Abstract
The coffee berry borer (CBB) is the most prevalent pest of coffee plantations. Within the Coffea genus, C. arabica is susceptible to CBB and C. liberica shows a lower susceptibility. Two EST libraries were constructed from the total RNA of C. arabica and C. liberica fruits artificially infested with CBBs for 24 h. Using 6000 clones sequenced per library, a unigene database was generated, obtaining 3634 singletons and 1454 contigs. For each contig, the proportion of sequences present in both species was determined and a differential gene expression between the species was detected. C. arabica displayed a higher relative expression of proteins involved in general stress responses, whereas C. liberica showed the induction of a higher number of insect defense proteins. In order to validate the results, quantifications through real-time PCR were done. A hevein-like protein, an isoprene synthase, a salicylic acid carboxyl methyltransferase and a patatin-like protein gene were highly upregulated in C. liberica at 24 and/or 48 h after insect infestation compared to C. arabica. The identification of metabolic pathways induced by this pest insect provides tools to take advantage of the genetic resources available for the control of CBB.
Introduction
Coffee is one of the most important world commodities. Colombia has the highest yield of Coffea arabica in the world, with a planted area of near 850,000 ha and an average production of 680,000 ton. Hypothenemus hampei (Ferrari) (Coleoptera: Curculionidae: Scolitidae), better known as the coffee berry borer (CBB), is the most prevalent pest of coffee plantations. In Colombia, most of the economic losses due to an insect are caused by the attack of the CBB. Regarding coffee plants, very little is known about the biochemical and molecular interactions between this insect and its only natural host, Coffea spp., especially concerning its relation to the coffee berry.
Coffee berries start a vulnerability period from 120 to 150 days after flowering, and as they reach ripeness, they become more attractive to the insect because of the release of volatiles (Mathieu et al. Citation1996, Citation1998; Ortiz et al. Citation2004; Toci and Farah Citation2008). The females penetrate the berry, feed and lay their eggs. The larvae hatch and feed on the seed endosperm, decreasing its quality and often causing the detachment of small fruits, weight loss of the berries and a decrease in production (Duque et al. Citation1997).
The life cycle and habitat of the insect complicate its control since most of its life cycle occurs inside the fruit; because of this, it is necessary to adopt an integrated pest control strategy that includes cultural, biological and chemical control (Bustillo Citation2005). In addition, the climate conditions in Colombia make possible for trees to have berries all the year around, therefore supplying a constant source of food for the development of insect populations.
A variety with reduced susceptibility or moderate resistance to the CBB could be part of integrated pest management practices. Varieties with high incomplete resistance or complete resistance to CBB would mean a great competitive advantage. These varieties can alleviate the balance of expenses of the coffee growers and the negative effects that chemical control practices may have in the environment and the families who inhabit within the farm premises. Therefore, as a strategy for improving the integrated CBB control, works have been initiated to add a genetic control to this practice. To reach this goal, it is necessary to identify contrasting genotypes: a genotype that is susceptible to CBB, with agronomic characteristics of interest such as quality and production, and a genotype that is resistant to the insect.
To date, genetic factors that confer resistance have not been detected within the Coffea genus. However, preliminary studies have shown differences in the susceptibility of some species to CBB (Villagran Citation1991). Romero and Cortina (Citation2004, Citation2007) reported a reduction of 30% in the insect's intrinsic growth rate when it fed on C. liberica fruits as compared to C. arabica var. Caturra fruits. These differences can be related to the presence of antimetabolic substances, the absence of appropriate nutrients or differences in the structure of the cell wall that directly affect the insect's digestion.
Based on these findings, differential expression studies began to characterize candidat e genes whose expression patterns correspond with the differences in the oviposition that the insects exhibited when they ate berries of C. liberica as opposed to C. arabica var. Caturra. Expression libraries of full-length cognate DNA (cDNA) copies from fruits infested with CBB for 24 h were constructed per each genotype (C. liberica and C. arabica var. Caturra) to determine transcriptional changes in the coffee berries that were induced by the attack of the insects and to compare the responses between these two genotypes under that condition. Based on the results using CBB berries 24, 48, and 72 h postinfestation, five candidate genes showing a significantly higher number of transcripts in the C. liberica library and previously reported in the literature as being involved in plant responses to insect attack were selected for expression validation by real-time PCR. Gene identification and the specific metabolic pathways of plant responses to insects in which they are involved complement and support coffee breeding efforts against the CBB.
Methodology
Biological material for the construction of cDNA libraries
Plants of the genotypes C. arabica var. Caturra and C. liberica (accession CCC1025) were selected from the Coffee Germplasm Bank at Naranjal, the Cenicafé Central Experimental Station at Chinchiná, Caldas, Colombia. Branches with healthy berries aged to approximately 150 days after flowering (DAF) were chosen from C. arabica and from 180 DAF for C. liberica because it takes longer time for this genotype to develop and reach the maturation stage (Medina et al. Citation1984; Eira et al. Citation2006).
An artificial infestation with CBBs in a proportion of three insects per berry was carried out. The infested branches were covered with entomological sleeves for 24 h. The berries penetrated by the CBB were collected and immediately stored in liquid nitrogen until their laboratory processing.
RNA isolation
Total RNA was extracted from each genotype using a kit for RNA isolation from plants (RNeasy® Plant Mini Kit; Qiagen, Chastworth, CA, USA). Buffers RLT and RLC, provided by the manufacturer, were used for C. arabica var. Caturra and C. liberica, respectively. The RNA integrity was assessed by denaturing agarose gels and quantified by fluorometry.
Real-time PCR
Total RNA (250–750 ng) was reverse transcribed to its cDNA using MMLV reverse transcriptase (Promega, Madison, WI, USA) for one hour at 42°C. The primers used were CDS III/3′ and SMART IV oligo; the former recognizes the poly(A) tail, and the latter covers the region 5′ of the transcript. The CDS III/3′ and 5′ primers were used to synthesize the second chain. The amplification conditions were the following: 95°C for 1 min, followed by cycles of 95°C for 15 s and 68°C for 6 min. For C. arabica, 22 cycles were performed; and for C. liberica, 20 cycles.
Construction of cDNA libraries
The construction of cDNA libraries was carried out following the manufacturer's instructions of the Creator SMART cDNA Library Construction Kit (Clontech; Palo Alto, CA, USA). After the synthesis of the double-stranded cDNA, it was digested with the enzyme Sfi I. The digestion product was fractioned according to size using Chroma Spin 400 columns (Becton Dickinson, Franklin Lakes, NJ, USA). The fractions that exhibited the largest sizes were selected and inserted in the transformation vector pDNR-Lib. Finally, Escherichia coli DH 10B cells were transformed with this construct by electroporation. The titer recombination percentage and the number of independent clones of the genomic library were determined.
cDNA sequencing
Sequencing of the cDNA libraries was performed in two phases by two different organizations. In the first phase, 3072 sequences per genotype (Group 1) were obtained from REXAGEN, Seattle, WA, USA. In the second phase, 2500 sequence reactions per genotype (Group 2) were carried out by MACROGEN, Korea. All of the sequencing reactions were initiated from the extreme 5′-end, using the universal M13F primer.
Sequence analysis
Sequence quality was assessed using the program Codon Code Aligner (version 3.5, 2009) and with a Phred quality value of 30 (Ewing et al. Citation1998). Vector and contaminant sequences were eliminated, as well as sequences smaller than 200 bp. The number of reads corresponding to full-length sequences or to partial sequences was determined through the program Target Identifier (Min et al. Citation2005). The sequences were assembled with CAP3 (Huang and Madem Citation1999) using the information of both genotypes to identify shared sequences. Unigenes that corresponded to grouped sequences (contigs) and to nongrouped ones (singletons) were generated. The proportion of sequences present from both genotypes was determined for each contig. Protein translation was conducted using the program ESTScan (version 2.0) (Iseli et al. Citation1999). The putative function of the unigenes was determined through comparisons among the protein databases available in InterProScan (version 4.0) (Zdobnov and Apweiler Citation2001). BLASTn and BLASTx comparisons were also made (Altschul et al. Citation1990).
Statistical analysis of the cDNA libraries
Once the representative categories were determined, a comparison of the relative abundance of sequences in the cDNA libraries was carried out between orthologs of both genotypes, including sequences with and without known function, using the R statistic proposed by Stekel et al. (Citation2000) and defined as R= − 2 log(L), where L is the ratio of the maximized likelihoods. For each gene a p value was calculated for the R statistic using the IDEG6 web tool (Romualdi et al. Citation2003).
