Abstract
The genetic basis of resistance to soil-borne cereal mosaic virus (SBCMV) in the Triticum turgidum L. var. durum cv. Neodur was analyzed in this study, using a linkage mapping approach. We performed phenotypic and molecular analyses of 146 recombinant inbred lines derived from the cross Cirillo (highly susceptible)×Neodur (highly resistant). A major quantitative trait locus (QTL) that explained up to 87% of the observed variability for symptom severity was identified on the short arm of chromosome 2B, within the 40-cM interval between the markers Xwmc764 and Xgwm1128, with wPt-2106 as the peak marker. Three minor QTLs were found on chromosomes 3B and 7B. Two markers coding for resistance proteins co-segregate with the major QTL on chromosome 2B and the minor QTL on chromosome 3B, representing potential candidate genes for the two resistance loci. Microsatellite markers flanking the major QTL were evaluated on a set of 25 durum wheat genotypes that were previously characterized for SBCMV resistance. The allelic composition of the genotypes at these loci, together with pedigree data, suggests that the old Italian cultivar Cappelli provided the SBCMV-resistance determinants to durum cultivars that have been independently bred in different countries over the last century.
Introduction
Soil-borne cereal mosaic virus (SBCMV), soil-borne wheat mosaic virus (SBWMV), and Chinese wheat mosaic virus (CWMV) are closely related furoviruses (Shirako et al. Citation2000; Torrance and Koenig Citation2005; Hariri and Meyer Citation2007) that can infect durum wheat (Triticum turgidum L. var. durum), common wheat (T. aestivum L. var. vulgare), and numerous other cultivated and spontaneous Gramineae (Canova Citation1964; Verchot-Lubicz Citation2005). These viruses are transmitted to the roots of their host plants by Polymyxa graminis Led., a soil-borne plasmodiophorid protist that can preserve its infectivity in soil for 10 years or more (Rao and Brakke Citation1969; Kendall and Lommel Citation1988). SBCMV is prevalent in Europe, CWMV in Asia, and SBWMV (the furovirus type member) in North America (Diao et al. Citation1999; Kanyuka et al. Citation2003; Ratti et al. Citation2005; Budge et al. Citation2008).
In Italy, SBCMV is widespread in the main wheat growing areas, and especially in the northern and central regions of Italy, where it causes grain yield losses often above 50% on susceptible cultivars of durum wheat (AABB genome) and common wheat (AABBDD genome) (Vallega et al. Citation2002; Rubies-Autonell et al. Citation2006). Despite this serious economic impact, the presence of SBCMV, SBWMV, and CWMV in the main wheat growing areas worldwide has gone essentially undetected for many decades. This has been mainly because visible symptoms induced by these pathogens, which include stunting and chlorotic mottling, were attributed to other causes (e.g. abiotic stress) (Kanyuka et al. Citation2003). To date, the only economically viable means of controlling SBCMV, SBWMV, and CWMV is the adoption of resistant cultivars (Kanyuka et al. Citation2003).
The durum wheat cultivars tested thus far in Italy show a wide and continuous array of reactions to SBCMV and only a few have proven to be highly resistant to SBCMV; of note, none of the nearly 200 cultivars assayed showed immunity to SBCMV infection (Maccaferri et al. Citation2005a, Citation2005b; Rubies-Autonell et al. Citation2006; Ratti et al. Citation2006; Vallega et al. Citation2006; Rubies-Autonell et al. Citation2008). Interestingly, a high proportion of the SBCMV-resistant cultivars identified were derived from the SBCMV-resistant cv. Edmore that was bred in the USA about 30 years ago. This thus suggests that this cultivar and its derivatives carry a major gene or gene-block for SBCMV resistance.
In bread wheat, the genetic analysis of furovirus resistance has identified up to three major loci (Shaalan et al. Citation1966; Dubey et al. Citation1970; Modawi et al. Citation1982; Merkle and Smith Citation1983; Barbosa et al. Citation2001; Bass et al. Citation2006). Sbm1 is located on chromosome 5D and has been shown to be effective against SBWMV in North America (Narasimhamoorthy et al. Citation2006) and against SBCMV in Europe (Bass et al. Citation2006).
In durum wheat, extensive surveys were performed to assess reactions to SBCMV in a wide panel of germplasm (Vallega et al. Citation1999; Ratti et al. Citation2006; Rubies-Autonell et al. Citation2008) and a major gene located on chromosome 2B has been recently identified in a segregating population, together with some genes that have small, but statistically significant effects on resistance to SBCMV (Ratti et al. Citation2009; Maccaferri et al. Citation2011). Apart these indications, little information is available on genetic determinants of SBCMV resistance in durum wheat and the identification of new sources of resistance and of molecular markers that are tightly linked to resistance genes is needed. Breeding for resistance to SBCMV based on phenotypic analysis is extremely disadvantageous, due to the difficulty to obtain spatially uniform field infections that are repeatable over time. The use of markers in marker-assisted selection (MAS) programs would, therefore, be extremely useful for the development of virus-resistant genotypes.
The main objectives of this study were to identify molecular markers linked to the locus specifying resistance to SBCMV in the durum wheat cv. Neodur (a cv. Edmore derivative), to validate these markers in a set of further cultivars previously scored for SBCMV resistance and susceptibility under field conditions and to find candidate genes for genetic determinants of resistance which might help in cloning the quantitative trait loci (QTLs) for resistance to SBCMV.
