected by reverse transcription-polymerase chain reaction (RT‒PCR). The result of fluorescence in situ hybridization (FISH) using the dual-color break-apart 5q32/PDGFRB-specific probe (Abbott Molecular. Inc.) was negative. To detect the underlying molecular aberrations, peripheral blood mononuclear cells were isolated, and total mRNA was extracted. Transcriptome sequencing was performed on the Illumina HiSeq 2000 instrument. A total of 19 gene fusion events were explored by SOAPfuse software [Citation2], among which the ETV6::ACSL6 fusion genes had the most span reads and junction reads, which corresponded to the main chromosomal abnormalities (Supplemental Table 1). Primers flanking the fusion gene breakpoint were designed based on the reads targeted by SOAPfuse software. RT‒PCR was then performed for confirmation (C) using the following primers: forward primer, 5'-TCGACGCCACTTCATGTTCC-3’; and reverse primer, 5'-CTCCTGTGTCTGCATCTTCTCCA-3’. The PCR product length was 117 bp. Subsequent Sanger sequencing of the PCR product (D) and BLAST comparison showed that the amplicon sequence was consistent with that of transcriptome sequencing. The breakpoint of the ETV6::ACSL6 fusion gene was located between exon 2 of ETV6 and exon 2 of ACSL6, and it was an in-frame fusion ((D, E)).
Case selection
We set the search deadline as January 2022. The search of English databases included MEDLINE and Embase, and the search of Chinese databases included the China National Knowledge Infrastructure Literature (CNKI), China Science and Technology Journal Database, and Wanfang Data Knowledge Service Platform. The search terms were ‘ETV6 and ACSL6’ and ‘Tel and ACS2’.
Results
Literature review
A total of 12 articles were retrieved that were distributed between 1999 and 2022, which reported a total of 19 patients. The detailed characteristics of these cases are summarized in [Citation3–14].
Table 1. Profiles of patients with ETV6::ACSL6 fusion.
Clinical presentation
The hematologic features of most of these cases presented as MPN or MDS/MPN, and four presented as CEL. Two patients were PV, and one of them transformed to AML (Case 6) [Citation6]. One case presented as MDS/MPN but was not detailed [Citation8]. Four cases presented as unspecified MPN, including two with eso-MPN and a baso-MPN. Two cases were diagnosed as MDS (Cases 1 and 7), but they may not be classified as MDS according to the current WHO classification criteria due to their elevated peripheral leukocytes. Of these two cases, the blast cells in Case 1 had reached AML standards (23% in peripheral blood and 30.8% in bone marrow), while the cytology features of Case 7 were not described in detail, so it was difficult to assess[Citation3, Citation7]. There were three cases of secondary AML, including the aforementioned Case 6. In addition, three cases were relapsed AML, and Case 3 was AEL [Citation3]. It is worth noting that according to the current WHO classification standard, Case 3 should be classified as CEL. Thus, there were five CEL cases (20%) and seven AML cases (35%).
Neoplasms harboring ETV6::ACSL6 were more common in males (n = 14) than in females (n = 6) (male-to-female ratio: 2.33:1), which was slightly more prominent than the ratio for myeloid/lymphoid neoplasms with PDGFRB rearrangement (2:1). This neoplasm occurred predominantly in middle-aged adults, and the median age of onset was 49. The age range (26—71 years) was slightly narrower than that of neoplasms with PDGFRB rearrangement (8—72 years), and at least 13 patients (65%) were between 35 and 65 years old.
Among the nine patients with documented blood counts, the WBC count was increased in all patients, with a median count of 28.2×109/L (range 11.2-133.63×109/L), which was more pronounced in cases of CEL and de novo AML (range 21.6-133.63× 109/L). Elevated peripheral eosinophil counts were a prominent feature documented in 14 of 16 patients (87.5%), with a median level of 42% (range 2.3-84%). Increased basophil counts were observed in five patients, but they were not recorded in a significant number of cases.