Real-time PCR assays
In order to validate the expression of some of the genes identified in the cDNA libraries, new coffee artificial infestations with CBBs in a proportion of three insects per fruit were carried out in the same plant material used for the cDNA libraries. For this, three adult trees planted in the field with berries in the same developmental stages that were used for the libraries were selected. In each tree, two branches were selected and they were covered with entomological sleeves: in one, CBBs were added in a 3:1 insect–berry relation, and in the other branch no insects were added (control treatment). In the first tree, the berries from the infested and noninfested branches were collected after 24 h. In the other trees, the berries were collected 48 and 72 h postinfestation. After collection, berries were stored in liquid nitrogen. The setup of this experiment was done in October of 2009.
The frozen berries were ground in a mortar. The powder obtained from two berries (1 g) was used. Total RNA extraction was performed using the RNeasy® Plant Mini Kit (Qiagen, Valencia, CA, USA). Samples were treated with RNase-free DNase I (Promega, Madison, WI, USA). Finally, RNA integrity and purity were evaluated using agarose gel electrophoresis and a TBS-380 fluorometer (Turner Biosystems, Sunnyvale, CA, USA).
The first-strand cDNA synthesis was performed with oligo(dT) and a Sensiscript kit (Qiagen, Valencia, CA, USA) using 50 ng of the total RNA as a template in a 25 µL reaction mixture. The cDNA samples were then diluted (1:25), and 2 µL of the dilution was used in a SYBR Green PCR. Gene-specific primers were designed (Primer3plus; Untergasser et al. Citation2007) using a stringent set of criteria: predicted melting temperature of 61–62°C, primer length of 19–23 nucleotides, guanine–cytosine contents of 40%–60% and PCR amplicon lengths of 70–150 bp.
The genes chosen for real-time PCR and the primers used for the amplification are shown in .
Table 1. List of primers used in real-time PCR.
PCR was carried out in an optical 96-well plate with chromo 4 (MJ Research, Waltham, MA, USA) sequence detection. The following is the standard thermal profile for all PCRs: 95°C for 10 min, 40 cycles at 95°C for 15 s, 60°C for 30 s, 72°C for 30 s and 72°C for 10 min.
The fluorescence signal was captured at the end of each cycle, and melting curve analysis was performed from 65 to 95°C, with data captured every 0.2°C during a 1-s hold. Data were analyzed using the Opticon monitor analysis software version 2.02 (MJ Research, Waltham, MA, USA). Quantification of each transcript in each cDNA source consisted of at least three independent technical replicates. The housekeeping gene for cyclophylin (Romano et al. Citation2004) was selected for reference expression.
Transcript abundance was normalized to cyclophilin by subtracting the Ct value of cyclophilin from the Ct value of the transcript, that is Ct = Cttranscript−Ctcyclophilin. The efficiency was estimated for each gene using absolute fluorescence data captured during the exponential phase of amplification of each reaction using the equation (1 + E) = 10(−1/slope) (Ramarkers et al. Citation2003). Efficiency values were taken into account in all subsequent calculations. The transcript abundance in control and treated samples was obtained from the equation (1 + E)(−Ct).
Results
Libraries analysis
Approximately 6000 clones per genotype were sequenced (6048 and 5952 from C. arabica and C. liberica, respectively) in two sequencing groups. shows and summarizes the distribution of the clones in each sequencing round and each genotype. Using this data set, 10,603 sequences were assembled, representing 88±5% of the total. The discovery rate of genes is related to the degree of redundancy of the clones obtained per genomic library (Susko and Roger Citation2004), reaching, in this case, a value of 63% for both genotypes and an average length of 534 bp per sequence.
Table 2. General information regarding the C. arabica var. Caturra and C. liberica clones obtained from the sequencing of the full-length cDNA libraries.
After quality examination, the percentage of full-length sequences for both genotypes was 52% and 31%, which are partial-length sequences exhibiting an open reading frame ().
Table 3. Full-length sequence description obtained from the cDNA libraries of C. arabica var. Caturra and C. liberica.
The assembling with CAP3 was performed with all of the sequences from both genotypes to find redundancies among them and differences in the number of clones expressed per gene or protein (). From the 5088 unigenes obtained from this assemblage, 1754 corresponded to contigs and 3634 to singletons, with an average size of 654 bp per unigene. The redundancy obtained from the assembly was 4.8 ESTs/contig. Forty percent of the unigenes (contigs and singletons) are unique sequences of C. arabica var. Caturra, 41% corresponded to unique sequences of C. liberica and 19% were contigs with sequences shared between both genotypes.
Table 4. Good-quality sequence assemblage corresponding to both genotypes.
Sequence translation into proteins using the ESTScan program was achieved for 75% of the unigenes, meaning that open reading frame candidates for these sequences were found. Once the amino acid sequence for each unigene was obtained, putative functions were assigned by making comparisons among the databases available in InterProScan. The information was complemented through BLASTx and BLASTn comparisons with the nonredundant GenBank databases. shows the unigene distribution obtained through this process. Of the total unigenes analyzed, 7% revealed a functional prediction (amino acids prediction), but they did not exhibit homology with the proteins reported in InterProScan and GenBank. Thirteen percent corresponded to hypothetical proteins that do not have previous experimental evidence for their assigned function (Eisenstein et al. Citation2000). Two percent of the sequences corresponded to ribosomal proteins, and 53% of the unigenes exhibited homology with a reported protein. Among the unigenes with known functions, several were reported with the same function. Therefore, groupings were carried out according to these annotations.
Figure 1. Distribution of the unigene annotations from the C. arabica var. Caturra and C. liberica assemblage.
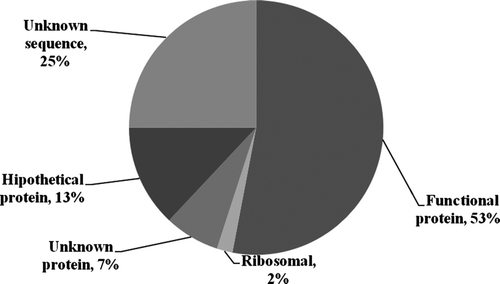
Once the unigenes were annotated, the protein assignments were classified on the basis of Gene Ontology (The Gene Ontology Consortium 2000) information. contains the categories with the highest representation in the annotated unigenes.
Table 5. Representative categories related to the annotation of the assembled unigenes for C. arabica var. Caturra and C. liberica.
Three expression groups were obtained after the statistical analysis of the cDNA libraries defined by p values less than or equal to 0.1: orthologs with similar expression patterns in both genotypes C. liberica and C. arabica var. Caturra (); proteins that are more abundantly expressed in C. arabica var. Caturra as compared to C. liberica (); and proteins with a higher expression in C. liberica ().
Table 6. Proteins from C. arabica var. Caturra and C. liberica with similar expression patterns (R p-value > 0.1).
Table 7. Proteins with higher expression in C. arabica with respect to C. liberica (R p-value < 0.1).
Table 8. Proteins with higher expression in C. liberica with respect to C. arabica (R p-value < 0.1).
Real-time PCR
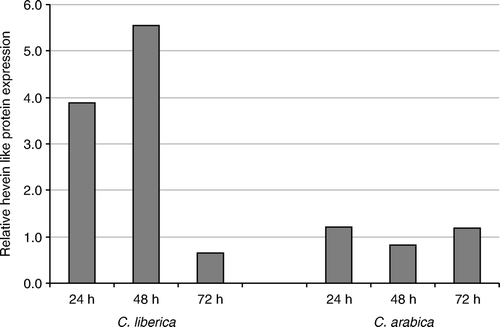
For the real-time PCR validation, the genes that corresponded to an insect defense function annotation were chosen. The real-time PCR amplification of the sequence CEN72529, annotated as a hevein-like protein, in each genotype and each infection treatment is shown in . For this sequence, there is an early overexpression in C. liberica after 24 h of the insect attack. CBB induces gene expression almost four times higher than the control noninfested berries, and the overexpression continues and increases at 48 h. The gene repression is observed only at 72 h postinfestation. On the contrary, in C. arabica, the level of gene expression is low when compared to the controls and there is no difference in the expression among the infestation times tested.
Figure 2. Relative expression of hevein-like protein gene CEN72529 by quantitative real-time PCR. Quantitative real-time PCR was performed with cDNA from green coffee berries at 24, 48, and 72 h after infestation of CBB in C. liberica and C. arabica.