Materials and methods
Plant material
A population of 146 F8 recombinant inbred lines (RILs) was obtained from a cross between durum wheat cultivars Neodur (a cv. Edmore derivative; highly resistant to SBCMV) and Cirillo (highly susceptible to SBCMV) by advancing random individual F2 plants to the F7 generation by single-seed descent. After the last selfing, each line was bulk harvested to provide seeds for field experiments and DNA extraction.
A group of 25 durum wheat cultivars besides Cirillo and Neodur () that were previously scored for SBCMV resistance under field conditions (Ratti et al. Citation2006) were genotyped using the microsatellite markers flanking the resistance QTL identified in this study.
Table 1. Analysis of durum wheat cultivars, previously scored for resistance and susceptibility under field conditions, with SSR markers flanking the QTL (Vallega et al. 2003; Ratti et al. 2006; Rubies-Autonell et al. 2008).
Evaluation of resistance to SBCMV
The RIL population and the two parents were evaluated for SBCMV resistance during the 2007–2008 growing season, in a field characterized by severe and uniform SBCMV infection, which is located at Cadriano (BO; 44°35′N 11°27′E; northern Italy). Genotypes were sown on 7 November 2007, in plots of 2.0 m2 that were distributed according to a randomised complete block design, with three replicates. Cultivar Grazia, well known for its high susceptibility to SBCMV (Vallega et al. Citation2003), as well as the two parents, was included in the field trial and inserted at regular intervals (every seven RILs) in each replication, to monitor infection levels and homogeneity throughout the trial field. The sowing density was 500 seeds m2. The previous crop had been durum wheat and standard agronomic management practices were applied during the growing season. Plants were not treated against fungal diseases; mild infections of Fusarium spp. occurred towards the end of the growing cycle.
Visual evaluation of SBCMV symptoms and enzyme-linked immunosorbent assay (ELISA) were used as indicators of resistance/susceptibility (Hunger and Sherwood Citation1985). Symptom severity (SS) was scored in the field on three dates in 2008, 11 March, 27 March, and 15 April (corresponding to growth stages 27, 30, and 36 in the Zadoks scale; Zadoks et al. Citation1974), on a 0 to 4 scale. Values from 0 to 1.0 indicated no symptoms or slight symptoms; from 1.1 to 2.0 mild mottling and stunting; from 2.1 to 3.0 mottling and stunting; and from 3.1 to 4.0 severe mottling and stunting with virus-killed plants (Vallega and Rubies-Autonell Citation1985).
The relative virus concentrations (E) were determined on homogenized extracts from leaves collected on 11 March and 15 April in 2008 (growth stages 30 and 36 in the Zadoks scale; Zadoks et al. Citation1974), using a double antibody sandwich enzyme-linked immunosorbent assay (DAS-ELISA), according to the procedure described by Vallega et al. (Citation2003). Extracts were obtained from the bulked basal halves of the second and third youngest leaves of 10 randomly chosen plants/plot, with the dead parts in the leaf samples removed. The bulked leaf samples from each plot were processed separately. The antiserum was prepared using SBCMV purified from infected plants of the durum wheat cv. Grazia. Infection by wheat spindle streak mosaic virus (WSSMV), a soil-borne pathogen that is also present in Italy (Rubies-Autonell and Vallega Citation1987), was excluded after ELISA and real-time PCR indexing. At maturity, genotypes were characterized for agro-morphological traits. Plant height (cm) was measured during the milk waxy maturation, from the ground to the tip of the ear (excluding awns) on five main culms per plot. The plots were harvested mechanically early in June and the grain yield (t ha−1) was determined. The thousand kernel weight and the test weight were measured and expressed as g and kg hl−1, respectively. The test weight measurements were obtained using a Shopper chondrometer equipped with a 1 L-sized container.
DNA extraction and linkage analysis
DNA from the parents and the RILs were extracted following the cetyltrimethylammonium bromide (CTAB)-based method (Hoisington et al. Citation1994). A total of 674 simple sequence repeat (SSR) and 50 sequence-tagged site (STS) markers (Xue et al. Citation2008) were initially used to screen for polymorphisms between cultivars Neodur and Cirillo. The SSRs used belong to the following marker groups: Gatersleben wheat microsatellites (GWM; Röder et al. Citation1998; M. Röder, personal communication); Wheat Microsatellite Consortium (WMC; Gupta et al. Citation2002); Beltsville Agricultural Research Centre (BARC; Song et al. Citation2005); Colorado wheat Expressed Sequence Tag (EST)-derived microsatellites (CWEM; Peng et al. Citation2005); Du Pont wheat (DuPw; Eujayl et al. Citation2002); Clermont-Ferrand D genome (CFD; www.graingenes.com) and Clermont-Ferrand A genome (CFA; Sourdille et al. Citation2003). PCR reactions were performed as reported by Marone et al. (Citation2009). The amplification products were analyzed using capillary electrophoresis (ABI3130), with multiplexing of different fluorescent dyes. Electropherograms were analyzed with GeneMapper version 4.0.