Cytogenetics and ETV6::ACSl6 variant summary
The vast majority of cases showed characteristic t(5;12)(q31;p13) chromosomal abnormalities or t(5;12)(q23-31;p13) (n = 2) and t(5;12)(q31-33;p13) (n = 1). Two cases harboring t(5;12) have been reported, but the authors supplied no detailed information about the translocation breakpoint. One case had a complex karyotype involving 5q and 12p [Citation9]. To date, a total of six ETV6::ACSL6 variants and three reciprocal variants of ACSL6::ETV6 have been reported, which are summarized in . The coexistence of two reciprocal chimeric genes was observed (n = 2). There were also cases in which two ETV6::ACSL6 variants coexisted (n = 2). The most commonly reported variant (n = 6) is ETV6::ACSL6, the breakpoint of which is flanked by exon 1 of ETV6 and exon 2 of ACSL6. Intron retention and truncated exons (or alternative splicing) may be found in some variants.
Figure 2. Schematic of all the variants of ETV6::ACSL6 and reciprocal fusion transcripts thus far. In total, six variants of ETV6::ACSL6 and three variants of ACSL6::ETV6 are exhibited. ACSL6 was previously known as ACS2. Yagasaki et al. cloned a transcript of ACS2, which was regarded as a template to analyze the sequence structure of the fusion transcripts that they identified [Citation3]. We utilized the Basic Local Alignment Search Tool (BLAST) program and demonstrated that the fusion transcripts could not be entirely aligned to any ACSL6 transcript. The upstream sequence of the 3’-UTR (upstream of 1957bp) of the fusion transcript was identical to ACSL6 transcript variant 4 (NM_001205248.2), except for the lack of both terminal exons in the latter. Hence, we selected ACSL6 transcript variant 4 as a template in this schematic. Red arrows represent the breakpoint site, and the horizontal dotted line indicates that the display of partial sequences is omitted. ‘Δ’ refers to a truncated exon. UTR, untranslated region; CDS, coding sequence.
![Figure 2. Schematic of all the variants of ETV6::ACSL6 and reciprocal fusion transcripts thus far. In total, six variants of ETV6::ACSL6 and three variants of ACSL6::ETV6 are exhibited. ACSL6 was previously known as ACS2. Yagasaki et al. cloned a transcript of ACS2, which was regarded as a template to analyze the sequence structure of the fusion transcripts that they identified [Citation3]. We utilized the Basic Local Alignment Search Tool (BLAST) program and demonstrated that the fusion transcripts could not be entirely aligned to any ACSL6 transcript. The upstream sequence of the 3’-UTR (upstream of 1957bp) of the fusion transcript was identical to ACSL6 transcript variant 4 (NM_001205248.2), except for the lack of both terminal exons in the latter. Hence, we selected ACSL6 transcript variant 4 as a template in this schematic. Red arrows represent the breakpoint site, and the horizontal dotted line indicates that the display of partial sequences is omitted. ‘Δ’ refers to a truncated exon. UTR, untranslated region; CDS, coding sequence.](/cms/asset/e008a0d1-99fd-49e6-afdb-19c126bad0a3/yhem_a_2117206_f0002_oc.jpg)
Therapy and outcome
Some patients were treated with mild drugs, e.g. hydroxyurea and interferon. In addition to patients with PV, one patient with CEL receiving hydroxyurea therapy showed an improvement in clinical symptoms, eosinophilia, and anemia [Citation12]. Seven patients received chemotherapy, and only two showed a significant response (complete or near complete response). Four patients received allogeneic stem cell transplantation; two lived >8 months or 15 months. Some patients were administered TKIs, including imatinib (n = 4) or sorafenib (n = 2), neither of which showed efficacy [Citation9, Citation11, Citation12]. Including the present case, there were eight patients who survived less than 1 year. ETV6::ACSL6 fusion genes were newly generated in four patients during the process of disease progression and transformation, and their survival time was less than 1 year after that point [Citation3, Citation6, Citation14]. Together, these findings indicate a poor prognosis for this disease entity. Only one patient with PV survived more than 42 months [Citation6].