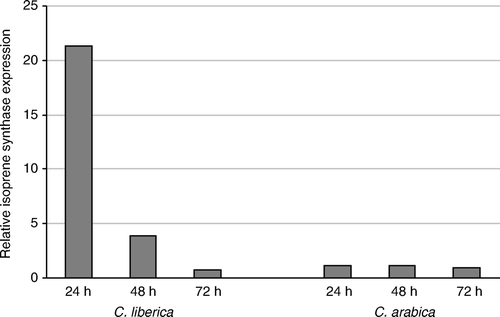
In the case of the sequence CEN71857, annotated as a putative isoprene synthase gene (), C. liberica showed a high overexpression at 24 h after infestation, with almost 20 times overproduction as compared to the control. This induction decreases at 48 h, and at 72 h, the levels are similar to the controls. In C. arabica, no overexpression of these genes is observed at any time.
Figure 3. Relative expression of isoprene synthase gene CEN71857 by quantitative real-time PCR. Quantitative real-time PCR was performed with cDNA from green coffee berries at 24, 48 and 72 h after infestation of CBB in C. liberica and C. arabica.
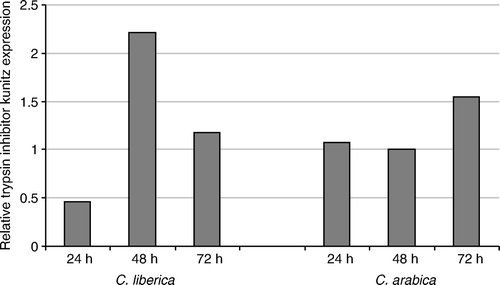
The sequence CEN73476, annotated as a putative trypsin inhibitor Kunitz gene (), has a relatively low expression compared to the other genes evaluated. In C. liberica, a low expression is observed at 24 h after infestation, which increases twice at 48 h and goes down again at 72 h. In C. arabica, the expression of this gene is similar at 24 and 48 h and increases at 72 h.
Figure 4. Relative expression of trypsin inhibitor Kunitz gene CEN73476 by quantitative real-time PCR. Quantitative real-time PCR was performed with cDNA from green coffee berries at 24, 48, and 72 h after infestation of CBB in C. liberica and C. arabica.
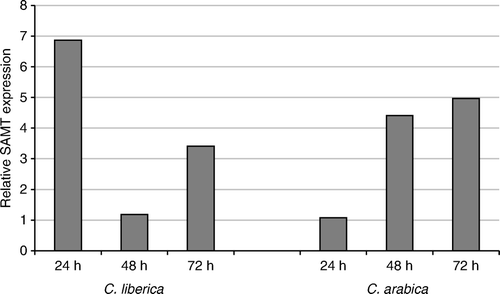
The validation of the sequence CEN73008 (), a putative S-adenosyl-l-methionine:salicylic acid carboxyl methyltransferase (SAMT), showed an overexpression of almost seven times in C. liberica at 24 h after infestation, with the levels decreasing at 48 h and induced again three times at 72 h. In C. arabica at 24 h the overproduction is lower than in C. liberica, but at 48 and 72 h the levels are higher than those in C. liberica at the same times. However, the overall highest expression is observed in C. liberica at 24 h.
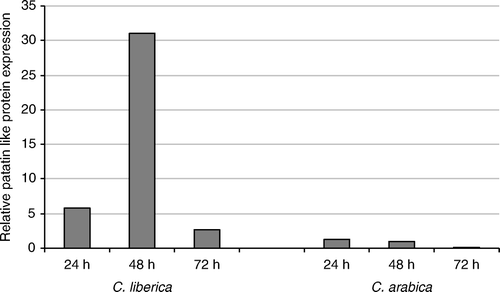
The expression pattern of sequence CEN73488 (), annotated as a patatin-like protein, is shown as a fivefold overexpression in C. liberica at 24 h as compared to the controls. After 48 h, the overproduction increased 30 times and the gene was then repressed at 72 h. It is the gene that overexpresses the most of all evaluated. On the contrary, C. arabica showed very low levels of expression at 24 and 48 h as compared to the controls, and at 72 h the expression is even lower.
Discussion
The Coffea genus is becoming the subject of increasing genomic research because of the importance of the crop around the world. Around 10,000 sequences coming from coffee seeds in an active transcriptional stage from two different genotypes – C. arabica and C. liberica – and induced by infestation with the CBB were obtained in this work. Previously, Salmona et al. (Citation2008) studied the transcriptional expression of 266 genes involved in seed maturation, but this is the first study involving gene expression of a coffee seed under an insect attack.
In general, most of the previous work focused on the plant response to phloem-feeding insects such as aphids and whiteflies (Voelckel et al. Citation2004; Thompson and Goggin Citation2006) or the response of plant leaves to chewing lepidoptera insects (see review by Wu and Baldwin Citation2010), but only a few studies focused in the response of plant seeds to a coleoptera.
For this study, coffee berries with 150 DAF from C. arabica and with 180 DAF from C. liberica were chosen. Those fruits are in an early active developmental stage, as defined by De Castro and Marracini (Citation2006). In this stage, genes that are involved not only in seed development but also in response to early insect infection can be identified. Previous research showed that during the first 24 h of infestation, the CBBs are found in position B, which means penetrating the pericarp (Bustillo Citation2006). De Castro and Maraccinni (2006) summarized that the pericarp is formed by the exocarp (peel) and the mesocarp, commonly referred to as the ‘true pulp’, rich in sugar and formed from the parenchyma cells with compact and dense cell walls in green fruits and the endocarp (also called parchment layer or ‘pergaminho’) that is a hard and lignified tissue. During this first 24 h of infection, the insects identify the berries and start a chewing of the tissue that forms the pericarp, penetrating the exocarp and the mesocarp without arriving to the endocarp. The oral secretions of the CBB that have been in contact with the tissue can act as elicitors that stimulate all of the major defense pathways, in a situation similar to the one reported in other insects (Felton and Korth Citation2000; Kahl et al. Citation2000). In addition to the chewing process, there is a mechanical stimulus caused by the insect in its effort to penetrate the pericarp that could be sensed by the cells of the pericarp in a fashion similar to the one reported in other crop plant tissues (Reymond et al. Citation2000, Citation2004). Here, a response in the coffee berry is induced because of the insect attack.
shows that a high number of sequences are involved in plant responses to stress from the 1836 total number of unigenes belonging to a specific functional category; almost 30% of the sequences, the highest number found, correspond to stress response. This suggests that the insect is capable of trigging an intense response, engaging more that 2000 clones from both genotypes in C. arabica (1106) and C. liberica (1070) related to stress. It is important, however, to point out that the berries are coming from plants located in the field and, in addition, to be exposed to the insect attack they may been sensing other biotic and abiotic stress stimuli. The second category in importance corresponds to the sequences related to general metabolism; this category accounts for 25% of the sequences. The other groups are related to seed development and sugar biosynthesis, which are abundant; this feature is expected to take into account that the coffee berries observed were still under development. A similar representation from this category was obtained in both genotypes. There is an additional group related to photosynthesis. This group confirms the active photosynthesis process produced in the fruits already reported by Geromel et al. (2006) and Salmona et al. (2008).
Our interest was to identify the differences between the sequences coming from each g enotype that can account for the differences in susceptibility against the CBB. On the basis of this, the proteins were grouped into three groups: (1) proteins with similar levels of expression on both genotypes (p>1) (), (2) proteins overexpressed in C. arabica (p<1) () and (3) proteins overexpressed in C. liberica (p<1) ().
The first group () includes proteins related to cellular maintenance, those involved in cellular elongation, fruit growth and development, as well as in fruit ripening. The results observed in this sequence group allowed comparisons between the assessed coffee species because the proteins related to cellular maintenance were expected to show a similar expression in both genotypes, as was observed.
Among them were found seed storage proteins highly expressed, such as albumins and lipid transfer proteins (Marion et al. Citation2007; Salmona et al. Citation2008), and NtEIG-E80, a protein-elicitor-inducible gene described in Nicotiana tabacum (Takemoto et al. Citation2003) whose function is photoassimilate-responsive and an eukaryotic initiation factor eIF5A-2 that supports growth (Feng et al. Citation2007) but also plays a regulatory role in the response of plants to sublethal osmotic and nutrient stresses (Ma et al. Citation2010), as well as lipid transferases, oleosins OLE3 protein and alpha-galactosidases genes involved in upstream fatty acid metabolism. Some of those proteins were reported in C. arabica seed developmental stage by Salmona et al. (Citation2008).