Diversity arrays technology (DArT) markers (Wenzl et al. Citation2004; Akbari et al. Citation2006) were generated by Triticarte Pty Ltd (Canberra, Australia; http://www.triticarte.com.au), which is a whole-genome profiling service laboratory. For the DArT assay, the DNA from the mapping population was subjected to PstI/TaqI digestion and size purification, and probed against the durum DArT array at the Triticarte laboratory. Individual genotypes were scored for the presence (1) or absence (0) of hybridization based on fluorescence signal intensities. A total of 208 PCR-based and 218 DArT markers were used to construct a linkage map using the Kosambi mapping function within the JoinMap 4 software (van Ooijen and Voorips Citation2004) considering a minimum limit of detection (LOD) score of 4.0. Four-hundred and fourteen markers were incorporated into the map, while the remaining 12 were considered as unlinked. The goodness of fit for all loci to an expected 1:1 segregation ratio was tested using chi-squared analysis. Linkage groups were assigned to chromosomes by comparing the marker positions with the previously published wheat maps (Blanco et al. Citation1998, Citation2004; Korzun et al. Citation1999; Elouafi et al. Citation2001; Nachit et al. Citation2001; Elouafi and Nachit Citation2004; Xue et al. Citation2008) and with the hexaploid wheat SSR consensus map (Somers et al. Citation2004).
Statistical analysis and QTL detection
Data from the field experiment were analyzed using an ANOVA test, the homogeneity of the phenotypic variance between replications was verified, and the means were separated by Fischer's protected least significant difference at p < 0.05 for all traits, to test the differences between each RIL and the two parents. All data were statistically analyzed using a statistical software package (Statistica, Statsoft Inc., Tulsa, OK, USA). QTL analysis was performed using the software package MapQTL® version 5.0 (Van Ooijen Citation2004): the LOD profiles from simple interval mapping (SIM) were inspected and the marker closest to each LOD peak was selected as the cofactor to perform the multiple QTL mapping (MQM) analysis further. Minor QTLs were searched by taking into consideration 64 lines with the unbroken Cirillo (susceptible) molecular haplotype at the major SBCMV resistance QTL to eliminate its effect. The markers used as cofactors were wPt-2106 on chromosome 2B, Xgwm376 and wPt-2720 on chromosome 3B, and wPt-2883 on chromosome 7B. The LOD significance threshold levels of the respective traits were calculated with the permutation test option provided in MapQTL, using 10,000 permutations.
Results
Phenotypic evaluation
Means of the parental lines, means and ranges of the RIL population, are reported for each trait in . ANOVA indicated that the differences between the parents and among the 146 RILs were significant for all traits considered (p<0.001). The SBCMV infection levels throughout the field trial were severe and uniform, as indicated by the limited range of the mean SS and the mean E values observed for the check cv. Grazia ().
Table 2. Phenotypic variations among the parental lines and RIL population for the agronomic traits evaluated under natural SBCMV infection in the field.
The mean SS of the two parents differed significantly in all field observations; indeed, the mean SS over the three scoring dates were 0.28 for the resistant cv. Neodur, and 3.10 for the susceptible cv. Cirillo, with the latter very similar to that of the susceptible check Grazia (2.87). The mean E values also confirmed the phenotypic differences observed between the two parents ().
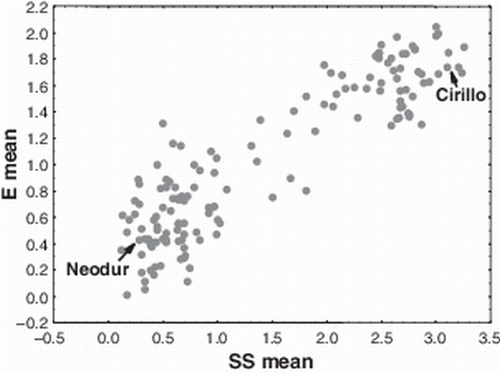
When the segregating population was considered, a wide range of reactions to SBCMV was observed among the RILs, both in terms of SS (0.1 to 3.25) and E values (0.0 to 2.05) (). The frequency distribution of the RILs (mean values over the three and two sampling days for SS and E values, respectively) was fitted with bimodal curves for both of these traits, which suggested a simple genetic basis for SBCMV resistance (). Indeed, as can be seen in , which shows the correlation between mean SS and mean E values (r=0.90, p<0.001), most of the RILs grouped into two clearly distinguishable clusters based on their responses in terms of both mean SS and mean E value. On the other hand, the two frequency distributions and the mean SS versus mean E values correlation suggest the presence of RILs with intermediate SBCMV reactions as well as a general tendency to express less extreme responses than their parents.
Figure 1. Frequency distributions of the average values over the three and two dates for SS (a) and E value (b), respectively, in the RIL population derived from the cross Cirillo×Neodur.
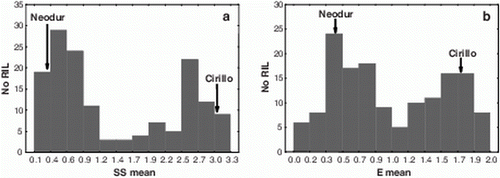
Figure 2. Correlation between mean SS and E values recorded in the population derived from the cross Cirillo×Neodur.
Based on the SS evaluation, 82 RILs were classified as resistant (SS=0.0–1.25), while 64 were classified as susceptible (SS=1.26–3.25). The resulting segregation ratio was statistically different from the expected 1:1 ratio (p< 0.05), based on chi-squared analysis, with more resistant genotypes than susceptible ones.