Discussion
The ETS variant 6 gene (ETV6), mapping to chromosome 12p13.2, belongs to the E-twenty-six (ETS) family of transcription factors. As a transcriptional repressor, ETV6 binds to the 5'-CCGGAAGT-3’ DNA sequence via its C-terminal DNA-binding domain and exerts functions in association with a plethora of corepressors, i.e. SIN3A, NCOR, and HDAC3. ETV6 is ubiquitously expressed in a broad spectrum of tissues, including bone marrow. ETV6 is essential for maintaining hematopoietic stem cell function and megakaryocyte development [Citation15, Citation16]. A variety of ETV6 germline or somatic aberrations have been reported in hematologic malignancies, such as mutations, deletions, rearrangements, and fusions, demonstrating its role in leukemogenesis [Citation17, Citation18]. Acyl-CoA synthetase long chain family member 6 (ACSL6) belongs to the long-chain acyl-CoA synthetase (ACSL) family. ACSL6 catalyzes the conversion of long-chain fatty acids to their active form, acyl-CoA, together with CoA and ATP. ACSL6 is essential for fatty acid metabolism and affects mitochondrial content, respiratory rates, and lipid oxidation. ACSL6 is highly expressed in the brain, testis, and bone marrow, and it has been found to be related to cell proliferation and apoptosis [Citation19–22].
A number of variants of ETV6::ACSL6 and reciprocals have been identified since 1999, comprising six ETV6::ACSL6 and three ACSL6::ETV6 transcripts thus far, in frame or out of frame. The ETV6 portion adjacent to the breakpoint varies from exons 1–3, and the corresponding residual of ACSL6 spans from exon 1 to the 3’-UTR. Here, we identified a novel variant in which exon 2 of ETV6 was fused to exon 2 of ACSL6 in frame. The main sequence of ACSL6 remained, which led to the retention of functional domains, while ETV6 retained only a short part of the 5’ terminus. For ACSL6, the mechanism of lipid biosynthetic processes in tumorigenesis remains unclear.
According to a literature review, compared with neoplasms with PDGFRB rearrangement (male-to-female ratio: 2:1) [Citation1], neoplasms with ETV6::ACSL6 had a higher male-to-female ratio (2.33:1). The median onset age of neoplasms with ETV6::ACSL6 was 49 years old, slightly older than neoplasms with PDGFRB rearrangement (late 40s) [Citation1]. All patients carrying ETV6::ACSL6 presented with myeloid tumors, whereas either myeloid or lymphoid neoplasms may occur in cases harboring PDGFRB rearrangement. The hematological features of neoplasms with ETV6::ACSL6 are predominantly those of chronic eosinophilic leukemia, while chronic myelomonocytic leukemia (usually with eosinophilia) is most often observed in neoplasms with PDGFRB rearrangement. Fewer patients carrying ETV6::ACSL6 present with other chronic myeloid disorders, such as PV, aCML, MPN (some with eosinophilia), and unspecified MDS/MPN. Acute myeloid leukemia occurs in a considerable proportion of these cases (35%), of which more than half are secondary or relapsed AML. All patients had elevated peripheral leukocyte levels, and the level in our case was the highest. Eosinophilia was found in most patients but was reported in a minority of patients with PDGFRB rearrangement [Citation23]. Similarly, basophilia is also common in these diseases, which is contrary to neoplasms with PDGFRB rearrangement [Citation1]. Bone marrow trephine biopsy of neoplasms with PDGFRB rearrangement shows an increase in mast cells, but this phenomenon was not mentioned in the literature concerning ETV6::ACSL6 fusion. We speculate that most cases did not undergo bone marrow biopsy, which prevented this determination.