Similarly, flower and seed developmental proteins were found: MADS-box protein (Favaro et al. Citation2003) as well as a catalase, peroxidase and polyphenol oxidase, proteins related to ethylene production (Constabel et al. Citation1995; Thaler et al. Citation1999; Ward et al. Citation2001; Montavo et al. Citation2007) that participate in fruit maturation. A ‘Senescence-associated Nodulin 1A’ (Birch et al. Citation1999), two types of SAM-dependent N-methyltransferases superfamily enzymes, which are involved in diverse metabolic activities (Moffatt and Weretilnyk Citation2001), and an S-adenosyl methionine synthetase 3 SAMS3, associated with methionine regulation (Goto et al. 2002). Ethylene-responsive transcription factor implicated in seed development (Bustamante-Porras et al. Citation2005, Citation2007; Pereira et al. Citation2005), but also, able to induce the chitinase activity triggering a defense response against pathogens (Dinesh et al. Citation2010).
The overexpression of proteins related to photosynthesis and photooxidative stress was also observed with the identification of aldolases (Taji el al. 2004) and redox enzyme glutaredoxin, which additionally participate in flower development and salicylic acid signaling (Rouhier et al. Citation2008).
A group of proteins is involved in plant responses to biotic and abiotic stresses, such as miraculin-type proteins, generally reported in injury or pathogen responses (Tsukuda et al. Citation2006); a hypersensitive response protein induced by diseases and osmotic stress (Jung et al. Citation2008); several pathogenesis-related proteins (Jami et al. Citation2008); two types of heat shock proteins: HSP90 and HSP70, which frequently function together in pathogenic defense signal transduction pathway (Kansaki et al. Citation2003; Sangster and Queitsch Citation2005); and dirigent protein and caffeoyl-CoA O-methyltransferase, which are involved in the induction of lignin for plant protection (Burlat et al. Citation2001; Ralph et al. Citation2006) and other genes reported to be induced by pathogens such as an Avr9/Cf-9 rapidly elicited protein (ACRE gene) that is immediately activated upon perception of the pathogen (Rowland et al. Citation2005). The induction of those genes suggests that both genotypes are able to sense the stress produced by the insects.
However, differential expression between the genotypes was also observed in response to the biotic and abiotic stresses present at the moment of collecting the fruits.
includes proteins that showed higher expression in C. arabica var. Caturra. Here are found proteins such as cysteine proteases involved in a large variety of function – plant growth and development – but also in senescence and programmed cell death, in accumulation of storage proteins in seeds, as well as in storage protein mobilization and in pathways signaling in response to biotic and abiotic stresses (Van der Hoorn and Jones Citation2004). Also, there were metallothioneins, cysteine-rich proteins that bind to heavy metals. The metallothioneins are upregulated during the development of fruits and they are a systemic response for abiotic stress (Moyle et al. Citation2005; De Nardi et al. Citation2006). Also, an embryo-specific protein 3 that participates in seed development (Girke et al. Citation2000), an auxin receptor F box protein that regulates auxin response, implicated in every aspect of plant growth and development (Dharmasiri et al. Citation2005), and a cupin family protein, which includes many of the storage proteins from higher plants and nutrient reservoir activity (Dunwell et al. Citation2000), were found.
Also noted is the overexpression of proteins related to photosynthesis and photooxidative stress with the induction of an early light-induced protein (Hutin et al. Citation2003).
In addition, a group of proteins involved in plant responses to biotic and abiotic stresses were observed to be upregulated; in this group, a glutathione S-transferase complex regulated for protective functions by environmental stimuli (Edwards et al. Citation2000), an auxin-repressed protein (ARP19) that allows salicylic acid inhibiting the pathogen growth in plants (Kim et al. Citation2007; Wang et al. Citation2007) and a mannose/glucose-specific lectin with antifungal activity in seeds (Kuku et al. 2009) were found.
In general, it seems that C. arabica responds to the CBB attack with a plethora of genes involved in general biotic and abiotic stress responses and they are not very specific to insect defense; the genes induced are more related to plant–pathogen interactions.
Unigenes with sequences corresponding only to C. arabica var. Caturra (data not shown) were identified; however, proteins of interest related to defense were not found in this group.
Regarding the group with the highest expression in C. liberica (), this protein set contained in general more sequences related to biotic and abiotic stress responses.
Among the proteins that the literature reports to respond to general metabolism were found: a CR9 protein involved in plant growth and development by regulating cell cycle (Alfenas-Zerbini et al. Citation2009), a succinate dehydrogenase related to mitochondrial respiration (Millar et al. Citation2004) and cytochrome B5 implicated in sugar transportation interviewing with flowering and fruit development (Fan et al. Citation2009).
Genes overexpressed by the presence of biotic and abiotic general stresses were also found in this group, such as an annexin related to drought, salt and oxidative stress responses (Huh et al. 2010) and also to the increasing message levels for several pathogenesis-related proteins (Jami et al Citation2008); a DEA1 protein induced by arachidonic acid that has a protective function against abiotic and biotic stresses including weather change, pathogens and pests (Weyman et al. Citation2006); a catechol O-methyltransferase that plays a key role in lignin biosynthesis and also in disease resistance (Lam et al. 2007); two types of cytochrome P450: the CYP73A1 and the CYP71E1, that responses to different environmental conditions, wounding and it is involved in plant–insect interactions (Batard et al. Citation1997; Ganjewala et al. Citation2010).
The genes induced by pathogen s such as a tyrosine aminotransferase involved in the plant defense response to pathogens attack (Lopukhina et al. Citation2001), and MYB transcriptor factor inducible within 1 day after fungal infection (Lee et al. Citation2001) were also identified.
In this group, there were proteins more specifically related to herbivory and insect attack response (i.e. genes related to the jasmonic acid pathway), such as a hevein-like protein (Reymond et al. Citation2000), isoprene synthetase (Loivamäki et al. Citation2008), a trypsin inhibitor Kunitz gene (Mondego et al. Citation2005, Citation2011), an S-adenosyl-l-methionine:salicylic acid carboxyl methyltransferase (Köllner et al. 2010) and patatin-like protein (Alidhai and Rydel Citation2010) that has been reported as specifically induced by insects. These genes induced or upregulated by insects that show overexpression in C. liberica, the genotype with lower susceptibility to CBB attack when compared to C. arabica var. Caturra, may explain the negative effect that this genotype causes on the CBB. Because of that, the expression of them was validated by real-time PCR.
The real-time PCR results showed three genes with very high expression in C. liberica at 24 or 48 h and low or none expression in C. arabica. The gene hevein-like protein was upregulated at 24 and 48 h after insect infestation (), results similar to the ones obtained by comparing the number of transcripts in the libraries (). This gene is involved in the salicylic acid pathway (Medeiros et al. Citation2010); it has antifungal and antibacterial activities, but also it is induced by methyl jasmonate treatment in plants (Kiba et al. Citation2003) and it can be induced by insect herbivory (Reymond et al. Citation2000; Falco et al. Citation2001). Similar results were obtained with the gene that encodes the isoprene synthase enzyme, which was upregulated almost 20 times over at 24 h. The isoprene synthase is involved in the production of isoprene terpene by the plants. The isoprene is produced by some plants to protect its photosynthetic apparatus from the high temperatures and oxidative stress (Ferrieri et al. Citation2005), but also, recently, the isoprene has been reported as an insect repellent (Laothawornkitkul et al. Citation2008a, Citationb; Loivamäki et al. Citation2008). The upregulation of isoprene and isoprene emission has been reported before in response to insect herbivory and jasmonic acid treatment (Ferrieri et al. Citation2005).
The other gene that was highly upregulated in C. liberica was the patatin-like protein, which showed between 5 and 30 times overproduction during the first 48 h of infestation; this result also corresponds to the ones obtained in the libraries. The patatin gene encodes a lipid acyl hydrolase protein that affects lipids metabolism (Renier et al. 2004) and inhibits the growth of coleoptera larvae (Strickland et al. Citation1995). When applied in appropriate levels in artificial diets, potato patatin is lethal to spotted cucumber beetle Diabrotica undecimpunctata larvae and will stunt the growth of survivors; so, maturation is prevented or severely delayed, resulting in no reproduction (Alidhai and Rydel Citation2010). So far this gene has not been described in coffee, which is an important founding, because it can cause similar effects in the CBB larvae. These tree genes are good candidates to be overexpressed in C. arabica using constitutive promoter and genetic transformation or transient expression experiments in order to confirm their function as related to CBB coffee defenses.