A number of agronomic traits whose expression is heavily affected by virus infection were also evaluated (). The mean grain yield of the resistant parental cv. Neodur (3.65 t ha−1) was about threefold higher than that of the susceptible cv. Cirillo (1.17 t ha−1). The mean grain yield of the susceptible check Grazia was 1.39 t ha−1. Neodur was also characterized by significantly higher values for plant height and test weight, with respect to Cirillo (). These differences probably reflect in part the decrease in productivity and in plant height due to the impact of SBCMV on the susceptible cultivar. Indeed, symptom scores above 3.0 were associated with mean grain yield and mean plant height reductions of 48.2 and 18.5%, respectively, when compared with the resistant genotypes (SS < 1).
Linkage mapping and QTL analysis
A total of 202 PCR-based and 212 DArT markers were assigned to 30 linkage groups. All chromosomes were represented; for four chromosomes a single linkage group was identified, while the remaining chromosomes were associated to two or three linkage groups. Globally, each chromosome was covered by a number of markers ranging from 8 to 51. The final map for QTL analysis consisted of 414 markers covering 1917 cM, with chromosome 6B having the greatest coverage, with 51 markers, covering about 183 cM.
This map was used to perform MQM analyses, which confirmed the presence of a major SBCMV resistant locus on the short arm of chromosome 2B within the 40-cM interval between markers Xwmc764 and Xgwm1128, with wPt-2106 as the peak marker (). The LOD values were very high, ranging from 19.4 to 37.8 for SS, and from 11.2 to 19.8 for E values, depending on the dates of the observations. In general, greater effects were seen for the third date for SS and on the first date for E values. The percentages of explained variability ranged from 51.8 to 87.6% for SS and from 32.9 to 49.1% for E values. A positive additive effect (from 0.6 to 1.23 in terms of SS score) was found for the Neodur allele, which was effective in increasing resistance ().
Figure 3. Partial genetic map of the region on chromosome 2B carrying the major QTL controlling SS and E value registered on 15 April, grain yield and plant height, and the corresponding curves of the LOD values obtained by MQM analysis. LOD significance threshold, calculated by permutation test, was 3.0 (p <0.05).
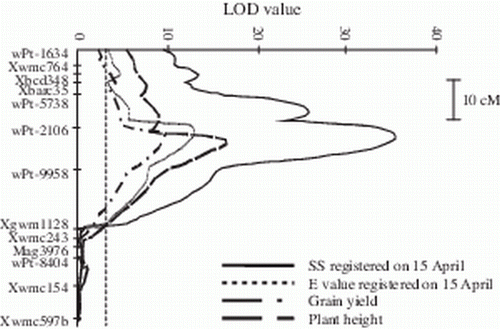
Table 3. QTLs detected in the Cirillo×Neodur RIL population for SS (11 March, 27 March and 15 April), E values (11 March and 15 April), grain yield and plant height by MQM analysis.
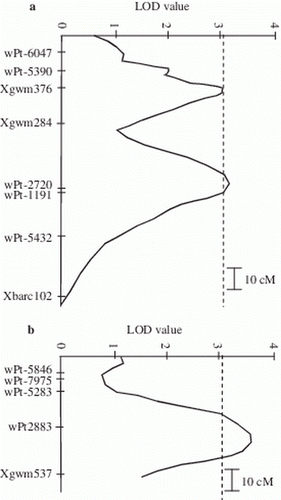
Minor QTLs were searched by taking into consideration 64 lines with the unbroken Cirillo (susceptible) molecular haplotype at the major SBCMV resistance QTL to eliminate its effect. Three significant minor QTLs were identified for SS3 (symptom severity evaluated on 15 April), two out of them located on the long arm of chromosome 3B (LOD values 3.1 and 3.2, peak markers Xgwm376 and wPt-2720, respectively) and one positioned on the short arm of chromosome 7B (LOD value 3.6, peak marker wPt-2883 – and ). They explained from 30.4 to 40.1% of observed variability in the 64 RILs. Also for these minor QTLs, the allele of Neodur was effective in increasing resistance.
Figure 4. Partial genetic map of the regions on chromosomes 3B (a) and 7B (b) carrying the minor QTLs for SS registered on 15 April, and the corresponding curves of the LOD values. LOD significance threshold, calculated by permutation test, was 3.1 (p <0.05).
QTL analysis was also carried out for the agronomic traits aiming to confirm the effect of virus susceptibility on agronomic traits. A QTL for grain yield and plant height was identified on chromosome 2BS, in coincidence with the QTL for resistance to SBCMV; the LOD values were 9.1 for grain yield (27.7% explained variability), and 13.9 for plant height (39.5% explained variability). As Cirillo and Neodur are characterized by similar yield performance and plant height in absence of virus infection as shown by the results of the national durum wheat yield trials (www.cerealicoltura.it), the negative additive effect for these traits indicated the tendency of the Cirillo allele for decreased grain yield and plant height due to its susceptibility to SBCMV. These data confirm the outcome of the correlation analysis, and indicate a strong effect of the susceptibility to SBCMV on grain yield and plant height, but not on test weight, for which no QTLs in the region of chromosome 2BS were identified ().