The median survival of patients with neoplasms with PDGFRB rearrangement was less than 2 years before the introduction of imatinib therapy [Citation1]. In the era of imatinib, the prognosis of patients with neoplasms with PDGFRB rearrangement significantly improved. In a report of 22 cases, the 5-year OS rate was 83% [Citation23]. Overall, the prognosis of patients with the ETV6::ACSL6 fusion gene was poor. A total of 12 deaths occurred during the follow-up period, of which eight patients generated ETV6::ACSL6 fusion at the onset of the disease, and the longest survival was 11 months. The remaining four patients developed ETV6::ACSL6 fusion during disease progression and transformation (AML), and the longest survival time was 1 month if measuring from the fusion development. Only one case of PV survived longer than 42 months. Due to the short follow-up period reported, we were unable to determine whether the remaining patients attained long-term survival. Combined chemotherapy reduced tumor load or even achieved CR in some patients, but the maintenance time was short.
One patient presenting with AML received TKI, and the remaining patients received chemotherapy. One MDS patient received Ara-C, STI571, and allo-SCT, and the survival was over 8 months. The therapeutic strategies for patients with chronic myeloid neoplasms are numerous, including hydroxyurea, interferon, glucocorticoid, TKI, chemotherapy, and allo-SCT. All cases receiving TKI did not respond to treatment, which may have been attributed to the absence of tyrosine kinase composition activity of the ETV6::ACSL6 chimeric protein [Citation24]. Most of the cases with CEL-NOS experienced a progressive clinical process. Hydroxyurea improved symptoms and blood count in only one patient with a moderate disease process [Citation12], while the other patients showed a poor response to hydroxyurea, interferon, and glucocorticoids. Our patient also exhibited a deteriorating disease course, and he had the highest leukocyte count and received the most courses of combined chemotherapy (7 cycles). The blood counts decreased temporarily but did not reach CR and then increased again after the myelosuppression stage. The patient did not receive TKI or allo-SCT. Case 17 also exhibited invasive CEL-NOS and survived disease-free during the 15-month follow-up period after allo-SCT, which was the longest survival period among invasive CEL-NOS cases [Citation13]. Two of the other three patients who received allo-SCT died 2 months after transplantation, and one patient with MDS lived for 8 months during the follow-up period.
Conclusion
In summary, we identified a novel ETV6::ACSL6 variant in a case of CEL. Neoplasms with ETV6::ACSL6 fusion are less common and have distinctive chromosomal abnormalities, and they present with hematological features of myeloid tumors, including chronic myeloid disorder, AML, and MDS, of which CEL is the most common. These characteristics make it difficult to distinguish them from neoplasms with PDGFRB rearrangement unless fusion genes are identified utilizing molecular biology techniques. However, the vast majority of cases of this disease entity present an aggressive course and poor prognosis with no response to TKI. Even intensive treatment is often ineffective. In the forthcoming fifth edition of the WHO classification, ‘myeloid/lymphoid neoplasms with eosinophilia and rearrangement of PDGFRA, PDGFRB, or FGFR1, or with PCM1-JAK2’ has been renamed ‘myeloid/lymphoid neoplasms with eosinophilia and tyrosine kinase gene fusion’, and several more tyrosine kinase fusions have been added [Citation25, Citation26]. ETV6::ACSL6 is distinct from this category due to a lack of tyrosine kinase activity. Currently, according to the WHO classification, neoplasms with ETV6::ACSL6 fusion would be dispersed among several categories, such as CEL-NOS. However, the disorders have unique characteristics, and isolating them to form a specific entity may be worthy of consideration.
Author contributions
ZS, XL, and JLZ conceived the project and designed the study. WYH contributed to participant data collection and data analysis. JY and XCY performed cell morphology diagnosis. FH and YQW helped with sample collection. ZS and XL were responsible for the molecular genetic tests and wrote the manuscript.
Ethics approval and consent to participate
Informed consent was obtained from all participating individuals. All procedures followed were in accordance with the ethical standards of the responsible committee on human experimentation (institutional and national) and with the Helsinki Declaration of 1975, as revised in 2008 (5).