There were another group of genes that showed upregulation in C. liberica and higher overexpression in this genotype as compared to C. arabica, but in C. arabica, induction of those genes was also observed, which is the case of S-adenosyl-l-methionine:salicylic acid carboxyl methyltransferase and the trypsin inhibitor Kunitz gene.
SAMT is an enzyme that converts salicylic acid to methyl salicylate, an important volatile emitted from plant herbivore damage, detectable after 2 h of insect feeding (Köllner et al. 2010). In coffee berries, SAMT showed high induction in C. liberica at 24 h as compared to C. arabica, results similar to the ones obtained with the libraries; however, the gene was also upregulated in C. arabica at 48 and 72 h postinfestation. C. liberica upregulated this gene earlier than C. arabica.
The trypsin inhibitor Kunitz gene showed higher upregulation in C. liberica at 48 h as compared to C. arabica at a similar time. However, this gene was also upregulated in C. arabica. The trypsin inhibitor Kunitz gene is already reported in coffee by Mondego et al. (2005, 2011); this is the same miraculin-like gene, CoMir. Structural protein modeling indicated that CoMir had structural similarities with the Kunitz STI proteins but suggested specific folding structures (Mondego et al. 2011). The gene is also upregulated after coffee leaf miner (Leucoptera coffella) oviposition in resistant plants of a progeny derived from crosses between C. racemosa (resistant) and C. arabica (susceptible) and downregulated during coffee leaf miner oviposition and larvae eclosion (herbivoria) in susceptible plants. In this experiment, C. liberica behaved in a way similar to that for the resistant genotype.
Finally, the genes that have been identified in this study increase our understanding of functional genomics. The genes overexpressed in the C. liberica genotype in response to the CBB attack can be turned into new tools to widen the genetic basis of the cultivable varieties in the effort to improve their resistance to pests and diseases as a complementary alternative for the integrated pest management strategy of the CBB control.
Acknowledgements
This research was cofinanced by the Scientific and Technical Cooperation Agreement between the Ministry of Agriculture and Rural Development and the National Federation of Coffee Growers of Colombia.
References
- Alfenas-Zerbini , P , Maia , IG , Fávaro , RD , Cascardo , JCM , Brommonschenkel , SH and Zerbini , FM . 2009 . Genome-wide analysis of differentially expressed genes during the early stages of tomato infection by a potyvirus . Mol Plant Microbe Interact. , 22 ( 3 ) : 352 – 361 .
- Alidhai MF , Rydel TJ. 2010 . Insect inhibitory lipid acyl hydrolases . US Patent 7,662,372 B2, filed February 13, 2006, and issued February 16, 2010 .
- Altschul , SF , Gish , W , Miller , W , Myers , EW and Lipman , DJ . 1990 . Basic local alignment search tool . J Mol Biol. , 215 ( 3 ) : 403 – 410 .
- Batard , Y , Schalk , M , Pierrel , MA , Zimmerlin , A , Durst , F and Werck-Reichhart , D . 1997 . Regulation of the cinnamate 4-hydroxylase (CYP73A1) in Jerusalem artichoke tubers in response to wounding and chemical treatments . Plant Physiol. , 113 : 951 – 959 .
- Bernards MA , Båstrup-Spohr L. 2008 . Phenylpropanoid metabolism induced by wounding and herbivory . In : Schaller A . Induced plant resistance to herbivory . Dordrecht , , The Netherlands : Springer . p. 189 – 211 .
- Birch , PRJ , Avrova , AO , Duncan , JM , Lyon , GD and Toth , RL . 1999 . Isolation of potato genes that are induced during an early stage of the hypersensitive response to Phytophthora infestans . Mol Plant Microbe Interact. , 12 : 356 – 361 .
- Boutrot , F , Guirao , A , Alary , R , Joudrier , P and Gautier , MF . 2005 . Wheat non-specific lipid transfer protein genes display a complex pattern of expression in developing seeds . Biochim Biophys Acta. , 1730 : 114 – 125 .
- Burlat , V , Kwon , M , Davin , LB and Lewis , NG . 2001 . Dirigent proteins and dirigent sites in lignifying tissues . Phytochemistry , 57 : 883 – 897 .
- Bustamante-Porras , J , Noirot , M , Campa , C , Hamon , S and de Kochko , A . 2005 . Isolation and characterization of a Coffea canephora ERF-like cDNA . Afri J Biotechnol. , 4 ( 2 ) : 157 – 159 .
- Bustamante-Porras J , Poncet V , Campa C , Noirot M , Hamon S , de Kochko A. 2007 . Characterization of three ethylene receptor genes in Coffea canephora Pierre: advances in plant ethylene research . Proceedings of the 7th International Symposium on the plant hormone ethylene . p. 53 – 56 . doi: 10.1007/978-1-4020-6014-4_13
- Bustillo , AE . 2005 . El papel del control biológico en el manejo integrado de la coffee berry borer del café, Hypothenemus hampei (Ferrari) (Coleoptera: Curculionidae: Scolytinae . Rev Acad Colomb Cienc. , 29 : 56 – 68 .
- Bustillo , AE . 2006 . Una revisión sobre la broca del café, Hypothenemus hampei (Coleoptera: Curculionidae. Scolytidae), en Colombia . Rev Colomb Entomol. , 32 : 101 – 116 .
- Chen , GQ , He , X and McKeon , TA . 2004 . 2S albumin gene expression in castor plant (Ricinus communis L.) . J Am Oil Chem Soc. , 81 : 867 – 872 .
- Constabel , CP , Bergey , DR and Ryan , CA . 1995 . Systemin activates synthesis of wound-inducible tomato leaf polyphenol oxidase via the octadecanoid defense signaling pathway . Proc Natl Acad Sci USA. , 22 : 407 – 411 .
- Coram , TE and Pang , ECK . 2005 . Isolation and analysis of candidate ascochyta blight resistance genes in chickpea. II. Microarray analysis of putative defence-related ESTs . Physiol Mol Plant Pathol. , 66 : 201 – 210 .
- De Castro , D and Marracini , P . 2006 . Cytology, biochemistry and molecular changes during coffee fruit development . Braz J Plant Physiol. , 18 ( 1 ) : 175 – 199 .
- De Nardi , B , Dreos , R , Del Terra , L , Martellossi , C , Asquini , E , Tornincasa , P , Gasperini , D , Pacchioni , B , Rathinavelu , R Pallavicini , A . 2006 . Diferential responses of Coffea arabica L. leaves and roots to chemically induced systemic acquired resistance . Genome. , 49 : 1594 – 1606 .
- Dewitte , W and Murray , JAH . 2003 . The plant cell cycle . Annu Rev Plant Biol. , 54 : 235 – 264 .
- Dharmasiri , N , Dharmasiri , S , Weijers , D , Lechner , E , Yamada , M , Hobbie , L , Ehrismann , JS , Jürgens , G and Estelle , M . 2005 . Plant development is regulated by a family of auxin receptor F box proteins . Dev Cell. , 1 : 109 – 119 .
- Dinesh , KP , SantaRam , A and Shivanna , MB . 2010 . Studies on the chitinase activity in coffee (Coffea Arabica L.) genetic resources in India . Res J Agric Biol Sci. , 6 ( 4 ) : 449 – 452 .
- Dunwell , JM , Khuri , S and Gane , PJ . 2000 . Microbial relatives of the seed storage proteins of higher plants: conservation of structure and diversification of function during evolution of the cupin superfamily . Microbiol Mol Biol Rev. , 64 ( 1 ) : 153 – 179 .
- Duque , OH , Chaves , CB and Hernández , SM . 1997 . Costos del manejo de la coffee berry borer Hypothenemus hampei (Ferrari) en lotes comerciales de café del departamento del Risaralda , Chinchiná , , Colombia : Federación Nacional de Cafeteros de Colombia, Centro Nacional de Investigaciones de Café – Cenicafé .
- Edwards , R , Dixon , DP and Walbot , V . 2000 . Plant glutathione S-transferases: enzymes with multiple functions in sickness and in health . Trends Plant Sci. , 5 ( 5 ) : 193 – 198 .
- Ehlting , J , Mattheus , N , Aeschliman , DS , Li , E , Hamberger , B , Cullis , IF , Zhuang , J , Kaneda , M , Mansfield , SD Samuels , L . 2005 . Global transcript profiling of primary stems from Arabidopsis thaliana identifies candidate genes for missing links in lignin biosynthesis and transcriptional regulators of fiber differentiation . Plant J. , 42 : 618 – 640 .