Identification of candidate genes for resistance to SBCMV in Neodur
The sequence information available for the DArT (www.triticarte.com.au) and the EST-derived SSR markers employed to construct the Cirillo×Neodur linkage map have allowed to establish a syntenic relationship between the durum wheat and the rice genome for the region corresponding to the QTL on wheat chromosome 2BS. A BLASTX search was carried out for the EST-based Xbcd348 and Mag3976 markers, positioned upstream and downstream the major QTL. The sequence of the first marker identified the rice locus Os04g0129500, located on the chromosome 4, and putatively coding for a Sec23/Sec24 zinc finger protein (E value 6e-16, with 100% identity on a segment of 45 aa). This result confirmed the syntenic correspondence previously found between the chromosome 4 of rice and the bin positioned in the telomeric region of the 2BS (2BS3-0.84-1.00) in wheat (http://wheat.pw.usda.gov/wEST/binmaps/wheat2_rice.html). The BLASTX search carried out with the Mag3976 marker as a query identified with good probability only a protein sequence of barley (BAJ96737, E value 5e-24, 81% identity in a region of 67 aa). The barley sequence was then used as a query in a BLASTP search to find the most similar genes in rice. Two rice sequences were identified, both with an E value 0.0, and one of them was localized on the chromosome 4 (Os04g0352400), nearly 15,000 kb far away from the Os04g0129500 locus (http://mips.helmholtz-muenchen.de/plant/rice/). Therefore, the region between the loci Os04g0129500 and Os04g0352400 on the chromosome 4 of rice might correspond to the region genetically delimited by the markers Xbcd348 and Mag3976 on the chromosome 2B in durum wheat ().
Figure 5. Partial genetic map of the region on chromosome 2B carrying the major QTL for SBCMV resistance and the correspondence with putative synthenic loci on rice chromosome 4. *This marker was mapped in the Meridiano×Claudio population by Maccaferri et al. (2011).
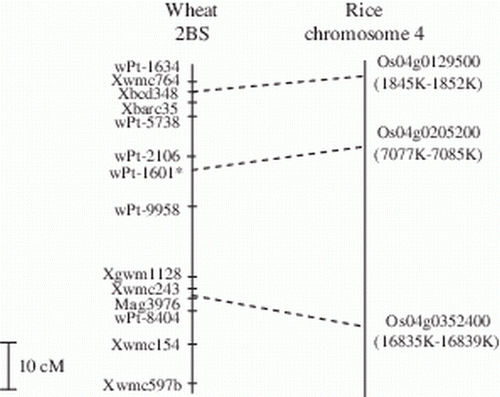
The nucleotide sequence of the QTL peak marker wPt-2106 on 2BS in the Cirillo×Neodur map is not available and other markers located in the same region were, therefore, investigated. An interesting result was achieved with the DArT marker (wPt-1601) positioned 1 cM apart from wPt-2106 by Maccaferri et al. (2011) in the durum wheat map Meridiano×Claudio. The BLASTX search performed using the 508 bp sequence of the wPt-1601 marker (http://www.diversityarrays.com/sequences.html) identified a barley sequence corresponding to a putative Nucleotide-Binding Site – Leucin-Rich Repeat (NBS-LRR) protein (BAJ96737). The E value in this alignment was not very high (2e-2, 41% identity for a region of 62 aa), probably because of the low quality of the sequence (10% of bases called as ‘n’); nevertheless, a BLASTP search carried out using the BAJ96737 barley sequence as query identified a locus on the chromosome 4 of rice, Os04g0205200, coding for a putatively NBS-LRR protein positioned within the interval Os04g0129500–Os04g0352400 ().
A similar approach was adopted for the minor QTLs identified in this work. A relevant result was obtained with the wPt-1191 marker, for which a 602 bp long sequence was available (http://www.diversityarrays.com/sequences.html). A BLASTN search performed against the draft assembly of the gene-rich regions of the bread wheat Chinese spring genome, available in the cerals database (cerealsDB – http://www.cerealsdb.uk.net/index.htm) has identified a 2873 bp long genomic contig which revealed to be highly similar to a barley protein (BAJ90725, E value 7e-72 with 78% identities on 196 aa following BLASTX search), encoding a putative NBS-LRR protein.
Validating of identified SSR markers linked to the major QTL on chromosome 2BS to predict resistance to SBCMV in durum wheat cultivars
The SSR markers flanking the QTL identified in the Cirillo×Neodur mapping population, Xbcd348 and Xbarc35 upstream and Xgwm1128 downstream were analyzed on a panel of durum wheat cultivars previously tested for resistance to SBCMV.
Xbcd348 showed a simple amplification pattern: two peaks of 330 and 334 bp were amplified in Neodur, while only the 330 bp fragment was amplified in Cirillo. All resistant cultivars shared the amplification profile of Neodur, except Ionio and AC Melita, which were characterized by a null allele and the same allele found in Cirillo, respectively. Six of the susceptible genotypes had the allele of Cirillo, while seven had the allele of Neodur. Ciccio was characterized by a null allele.
