ABSTRACT
Introduction
Risk stratification is essential for treatment decision in myelodysplastic neoplasms (MDS). Molecular international prognostic scoring system (M-IPSS) has been recently developed combining somatic mutations and clinical information being used in the revised international prognostic scoring system (R-IPSS).
Objective
We aimed to explore the performances of M-IPSS and R-IPSS in Thai patients with MDS.
Method
MDS patients were stratified into risk categories using R-IPSS and M-IPSS scores. The performance of both models were evaluated for prognostic prediction.
Results
One hundred and sixty-two MDS patients were recruited from the multicenter study. Survival analysis revealed that both R-IPSS and M-IPSS were good prediction models with the Concordance Index (C-index) of 0.71 (95% Confidence interval [CI] 0.64–0.78) and 0.75 (95% CI 0.69–0.80), respectively (p = 0.22). Comparing the two, 13 of 162 (8%) cases were re-staged between lower and higher risks which would have affected treatment decisions.
Conclusion
Our study showed that R-IPSS score can be used for risk stratification in most Thai patients. A prediction model using somatic mutations specifically in Asian patients should be formulated in the future.
KEYWORDS:
Introduction
Myelodysplastic neoplasms (MDS) are heterogeneous diseases with variable clinical courses ranging from indolent to rapid progression to acute myeloid leukemia within months. A revised international prognostic scoring system (R-IPSS) based on clinical factors has been applied for disease prognostication for 10 years [Citation1]. Subsequently, somatic gene mutations have been extensively studied showing high impact on clinical outcomes. Incorporating genetic information into R-IPSS, termed molecular international prognostic scoring system (M-IPSS), resulted in better prognostic determination [Citation2,Citation3]. However, most of the studied population comprised mainly Western ethnicity. Therefore, we aimed to study the performance of M-IPSS in predicting survival and compared to that of R-IPSS in Asian patients with MDS.
Materials and methods
Patients and study design
Adult MDS patients were enrolled from January 2017 to June 2022. The four participating medical centers included King Chulalongkorn Memorial Hospital in Bangkok, Ramathibodi Hospital in Bangkok, Maharaj Nakorn Chiang Mai in Chiang Mai Province, and Srinakarin Hospital in Khon Kaen Province. MDS was diagnosed by bone marrow aspiration, biopsy and cytogenetic studies according to the WHO 2022 classification after reviewing by both hematologists and hematopathologists at each participating center [Citation4].
All MDS patients were stratified according to the R-IPSS and M-IPSS. Therapy-related and secondary myeloid neoplasms were excluded. We collected clinical information including baseline characteristics, MDS subtypes, gene mutations, cytogenetic studies, and survival outcomes. This study was approved by the Institutional Review Board, Faculty of Medicine, Chulalongkorn University, Bangkok, Thailand.
Targeted next-generation sequencing
For targeted sequencing, DNA from bone marrow or peripheral blood was extracted using Gentra Puregene Blood Kits (Qiagen). Seventy-eight samples (48%) were processed and sequenced with an IONS5 sequencer using a customized 40-gene myeloid panel (Supplementary Table 1). Extracted DNA was prepared using the Ion ChefTM system. Variants were analyzed using Ion ReporterTM software as previously described [Citation5]. Eighty four samples (52%) were processed and sequenced by QIAGEN GeneReader using a customized 141-gene myeloid panel (Supplementary Table 2). Variants were analyzed using Ingenuity software as previously described [Citation6]. Selected variants were validated by conventional Sanger sequencing.
Statistical analysis
Categorical parameters were reported as frequencies and percentages. Survival rate was calculated by Kaplan-Meier and log rank test was used to compare between groups. The Hazard ratio with 95% confidence interval (95% CI) was calculated by Cox regression. Comparing the predictive powers for R-IPSS and M-IPSS of mortality rate using Harrell’s C. P-values lower than 0.05 were considered statistically significant. All statistical analyses were performed using STATA version 15.1.
Results
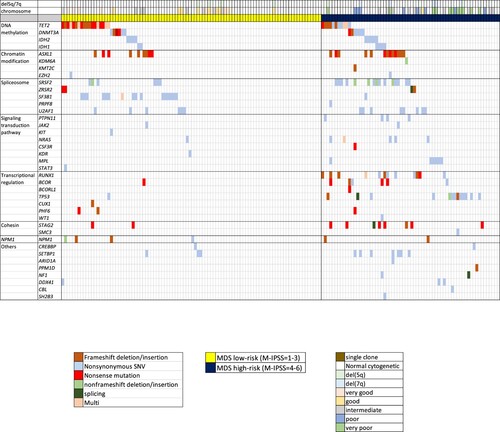
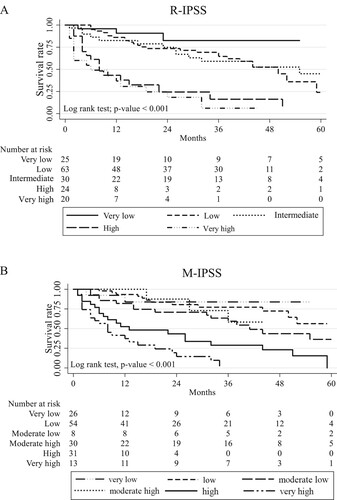
A total of 162 MDS patients were included in this study. Subtypes of MDS according to the WHO classification 2022 [Citation4] and somatic mutations are shown in and Supplementary Table 3 and 4. Of note, both FLT3-ITD and TKD mutations were assessed but not found in this cohort. MDS with biallelic TP53 inactivation was detectable in 8 of 14 cases with TP53 mutations (57%). Of 17 cases with SF3B1 mutations, 6 cases (35%) were detectable alone whereas the rest were associated with other genetic abnormalities ( and Supplementary Table 4). The frequency in each stratum and hazard ratios of R-IPSS and M-IPSS were displayed in . Survival analysis performances using Concordance Index (C-index) in R-IPSS and M-IPSS were 0.71 (95% Confidence interval [CI] 0.64–0.78) and 0.75 (95% CI 0.69–0.80), respectively (p = 0.22). The median follow-up time of patients in this cohort was 20 months (range 1–108 months). Survival analysis according to the R-IPSS and M-IPSS was demonstrated in . Using R-IPSS, low-risk MDS was defined by score of 3.5 or lower, while patients with R-IPSS >3.5 were categorized as high-risk MDS [Citation7]. In M-IPSS, scores of 1–3 were classified as low-risk, while scores of 4–6 were high-risk MDS. When R-IPSS was re-classified into M-IPSS score, 13 of the 162 cases (8%) were changed between low-risk and high-risk MDS. Four cases were downstaged from high-risk (R-IPSS) to low-risk MDS (M-IPSS), while 9 cases were upstaged from low-risk (R-IPSS) to high-risk MDS (M-IPSS).