- Eira , MTS , da Silva , EAA , de Castro , RD , Dussert , S , Walters , C , Bewley , JD and Hilhorst , HWM . 2006 . Coffee seed physiology . Braz J Plant Physiol. , 18 ( 1 ) : 149 – 163 .
- Eisenstein , E , Gilliland , GL , Herzberg , O , Moult , J , Orban , J , Poljak , RJ , Banerjei , L , Richardson , D and Howard , AJ . 2000 . Biological function made crystal clear—annotation of hypothetical proteins via structural genomics . Curr Opin Biotechnol. , 11 ( 1 ) : 25 – 30 .
- Ewing , B , Hillier , L , Wendl , MC and Green , P . 1998 . Base-calling of automated sequencer traces using phred. I. Accuracy assessment . Genome Res. , 8 ( 3 ) : 175 – 185 .
- Falco , MC , Marbach , PAS , Pompermayer , P , Lopes , FCC and Silva-Filho , MC . 2001 . Mechanisms of sugarcane response to herbivory . Genet Mol Biol. , 24 : 113 – 122 .
- Fan , RC , Peng , CC , Xu , YH , Wang , XF , Li , Y , Shang , Y , Du , SY , Zhao , R , Zhang , XY Zhang , LY . 2009 . Apple sucrose transporter SUT1 and sorbitol transporter SOT6 interact with cytochrome b5 to regulate their affinity for substrate sugars . Plant Physiol. , 150 : 1880 – 1901 .
- Favaro , R , Pinyopich , A , Battaglia , R , Kooiker , M , Borghi , L , Ditta , G , Yanofsky , MF , Kater , MM and Colombo , L . 2003 . MADS-box protein complexes control carpel and ovule development in Arabidopsis . Plant Cell. , 15 : 2603 – 2611 .
- Felton , GW and Korth , KL . 2000 . Trade-offs between pathogen and herbivore resistance . Curr Opin Plant Biol. , 3 : 309 – 314 .
- Feng , H , Chen , Q , Feng , J , Zhang , J , Yang , X and Zuo , J . 2007 . Functional characterization of the Arabidopsis eukaryotic translation initiation factor 5A-2 that plays a crucial role in plant growth and development by regulating cell division, cell growth, and cell death . Plant Physiol. , 144 : 1531 – 1545 .
- Ferrieri , RA , Gray , DW , Babst , BA , Schueller , MJ , Schlyer , DJ , Thorpe , MR , Orians , CM and Lerdau , M . 2005 . Use of carbon-11 in Populus shows that exogenous jasmonic acid increases biosynthesis of isoprene from recently fixed carbon . Plant Cell Environ. , 28 : 591 – 602 .
- Feurtado , JA , Banik , M and Bewley , JD . 2001 . The cloning and characterization of α-galactosidase present during and following germination of tomato (Lycopersicon esculentum Mill.) seed . J Exp Bot. , 52 : 1239 – 1249 .
- Ganjewala D , Kumar S , Devi A , Ambika K. 2010 . Advances in cyanogenic glycosides biosynthesis and analyses in plants: a review . Acta Biol Szeged . 54 1 : 1 – 14 . Available from: http://www.sci.u-szeged.hu/ABS
- Geromel , C , Ferreira , LP , Guerreiro , SM , Cavalari , AA , Pot , D , Pereira , LF , Leroy , T , Vieira , LG , Mazzafera , P and Marraccini , P . 2006 . Biochemical and genomic analysis of sucrose metabolism during coffee (Coffea arabica) fruit development . J Exp Bot. , 57 : 3243 – 3258 .
- Giovane , A , Servillo , L , Balestrieri , C , Raiola , A , D'Avino , R , Tamburrini , M , Clardiello , MA and Camardella , L . 2004 . Pectin methylesterase inhibitor . Biochim Biophys Acta. , 1696 ( 2 ) : 245 – 252 .
- Girke , T , Todd , J , Ruuska , S , White , J , Benning , C and Ohlrogge , J . 2000 . Microarray analysis of developing Arabidopsis seeds . Plant Physiol. , 124 : 1570 – 1581 .
- Goto , D , Ogi , M , Kijima , F , Kumagai , T , van Werven , F , Onouchi , H and Naito , S . 2002 . A single-nucleotide mutation in a gene encoding S-adenosylmethionine synthatase is associated with methionine over-accumulation phenotype in Arabidopsis thaliana . Genes Genet Syst. , 77 : 89 – 95 .
- Huang , X and Madem , A . 1999 . CAP3: a DNA sequence assembly program . Genome Res. , 9 ( 9 ) : 868 – 877 .
- Huh , SM , Noh , EK , Kim , HG , Jeon , BW , Bae , K , Hu , HC , Kwak , JM and Park , OK . 2010 . Arabidopsis annexins AnnAt1 and AnnAt4 interact with each other and regulate drought and salt stress responses . Plant Cell Physiol. , 51 ( 9 ) : 1499 – 1514 .
- Hutin , C , Nussaume , L , Morse , N , Moya , I , Kloppstech , K and Havaux , M . 2003 . Early light-induced proteins protect Arabidopsis from photooxidative stress . Proc Natl Acad Sci USA. , 100 : 4921 – 4926 .
- Iseli C , Jongeneel CV , Bucher P. 1999 . ESTScan: a program for detecting, evaluating, and reconstructing potential coding regions in EST sequences . ISMB-99 Proceedings . p. 138 – 148 .
- Jami , SK , Clark , GB , Turlapati , SW , Handley , C , Roux , SJ and Kirti , PB . 2008 . Ectopic expression of an annexin from Brassica juncea confers tolerance to abiotic and biotic stress treatments in transgenic tobacco . Plant Physiol Biochem. , 46 : 1019 – 1030 .
- Jung , HW , Lim , CW , Lee , SC , Choi , HW , Hwang , CH and Hwang , BK . 2008 . Distinct roles of the pepper hypersensitive induced reaction protein gene CaHIR1 in disease and osmotic stress, as determined by comparative transcriptome and proteome analyses . Planta. , 227 ( 2 ) : 409 – 425 .
- Kahl , J , Siemens , DH , Aerts , RJ , Gäbler , R , Kühnemann , F , Preston , CA and Baldwin , IT . 2000 . Herbivore-induced ethylene suppresses a direct defense but not a putative indirect defense against an adapted herbivore . Planta. , 210 : 336 – 342 .
- Kansaki , H , Saito , H , Ito , A , Fujisawa , S , Kamoun , S , Katou , S , Yoshioka , H and Terauchi , R . 2003 . Cytosolic HSP90 and HSP70 are essential components of INF1-mediated hypersensitive response and non-host resistance to Pseudomonas cichorii in Nicotiana benthamiana . Mol Plant Pathol. , 4 ( 5 ) : 383 – 389 .
- Kiba , A , Saitoh , H , Nishihara , M , Omiya , K and Yamamura , S . 2003 . C-terminal domain of a hevein-like protein from Wasabia japonica has potent antimicrobial activity . Plant Cell Physiol. , 44 ( 3 ) : 296 – 303 .
- Kim , JI , Sharkhuu , A , Jin , JB , Li , P , Jeong , JC , Baek , D , Lee , SY , Blakeslee , JJ , Murphy , AS Bohnert , HJ . 2007 . yucca6, a dominant mutation in Arabidopsis, affects auxin accumulation and auxin-related phenotypes . Plant Physiol. , 145 : 722 – 735 .
- Köllner TG , Lenk C , Zhao N , Seidl-Adams I , Gershenzon J , Chen F , Degenhardt J , 2010 . Herbivore-induced SABATH methyltransferases of maize that methylate anthranilic acid using S-adenosyl-L-methionine . Plant Physiol . 153 : 1795 – 1807
- Kuku A , Odekanyin O , Adentran K , Adewusi M , Olonade T . 2009 . Purification of a mannose/glucose-specific lectin with antifungal activity from pepper seeds (Capsicum annuum) . Afr Jof Biochem Res 3 6 : 272 – 278 .
- Lam KC , Ibrahim RK , Behdad B , Dayanandan S. 2007 . Structure, function, and evolution of plant O-methyltransferases . Genome . 50 11 : 1001 – 1013 . doi: 10.1139/G07-077
- Laothawornkitkul , J , Paul , N , Vickers , C , Possell , M , Mullineaux , P , Hewitt , N and Taylor , J . 2008a . The role of isoprene in insect herbivory . Plant Signal Behav. , 12 : 1141 – 1142 .