The second marker, Xbarc35, produced a very complex profile with three or four different peaks amplified in each genotype (368 bp, 371 bp, and 374 bp in Neodur; 359 bp, 377 bp, 382 bp, and 391 bp in Cirillo). All of the resistant cultivars except Ionio, Cappelli, and AC Melita showed the amplification profile of Neodur. Among the susceptible varieties, Grazia and Varano shared a common amplification profile with Cirillo; Karel and Fortore showed the same peaks found in Neodur and the remaining cultivars were characterized by different combinations of peaks of 361, 371, 377, and 379 bp.
The third marker, Xgwm1128, yielded a peak of 158 bp in Neodur, and of 145 bp in Cirillo. The allele of Neodur at the locus Xgwm1128 was identified in all resistant cultivars except Capeiti and San Carlo, where the Cirillo allele was found, and Saragolla was characterized by a different allele of 148 bp. All susceptible cultivars were characterized by the Cirillo allele, except Ciccio, Karel, and Varano, which showed null alleles, and Messapia showed a different allele (154 bp). Due to their simpler amplification profiles compared to Xbarc35, only the patterns of Xbcd348 and Xgwm1128 are reported in .
Discussion
Obtaining reliable data on reactions to SBCMV is dependent on an adequate distribution of SBCMV within the experimental field and on the appropriate environmental conditions for infection. Although they refer to one year, the field evaluation of the Cirillo×Neodur segregating population was carried out under optimal conditions. The uniform distribution of SBCMV throughout the field was confirmed by replicating genotypes, as well as by monitoring of the widespread planting of the susceptible check (cv. Grazia). Thus, all genotypes were subjected to approximately the same pathogen pressure under the same conditions and at the same developmental stage. Indeed, the parents of the segregating population and lines were characterized by nearly the same phenology, with Neodur slightly later than Cirillo in terms of flowering date (data not shown). Finally, the visual scoring of SS was accompanied by the evaluation of the SBCMV levels according to the presence of the viral coat-protein, as revealed by ELISA.
The correlation between the mean SS and the mean E values was high and statistically significant in the RIL population, which indicates that the visual score of SS in the field was generally confirmed by the laboratory tests. Nevertheless, a few resistant genotypes, including cv. Neodur, were characterized by intermediate E values, and hence of virus concentration, in their leaves, as 1.0 to 1.5, and a clear resistance in terms of field evaluation (SS < 1). The occurrence of some intermediate phenotypes following the visual scoring and ELISA evaluation was also clear from the correlation plot (), which suggests the presence of a major gene and of other genes with minor effects that contribute to SBCMV resistance in the Cirillo×Neodur segregating population.
The major gene identified in the present study explained up to 87.6% of the observed variability for resistance/susceptibility to SBCMV in terms of SS, and up to 49.1% in terms of E value. These are relevant results, although a certain overestimation is possible since the phenotypic data are from one environment only. This gene thus represents a very good target for the resistant breeding programs.
The locus identified in the present study might correspond to a major QTL that was reported in a preliminary study in the same chromosomal region in bread wheat (Sbm2) by Bayles et al. (Citation2007) and, in a recent work, in the durum wheat cv. Meridiano by Maccaferri et al. (2011). Considering the common peak marker, wPt-2106, and the common resistant ancestor (Cappelli) of the cultivars Neodur and Meridiano, the gene for resistance to SBCMV in Neodur might be the same as that identified in Meridiano, even if more than one gene might be located in this region, as suggested by the predominance of resistant genotypes with respect to susceptible ones observed in the Cirillo×Neodur population as well as in the population studied by Maccaferri et al. (2011).
Three minor QTLs were also identified for SS3 on chromosomes 3B and 7B, all contributed by Neodur. Also these regions could correspond to those identified by Maccaferri et al. (2011) in the Meridiano×Claudio population based on common molecular markers, with similar LOD and percentage of explained variability values.
Since EST-derived SSR and STS markers were utilized to develop the Cirillo×Neodur map, and the sequence of 2000 wheat DArT clones has recently been released (http://www.diversityarrays.com/sequences.html), a BLAST search was carried out to find candidate genes co-mapping with the resistant QTLs. Based on sequence similarity and colinearity data between wheat and rice, we propose that the markers wPt-1191 and wPt-1601 belong to genes coding for NBS-LRR proteins and might represent candidate genes for one of the two minor QTLs on chromosome 3B and for the major QTL on 2BS, respectively. Many members of NBS-LRR class of resistance genes have been shown to play a key role in plant resistance against viruses. Encoded proteins operate through a ‘gene-for-gene’ recognition of pathogen avr factors, which for viruses consists of a number of virus gene products. Interestingly, all the virus resistance NBS-LRR gene products lack a transmembrane domain, consistent with the intracellular lifestyle of viruses and the location of the avr products (Maule et al. Citation2007). NBS-LRR genes often occur in clusters in resistance loci (Lehmann Citation2002); other two sequences coding for NBS-LRR proteins (Os04g0219600 and Os04g0312000) are indeed present in the rice region of the chromosome 4 corresponding to the major QTL for SBCMV resistance on wheat 2BS. Even if the identified genes are not involved in resistance to SBCMV, they are strictly associated with the factor responsible of resistance, representing, therefore, a good starting point for the fine mapping and cloning of resistance QTLs based on co-linearity information to the rice genome.