Table 1. The frequencies of patients, hazard ratios (HR) with 95% confidence interval (CI) and Concordance Index (C-index) as classified by R-IPSS score and M-IPSS score.
Discussion
Our data demonstrated that R-IPSS and M-IPSS were the good models for survival prediction in Thai patients. The performances of R-IPSS and M-IPSS scores as determined by C-index were not statistically different. The failure to significantly improve the performance by adding genetic information in M-IPSS may be partly due to the differences in somatic mutations in Thai MDS compared with the Western patients [Citation8], such as the lower frequency of SF3B1 mutations (10.5% vs 24% respectively). The distribution of our patients in each M-IPSS subgroup was similar to that of the Western cohort except for the lower number in moderate high subgroup (M-IPSS = 4). Survival analysis using R-IPSS in very low, low, and intermediate groups was comparable to M-IPSS scoring system in very low, low, moderate-low and moderate-high groups. From our cohort, survival analysis using R-IPSS in very high-risk (R-IPSS = 5) was not significantly different from the high-risk group (R-IPSS = 4) (p = 0.32). On the other hand, M-IPSS scoring system significantly discriminated very high-risk group (M-IPSS = 6) from the high-risk group (M-IPSS = 5) (p = 0.045).
In our study, 8% of the patients were re-stratified between low and high-risk MDS which were in concordance with the previous study [Citation3] that showed 7% had changed more than one risk stratum. This re-stratification might affect appropriate treatment decisions in those patients. A limitation of this study is the inability to determine the loss of heterozygosity in all cases with TP53 mutation because it is not a routine investigation in our country. In addition, certain genes were not included in our panel, such as MLL-PTD associated with poor prognosis [Citation9]. This may also impact the score calculation. Lower number of patients and shorter duration of follow-up might be other factors affecting non-significant survival rate, especially in the lower risk group.
In conclusion, R-IPSS could be used for risk stratification in a majority of MDS patients. M-IPSS showed better prognostic classification between high and very high-risk groups. Model development using genetic information in Asian patients is warranted.
Author contributions
C.P. collected samples and clinical data, analyzed data, wrote the manuscript, and conceptualized the overall research. P.N., T.R., T.L., provided samples and clinical data. S.K. extracted DNA and performed targeted NGS. P.R. supervised, conceptualized the research, and wrote the manuscript. All authors read and approved the final manuscript.
Supplemental Material
Download MS Excel (35.6 KB)Supplemental Material
Download MS Word (13.4 KB)Supplemental Material
Download MS Excel (16.7 KB)Supplemental Material
Download MS Excel (15.7 KB)Acknowledgements
Authors thank Miss Jiratchaya Sophonphan for the statistical analysis.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Additional information
Funding
References
- Greenberg PL, Tuechler H, Schanz J, et al. Revised international prognostic scoring system for myelodysplastic syndromes. Blood. 2012;120:2454–2465.
- Nazha A, Narkhede M, Radivoyevitch T, et al. Incorporation of molecular data into the revised international prognostic scoring system in treated patients with myelodysplastic syndromes. Leukemia. 2016;30:2214–2220.
- Bernard E TH, Greenberg PL, Hasserjian RP, et al. Molecular international prognostic scoring system for myelodysplastic syndromes. NEJM EVID. 2022;1:1–14.
- Khoury JD, Solary E, Abla O, et al. The 5th edition of the World Health Organization classification of haematolymphoid tumours: myeloid and histiocytic/dendritic neoplasms. Leukemia. 2022;36:1703–1719.
- Ferrone C, Wong H, Semenuk L, et al. Validation and clinical impact of the Oncomine myeloid targeted DNA and RNA ion semiconductor sequencing assay. Blood. 2018;132:5523.
- Atli EI, Gurkan H, Atli E, et al. The importance of targeted next-generation sequencing usage in cytogenetically Normal myeloid malignancies. Mediterr J Hematol Infect Dis. 2021;13:e2021013.
- Greenberg PL, Stone RM, Al-Kali A, et al. NCCN guidelines(R) insights: myelodysplastic syndromes, version 3. 2022. J Natl Compr Canc Netw. 2022;20:106–117.
- Papaemmanuil E, Gerstung M, Malcovati L, et al. Clinical and biological implications of driver mutations in myelodysplastic syndromes. Blood. 2013;122:3616–3627; quiz 99.
- Patel JP, Gonen M, Figueroa ME, et al. Prognostic relevance of integrated genetic profiling in acute myeloid leukemia. N Engl J Med. 2012;366:1079–1089.