- Laothawornkitkul , J , Paul , N , Vickers , C , Possell , M , Taylor , J , Mullineaux , P and Hewitt , N . 2008b . Isoprene emissions influence herbivore-feeding decisions . Plant Cell Environ. , 31 : 1410 – 1415 .
- Lee , MW , Qi , M and Yang , Y . 2001 . A novel jasmonic acid-inducible rice myb gene associates with fungal infection and host cell death . Mol Plant Microbe Interact. , 14 ( 4 ) : 527 – 535 .
- Lin , C , Mueller , LA , McCarthy , J , Crouzillat , D , Pe′tiard , V and Tanksley , SD . 2005 . Coffee and tomato share common gene repertoires as revealed by deep sequencing of seed and cherry transcripts . Theor Appl Genet. , 112 : 114 – 130 .
- Liu , Y , Ahn , JE , Datta , S , Salzman , RA , Moon , J , Huyghues-Despointes , B , Pittendrigh , B , Murdock , LL , Koiwa , H and Zhu-Salzman , K . 2005 . Arabidopsis vegetative storage protein is an anti-insect acid phosphatase . Plant Physiol. , 139 : 1545 – 1556 .
- Loivamäki , M , Mumm , R , Dicke , M and Schnitzler , JP . 2008 . Isoprene interferes with the attraction of bodyguards by herbaceous plants . Proc Natl Acad Sci USA. , 105 ( 45 ) : 17430 – 17435 .
- Lopukhina , A , Dettenberg , M , Weiler , EW and Holländer-Czytko , H . 2001 . Cloning and characterization of a coronatine-regulated tyrosine aminotransferase from Arabidopsis . Plant Physiol. , 126 : 1678 – 1687 .
- Ma F , Liu Z , Wang TW , Hopkins MT , Peterson C , Thompson JE. 2010 Arabidopsis eIF5A3 influences growth and the response to osmotic and nutrient stress . Pant Cell Environ . 33 10 : 1682 – 1696 . doi: 10.1111/j.1365-3040.2010.02173.x
- Marion , D , Bakan , B and Elmorjani , K . 2007 . Plant lipid binding proteins: properties and applications . Biotechnol Adv. , 25 ( 2 ) : 195 – 197 .
- Mathieu , F , Malosse , CA and Frérot , B . 1996 . Comparative headspace analysis of fresh red coffee berries from different cultivated varieties of coffee trees . J High Resolut Chromatogr. , 19 : 298 – 300 .
- Mathieu , F , Malosse , CA and Frérot , B . 1998 . Identification of the volatile components released by fresh coffee berries at different stages of ripeness . J Agric Food Chem. , 46 ( 3 ) : 1106 – 1110 .
- McCarthy , AA and McCarthy , JG . 2007 . The structure of two N-methyltransferases from the caffeine biosynthetic pathway . Plant Physiol. , 144 : 879 – 889 .
- Medeiros FCL , Resende MLV , Medeiros FHV , Zhang HM . Paré PW . Forthcoming 2010 . Defense gene expression induced by a coffee-leaf extract formulation in tomato . Physiol Mol Plant P . doi: 10.1016/j.pmpp.2009.11.004
- Medina Filho , HP , Carvalho , A , Sondahl , MR , Fazuoli , LC and Costa , WM . 1984 . Coffee breeding and related evolutionary aspects . Plant Breed Rev. , 2 : 157 – 193 .
- Millar , AH , Day , DA and Whelan , J . 2004 . “ Mitochondrial biogenesis and function in Arabidopsis ” . In The Arabidopsis book , Edited by: Somerville , CR and Meyerowitz , EM . Rockville , MD : American Society of Plant Biologists .
- Min XJ , Butler G , Storms R , Tsang A. 2005 . TargetIdentifier: a web server for identifying full-length cDNAs from EST sequences . Nucleic Acids Res . 33 Web Server Issue : W669 – W672 . Available from: https://fungalgenome.concordia.ca/tools/TargetIdentifier.html
- Moffatt , BA and Weretilnyk , EF . 2001 . Sustaining S-adenosyl-l-methionine-dependent methyltransferase activity in plant cells . Physiol Plant. , 113 : 435 – 442 .
- Mondego JM , Guerreiro-Filho O , Bengtson MH , Drummond RD , Felix JM , Duarte MP , Ramiro D , Maluf MP , Sogayar MC , Menossi M . 2005 . Isolation and characterization of Coffea genes induced during coffee leaf miner (Leucoptera coffeella) infestation. Plant Sci . 169 : 351 – 360 .
- Mondego JM , Duarte MP , Kiyota E , Martínez L , de Camargo SR , De Caroli FP , Alves BS , Guerreiro SM , Oliva ML , Guerreiro-Filho O , et al. . 2011 . Molecular characterization of a miraculin-like gene differentially expressed during coffee development and coffee leaf miner infestation . Planta . 233 1 : 123 – 137 . doi: 10.1007/s00425-010-1284-9 .
- Montavo , P , Kukic , KR and Bortlik , K . 2007 . A simple method to measure effective catalase activities: optimization, validation, and application in green coffee . Anal Biochem. , 360 ( 2 ) : 207 – 215 .
- Moore , S , Payton , P , Wright , M , Tanksley , S and Giovannoni , J . 2005 . Utilization of tomato microarrays for comparative gene expression analysis in the Solanaceae . J Exp Bot. , 56 : 2885 – 2895 .
- Moyle , R , Fairbairn , DJ , Ripi , J , Crowe , M and Botella , JR . 2005 . Developing pineapple fruit has a small transcriptome dominated by metallothionein . J Exp Bot. , 56 ( 409 ) : 101 – 112 .
- Ortiz , A , Ortiz , A , Vega , F and Posada , F . 2004 . Volatile composition of coffee berries at different stages of ripeness and their possible attraction to the coffee berry borer Hypothenemus hampei (Coleoptera: Curculionidae) . J Agric Food Chem. , 52 : 5914 – 5918 .
- Passardi , F , Cosio , C , Penel , C and Dunand , C . 2005 . Peroxidases have more functions than a Swiss army knife . Plant Cell Rep. , 24 : 255 – 265 .
- Pereira , LFP , Galvao , RM , Kobayashi , A , Cacao , SM and Esteves-Vieira , LG . 2005 . Ethylene production and acc oxidase gene expression during fruit ripening of Coffea arabica L. BRAZ . J Plant Physiol. , 17 ( 3 ) : 283 – 289 .
- Rajangam , AS , Kumar , M , Aspeborg , H , Guerriero , G , Arvestad , L , Pansri , P , Brown , CJ , Hober , S , Blomqvist , K Divne , C . 2008 . MAP20, a micro tubule-associated protein in the secondary cell walls of hybrid aspen, is a target of the cellulose synthesis inhibitor 2,6-dichlorobenzonitrile . Plant Physiol. , 148 : 1283 – 1294 .
- Ralph , S , Park , JY , Bohlmann , J and Mansfield , SD . 2006 . Dirigent proteins in conifer defense: gene discovery, phylogeny, and differential wound- and insect-induced expression of a family of DIR and DIR-like genes in spruce (Picea spp.) . Plant Mol Biol. , 60 : 21 – 40 .
- Ramachandran , S , Christensen , HE , Ishimaru , Y , Dong , CH , Chao-Ming , W , Cleary , AL and Chua , NH . 2000 . Profilin plays a role in cell elongation, cell shape maintenance, and flowering in Arabidopsis . Plant Physiol. , 124 : 1637 – 1647 .
- Ramarkers , C , Ruijter , JM and Dprez , RHL . 2003 . Moorman, AFM. Assumption-free analysis of quantitative real-time pymerase chain reaction (PCR) data . Neurosci Lett. , 339 : 62 – 66 .
- Renier A , Van der Hoorn L , Leeuwenburgh MA , Bogyo M , Joosten MH , Peck SC . 2004 . Activity profiling of papain-like cysteine proteases in plants. Plant Physiol . 135 : 1170 – 1178 .
- Reymond , P , Bodenhausen , N , Van Poecke , RM , Krishnamurthy , V , Dicke , M and Farmer , EE . 2004 . A conserved transcript pattern in response to a specialist and a generalist herbivore . Plant Cell. , 16 : 3132 – 3147 .