The association between molecular markers and genes for resistance to various pathogens identified in a particular RIL population might not be conserved when analyzed in different genetic backgrounds (Bass et al. Citation2006). The diagnostic value of the Xbcd348 and Xgwm1128 markers associated with resistance to SBCMV was tested in a panel of 13 resistant and 14 susceptible durum wheat cultivars (). The Xbcd348 marker was effective in predicting resistance (all resistant genotypes except one showed the same allele carried by Neodur), although it was less effective in indicating susceptibility. When different genetic backgrounds were tested with the Xgwm1128 marker, 10 cultivars carrying the allele of Neodur at this locus had previously been described as resistant, and only one cultivar carrying the Neodur allele had been classified as susceptible. When both of the Xbcd348 and Xgwm1128 markers were considered, nearly all varieties showing the same allele as Neodur at the two loci had the resistant phenotype. The only exception was Agridur, which is a cultivar derived from Edmore, where a double recombination event broke the linkage between the resistance gene and the flanking markers. The distance between these markers justifies the occurrence of double recombination events, and alleles similar to cv. Neodur are expected to be found when sequence-specific markers will be developed from the sequence of the DArT markers positioned at the peak of the QTL. The scarcity of PCR-based molecular markers in the region where the major QTL for resistance was identified in the present study is a common feature of a number of linkage maps that have been developed in both bread and durum wheat (Huynh et al. Citation2008; Mantovani et al. Citation2008; Suprayogi et al. Citation2009; Genc et al. Citation2010); therefore, although not optimal, the Xbcd348 and Xgwm1128 markers identified in the present study represent a useful panel for the prediction of resistance/susceptibility to SBCMV in durum wheat.
An interesting question concerns the origin of the resistance to SBCMV identified in the present study and its occurrence in the other cultivars that are all characterized by a common ancestor. Based on the pedigree data reported in , Cappelli (resistant to SBCMV) is a common ancestor for all of the resistant genotypes analyzed here, except for San Carlo, which might be characterized by a different resistance locus. Cappelli also shares the same alleles as Neodur at the SBCMV-resistant flanking loci. Therefore, it is likely that Cappelli represents the original source of the resistance to SBCMV in durum wheat. In these terms, the African population from which Cappelli was selected at the beginning of the twentieth century might have provided the resistance to SBCMV to the derivate genotypes bred independently in Italy, USA, Mexico, Canada, and France. The wide use of Cappelli during the first half of the twentieth century in the Italian (and worldwide) breeding programs has been well described (Bozzini et al. Citation1998; Maccaferri et al. Citation2003). A large portion of the molecular variation detected among modern cultivars (41.5%) can be traced back to alleles present in the ancient founder Cappelli (Maccaferri et al. Citation2003). Furthermore, bread and durum genotypes probably share the source of this resistance, considering that the resistant locus Sbm2 was mapped on chromosome 2BS near the Xbarc35 marker (Bayles et al. Citation2007), the same marker that is associated with SBCMV resistance in the Cirillo×Neodur population.
In conclusion, the Xbcd348 marker, in combination with Xgwm1128, represents a useful tool for recovering resistance in crosses derived from Neodur, and predicting resistance/susceptibility in durum wheat varieties. In perspective, the availability of information about DArT sequences can be used to develop new PCR-based markers that are more effective for MAS for SCBMV resistance in wheat.
Acknowledgements
This study was supported by the Ministero dell'Università e della Ricerca (MiUR) of Italy, as special grant AGROGEN.
References
- Akbari , M , Wenzl , P , Caig , V , Carlig , J , Xia , L , Yang , S , Grzegorz , U , Volker , M , Lehmensiek , A Kuchel , H . 2006 . Diversity arrays technology (DArT) for high-throughput profiling of the hexaploid wheat genome . Theor Appl Genet. , 113 : 1409 – 1420 .
- Barbosa , MM , Goulart , LR , Prestes , AM and Juliatti , FC . 2001 . Genetic control of resistance to soil-borne wheat mosaic virus in Brazilian cultivars of Triticum aestivum L. Thell . Euphytica. , 122 : 417 – 422 .
- Bass , C , Hendley , R , Adams , MJ , Hammond-Kosack , KE and Kanyuka , K . 2006 . The Sbm1 locus conferring resistance to soil-borne cereal mosaic virus maps to a gene-rich region on 5DL in wheat . Genome. , 49 : 1140 – 1148 .
- Bayles R, O'Sullivan D, Lea V, Freeman S, Budge G, Walsh K. 2007 . Controlling soil-borne cereal mosaic virus in the UK by developing resistant wheat cultivars . HGCA Project 2616. HGCA Crop Res News. 32:PR418 .
- Blanco , A , Bellomo , MP , Cenci , A , De Giovanni , C , D'Ovidio , R , Iacono , E , Laddomada , B , Simeone , R , Tanzarella , OA and Porceddu , E . 1998 . A genetic linkage map of durum wheat . Theor Appl Genet. , 97 : 721 – 728 .
- Blanco , A , Simeone , R , Cenci , A , Gadaleta , A , Tanzarella , OA , Porceddu , E , Salvi , S , Tuberosa , R , Figliuolo , G Spagnoletti , P . 2004 . Extension of the ‘Messapia x dicoccoides’ linkage map of Triticum turgidum (L.) . Thell Cell Mol Biol Lett. , 9 : 529 – 541 .