Acknowledgments
The authors would like to thank the patient who participated in this study and to express gratitude to Pro. Xinxin Cao and Dr. Xia Wu for reviewing this manuscript.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Additional information
Funding
References
- Bain BJ, Horny H-P, Arber DA, et al. Myeloid/lymphoid neoplasms with eosinophilia and rearrangements of PDGFRA, PDGFRB or FGFR1, or with PCM1-JAK2. In: Swerdlow SH, Campo E, Harris NL, Jaffe ES, Pileri SA, Stein H, Thiele J, editor. WHO classification of tumours of haematopoietic and lymphoid tissues., revised 4th edition. Lyon: International Agency for Research on Cancer (IARC); 2017. p. 72–79.
- Jia W, Qiu K, He M, et al. SOAPfuse: an algorithm for identifying fusion transcripts from paired-end RNA-Seq data. Genome Biol. 2013;14:R12), doi:10.1186/gb-2013-14-2-r12.
- Yagasaki F, Jinnai I, Yoshida S, et al. Fusion ofTEL/ETV6 to a novelACS2 in myelodysplastic syndrome and acute myelogenous leukemia with t(5;12)(q31;p13). Genes, Chromosomes and Cancer. 1999;26:192–202.
- Cools J, Mentens N, Odero MD, et al. Evidence for position effects as a variantETV6-mediated leukemogenic mechanism in myeloid leukemias with a t(4;12)(q11-q12;p13) or t(5;12)(q31;p13). Blood. 2002;99:1776–1784. doi:10.1182/blood.V99.5.1776.
- Wlodarska I, La Starza R, Baens M, et al. Fluorescence In situ hybridization characterization of New translocations involving TEL (ETV6) in a wide spectrum of hematologic malignancies. Blood. 1998;91:1399–1406. doi:10.1182/blood.V91.4.1399.
- Murati A, Adélaïde J, Gelsi-Boyer V, et al. T(5;12)(q23-31;p13) with ETV6-ACSL6 gene fusion in polycythemia vera. Leukemia. 2006;20:1175–1178. doi:10.1038/sj.leu.2404194.
- Katsura Y, Suzukawa K, Nanmoku T, et al. Myelodysplastic syndrome accompanied by basophilia and eosinophilia with t(5;12)(q31;p13). Cancer Genet Cytogenet 2007;178:85–88. doi:10.1016/j.cancergencyto.2007.05.020.
- Erben P, Gosenca D, Muller MC, et al. Screening for diverse PDGFRA or PDGFRB fusion genes is facilitated by generic quantitative reverse transcriptase polymerase chain reaction analysis. Haematologica. 2010;95:738–744. doi:10.3324/haematol.2009.016345.
- Gosenca D, Erben P, Haferlach C, et al. Clinical and molecular heterogeneity of eosinophilia-associated myeloproliferative neoplasms with a translocation t(5;12). Blood. 2009;114:4983), doi:10.1182/blood.V114.22.4983.4983.
- Haferlach C, Bacher U, Schnittger S, et al. ETV6 rearrangements are recurrent in myeloid malignancies and are frequently associated with other genetic events. Genes, Chromosomes and Cancer. 2012;51:328–337. doi:10.1002/gcc.21918.
- De Luca-Johnson J, Ninfea JIR, Pearson L, et al. Myeloid neoplasms with t(5;12) and ETV6-ACSL6 gene fusion, potential mimickers of myeloid neoplasm with PDGFRB rearrangement: case report with imatinib therapy and review of the literature. Case Rep Med. 2016;2016:1), doi:10.1155/2016/8324791.
- Su RJ, Jonas BA, Welborn J, et al. Chronic eosinophilic leukemia, NOS with t(5;12)(q31;p13)/ETV6-ACSL6 gene fusion: A novel variant of myeloid proliferative neoplasm with eosinophilia, NOS with t(5;12)(q31;p13)/ETV6-ACSL6 gene fusion: a novel variant of myeloid proliferative neoplasm with eosinophilia. Hum Pathol (N Y). 2016;5:6–9. doi:10.1016/j.ehpc.2015.10.001.