- Reymond , P , Weber , H , Damond , M and Farmer , EE . 2000 . Differential gene expression in response to mechanical wounding and insect feeding in Arabidopsis . Plant Cell. , 12 : 707 – 720 .
- Richmond , TA and Somerville , CR . 2000 . The cellulose synthase superfamily . Plant Physiol. , 124 : 495 – 498 .
- Romano , P , Horton , P and Gray , J . 2004 . The Arabidopsis cyclophilin gene family . Plant Physiol. , 134 : 1268 – 1282 .
- Romero , JV and Cortina , H . 2004 . Fecundidad y Ciclo de Vida de la coffee berry borer Hypothenemus hampei F. (Coleoptera: Curculionidae: Scolytinae) en introducciones silvestres de café. Rev . Cenicafé. , 55 : 221 – 231 .
- Romero , JV and Cortina , H . 2007 . Tablas de vida de Hypothenemus hampei (Coleoptera: Curculionidae: Scolytinae) sobre tres introducciones de café . Rev Colom Entomol. , 33 : 10 – 16 .
- Romualdi , C , Bortoluzzi , S , D'Alessi , F and Danieli , GA . 2003 . IDEG6: a web tool for detection of differentially expressed genes in multiple tag sampling experiments . Physiol Genomics. , 12 : 159 – 162 .
- Rouhier N , Lemaire SD , Jacquot JP. 2008 . The role of glutathione in photosynthetic organisms: emerging functions for glutaredoxins and glutathionylation . Annu Rev Plant Biol . 59 : 143 – 166 . doi: 10.1146/annurev.arplant.59.032607.092811
- Rowland , O , Ludwig , AA , Merrick , CJ , Baillieul , F , Tracy , FE , Durrant , WE , Fritz-Laylin , L , Nekrasov , V , Sjölander , K Yoshioka , H . 2005 . Functional analysis of Avr9/Cf-9 rapidly elicited genes identifies a protein kinase, ACIK1, that is essential for full Cf-9–dependent disease resistance in tomato . Plant Cell. , 17 ( 1 ) : 295 – 310 .
- Salcedo , G , Sanchez- Monge , R , Barber , D and Diaz-Perales , A . 2007 . Plant nonspecific lipid transfer proteins: an interface between plant defence and human allergy . Biochim Biophys Acta. , 1771 : 781 – 791 .
- Salmona , J , Dussert , S , Descroix , F , de Kochko , A , Bertrand , B and Joet , T . 2008 . Deciphering transcriptional networks that govern Coffea arabica seed development using combined cDNA array and real-time RT-PCR approaches . Plant Mol Biol. , 66 : 105 – 124 .
- Sangster , TA and Queitsch , C . 2005 . The HSP90 chaperone complex, an emerging force in plant development and phenotypic plasticity . Curr Opinion Plant Biol. , 8 : 86 – 92 .
- Schrader , M and Fahimi , HD . 2006 . Growth and division of peroxisomes . Int Rev Cytol. , 255 : 237 – 290 .
- Schuler , MA and Werck-Reichhart , D . 2003 . Functional genomics of P450s . Ann Rev Plant Biol. , 54 : 629 – 667 .
- Simkin , AJ , Gaffe , J , Alcaraz , JP , Carde , JP , Bramley , PM , Fraser , PD and Kuntz , M . 2007 . Fibrillin influence on plastid ultrastructure and pigment content in tomato fruit . Phytochemistry. , 68 : 1545 – 1556 .
- Stekel , DJ , Git , Y and Falciani , F . 2000 . The comparison of gene expression from multiple cDNA libraries . Genome Res. , 10 : 2055 – 2061 .
- Strickland , JA , Orr , GL and Walsh , TA . 1995 . Inhibition of diabrotica larval growth by patatin, the lipid acyl hydrolase from potato tubers . Plant Physiol. , 109 ( 2 ) : 667 – 674 .
- Susko , E and Roger , AJ . 2004 . Estimating and comparing the rates of gene discovery and expressed sequence tag (EST) frequencies in EST surveys . Bioinformatics. , 20 ( 14 ) : 2279 – 2287 .
- Taji , T , Seki , M , Satou , M , Sakurai , T , Kobayashi , M , Ishiyama , K , Narusaka , Y , Narusaka , M , Zhu , JK and Shinozaki , K . 2004 . Comparative genomics in salt tolerance between Arabidopsis and Arabidopsis-related halophyte salt cress using Arabidopsis microarray . Plant Physiol. , 135 : 1 – 13 .
- Takemoto , D , Jones , DA and Hardham , AR . 2003 . GFP-tagging of cell components reveals the dynamics of subcellular re-organization in response to infection of Arabidopsis by oomycete pathogens . Plant J. , 33 : 775 – 795 .
- Thaler , JS , Fidantsef , AL , Duffey , SS and Bostock , RM . 1999 . Trade-offs in plant defense against pathogens and herbivores: a field demonstration of chemical elicitors of induced resistance . J Chem Ecol. , 25 : 1597 – 1609 .
- The Gene Ontology Consortium . 2000 . Gene ontology: tool for the unification of biology . Nat Genet . 25 : 25 – 29 . Available from: http://www.geneontology.org/
- Thompson , GA and Goggin , FL . 2006 . Transcriptomics and functional genomics of plant defence induction by phloem-feeding insects . J Exp Bot. , 57 ( 4 ) : 755 – 766 .
- Thompson , JE , Hopkins , MT , Taylor , C and Wang , TW . 2004 . Regulation of senescence by eukaryotic translation initiation factor 5A: implications for plant growth and development . Trends Plant Sci. , 9 : 174 – 179 .
- Toci , AT and Farah , A . 2008 . Volatile compounds as potential defective coffee beans’ markers . Food Chem. , 108 : 1133 – 1141 .
- Tsukuda , S , Gomi , K , Yamamoto , H and Akimitsu , K . 2006 . Characterization of cDNAs encoding two distinct miraculin-like proteins and stress-related modulation of the corresponding mRNAs in Citrus jambhiri Lush . Plant Mol Biol. , 60 ( 1 ) : 125 – 136 .
- Untergasser A , Nijveen H , Rao X , Bisseling T , Geurts R , Leunissen JAM. 2007 . Primer3Plus, an enhanced web interface to Primer3 . Nucleic Acids Res . 35 Web Server Issue : W71 – W74 . doi: 10.1093/nar/gkm306
- Van der Hoorn , RA and Jones , JD . 2004 . The plant proteolytic machinery and its role in defence . Curr Opin Plant Biol. , 7 : 400 – 407 .
- Villagran , GW . 1991 . Atractividad relativa y susceptibilidad de varias especies y cultivares de café Coffea spp. Broca del fruto Hypothenemus hampei Ferr. en condiciones de laboratorio [dissertation] , Guatemala : Universidad de San Carlos de Guatemala .
- Voelckel , C , Weisser , WW and Baldwin , IT . 2004 . An analysis of plant–aphid interactions by different microarray hybridization strategies . Mol Ecol. , 13 ( 10 ) : 3187 – 3195 .
- Wang D , Pajerowska-Mukhtar K , Culler AH , Dong X . 2007 . Salicylic acid inhibits pathogen growth in plants through repression of the auxin signaling pathway. Curr Biol . 17 20 1784 – 1790 . doi: 10.1016/j.cub.2007.09.025
- Ward , G , Hadar , Y , Bilkis , I , Konstantinovsky , L and Dosoretz , CG . 2001 . Initial steps of ferulic acid polymerization by lignin peroxidase . J Biol Chem. , 276 : 18734 – 18741 .
- Weyman , PD , Pan , Z , Feng , Q , Gilchrist , DG and Bostock , RM . 2006 . A circadian rhythm regulated tomato gene is induced by arachidonic acid and Phytophthora infestans infection . Plant Physiol. , 140 : 235 – 248 .
- Williams , LE and Mills , RF . 2005 . P1B-ATPases—an ancient family of transition metal pumps with diverse functions in plants . Trends Plant Sci. , 10 : 491 – 502 .
- Wong , K , Suchard , MA and Huelsenbeck , JP . 2008 . Alignment uncertainty and genomic analysis . Science. , 319 ( 5862 ) : 473 – 476 .
- Wu , J and Baldwin , IT . 2010 . New insights into plant responses to the attack from insect herbivores . Annu Rev Genet. , 44 : 1 – 24 .
- Zdobnov , EM and Apweiler , R . 2001 . InterProScan—an integration platform for the signature-recognition methods in InterPro . Bioinformatics. , 17 ( 9 ) : 847 – 848 .