- Wu X, Cai H, Qiu Y, et al. ETV6-ACSL6 fusion gene in myeloid neoplasms: clinical spectrum, current practice, and outcomes. Orphanet J Rare Dis. 2020;15:192), doi:10.1186/s13023-020-01478-6.
- Baldazzi C, Luatti S, Marzocchi G, et al. T(5;12)(q31;p13)/ETV6::ACSL6 and t(6;9)(p23;q34)/DEK::NUP214 concurrence in acute myeloid leukemia: an unusual association of two rare abnormalities. Cancer Genet. 2022;35–39:35–39. doi:10.1016/j.cancergen.2021.12.006.
- Di Paola J, Porter CC. ETV6-related thrombocytopenia and leukemia predisposition. Blood. 2019;134:663–667. doi:10.1182/blood.2019852418.
- Hock H, Shimamura A. ETV6 in hematopoiesis and leukemia predisposition. Semin Hematol 2017;54:98–104. doi:10.1053/j.seminhematol.2017.04.005.
- Melazzini F, Palombo F, Balduini A, et al. Clinical and pathogenic features of ETV6-related thrombocytopenia with predisposition to acute lymphoblastic leukemia. Haematologica. 2016;101:1333–1342. doi:10.3324/haematol.2016.147496.
- Feurstein S, Godley LA. Germline ETV6 mutations and predisposition to hematological malignancies. Int J Hematol 2017;106:189–195. doi:10.1007/s12185-017-2259-4.
- Nakahara K, Ohkuni A, Kitamura T, et al. The sjögren-larsson syndrome gene encodes a hexadecenal dehydrogenase of the sphingosine 1-phosphate degradation pathway. Mol Cell. 2012;46:461–471. doi:10.1016/j.molcel.2012.04.033.
- Ohkuni A, Ohno Y, Kihara A. Identification of acyl-CoA synthetases involved in the mammalian sphingosine 1-phosphate metabolic pathway. Biochem Biophys Res Commun 2013;442:195–201. doi:10.1016/j.bbrc.2013.11.036.
- Teodoro BG, Sampaio IH, Bomfim LHM, et al. Long-chain acyl-CoA synthetase 6 regulates lipid synthesis and mitochondrial oxidative capacity in human and rat skeletal muscle. J. Physiol. (Lond.). 2017;595:677–693. doi:10.1113/JP272962.
- Shishikura K, Kuroha S, Matsueda S, et al. Acyl-CoA synthetase 6 regulates long-chain polyunsaturated fatty acid composition of membrane phospholipids in spermatids and supports normal spermatogenic processes in mice. FASEB J. 2019;33:14194–14203. doi:10.1096/fj.201901074R.
- Jawhar M, Naumann N, Schwaab J, et al. Imatinib in myeloid/lymphoid neoplasms with eosinophilia and rearrangement of PDGFRB in chronic or blast phase. Ann Hematol 2017;96:1463–1470. doi:10.1007/s00277-017-3067-x.
- De Braekeleer E, Douet-Guilbert N, Morel F, et al. ETV6 fusion genes in hematological malignancies: a review. Leuk Res 2012;36:945–961. doi:10.1016/j.leukres.2012.04.010.
- Khoury JD, Solary E, Abla O, et al. The 5th edition of the world health organization classification of haematolymphoid tumours: myeloid and histiocytic/dendritic neoplasms. Leukemia. 2022;36:1703–1719. doi:10.1038/s41375-022-01613-1.
- Arber DA, Orazi A, Hasserjian RP, et al. International consensus classification of myeloid neoplasms and acute leukemia: integrating morphological, clinical, and genomic data. Blood. 2022. doi:10.1182/blood.2022015850