- Bozzini , A , Corazza , L , D'Egidio , MG , Di Fonzo , N , La Fiandra , D , Pogna , NE and Poma , I . 1998 . “ Durum wheat (Triticum turgidum spp. durum) ” . In Italian contribution to plant genetics and breeding , Edited by: Scarascia Mugnozza , GT and Pagnotta , MA . 181 – 194 . Italy : Viterbo .
- Budge , GE , Ratti , C , Rubies-Autonell , C , Lockley , D , Bonnefoy , M , Vallega , V , Pietravalle , S and Henry , CM . 2008 . Response of UK winter wheat cultivars to soil-borne cereal mosaic and wheat spindle streak mosaic viruses across Europe . Eur J Plant Pathos. , 120 : 259 – 272 .
- Canova , A . 1964 . Ricerche sulle malattie da virus delle graminacee. I Mosaico del Frumento trasmissibile attraverso il terreno . Phytopatol Medit. , 3 : 86 – 94 .
- Diao , A , Chen , J , Ye , R , Zheng , T , Yu , S , Antoniw , JF and Adams , MJ . 1999 . Complete sequence and genome properties of Chinese wheat mosaic virus, a new furovirus from China . J Gen Virol. , 80 : 1141 – 1145 .
- Dubey , SN , Brown , CM and Hooker , AL . 1970 . Inheritance of field reaction to soil-borne wheat mosaic virus . Crop Sci. , 10 : 93 – 95 .
- Elouafi , I , Nachit , MM and Martín , LM . 2001 . Identification of a microsatellite on chromosome 7B showing a strong linkage with yellow pigment in durum wheat (Triticum turgidum L. var. durum) . Hereditas. , 135 : 255 – 261 .
- Elouafi , I and Nachit , MM . 2004 . A genetic linkage map of the Durum x Triticum dicoccoides backcross population based on SSRs and AFLP markers, and QTL analysis for milling traits . Theor Appl Genet. , 108 : 401 – 413 .
- Eujayl , I , Sorrells , ME , Baum , M , Wolters , P and Powell , W . 2002 . Isolation of EST-derived microsatellite markers for genotyping the A and B genomes of wheat . Theor Appl Genet. , 104 : 399 – 407 .
- Genc , Y , Oldach , K , Verbyla , AP , Lott , G , Hassan , M , Tester , M , Wallwork , H and McDonald , GK . 2010 . Sodium exclusion QTL associated with improved seedling growth in bread wheat under salinity stress . Theor Appl Genet. , 121 : 877 – 894 .
- Gupta , PK , Balyan , HS , Edwards , KJ , Isaac , P , Korzun , V , Röder , M , Gautier , MF , Joudrier , P , Schlatter , AR Dubcovsky , J . 2002 . Genetic mapping of 66 new microsatellite (SSR) loci in bread wheat . Theor Appl Genet. , 105 : 413 – 422 .
- Hariri , D and Meyer , M . 2007 . A new furovirus infecting barley in France closely related to the Japanese soil-borne wheat mosaic virus . Eur J Plant Pathol. , 118 : 1 – 10 .
- Hoisington D, Khairallah M, González-de-León D. 1994 . Laboratory protocols: CIMMYT Applied Molecular Genetics Laboratory , 1st ed. . Mexico City : CIMMYT .
- Hunger , RM and Sherwood , JL . 1985 . Use of symptomatology and virus concentration for evaluating resistance to wheat soil-borne mosaic virus . Plant Dis. , 69 : 848 – 850 .
- Huynh , BL , Wallwork , H , Stangoulis , JCR , Graham , RD , Willsmore , KL , Olson , S and Mather , DE . 2008 . Quantitative trait loci for grain fructan concentration in wheat (Triticum aestivum L.) . Theor Appl Genet. , 117 : 701 – 709 .
- Kanyuka , K , Ward , E and Adams , MJ . 2003 . Polymyxa graminis and the cereal viruses it transmits: a research challenge . Mol Plant Pathol. , 4 : 1 – 14 .
- Kendall , TL and Lommel , SA . 1988 . “ Fungus-vectored viruses of wheat in Kansas ” . In Viruses with fungal vectors , Edited by: Cooper , JI and Asher , MJC . 37 – 60 . Lavenham : The Lavenham Press Ltd .
- Korzun , V , Röder , MS , Wendehake , K , Pasqualone , A , Lotti , C , Ganal , MW and Blanco , A . 1999 . Integration of dinucleotide microsatellites from hexaploid bread wheat into a genetic linkage map of durum wheat . Theor Appl Genet. , 98 : 1202 – 1207 .
- Lehmann , P . 2002 . Structure and evolution of plant disease resistance genes . J Appl Genet. , 43 : 403 – 14 .
- Maccaferri , M , Sanguineti , MC , Donini , P and Tuberosa , R . 2003 . Microsatellite analysis reveals a progressive widening of the genetic basis in the elite durum wheat germplasm . Theor Appl Genet. , 107 : 783 – 797 .
- Maccaferri M, Ratti C, Tuberosa R, Rubies-Autonell C, Vallega V, Sanguineti MC. 2005a . A durum wheat germplasm collection suitable for gene discovery via association mapping. Proceedings of the International Symposium on Genomics-based Plant Germplasm Research . April, 2005 Bejing, China. 47 – 57 .
- Maccaferri , M , Sanguineti , MC , Noli , E and Tuberosa , R . 2005b . Population structure and long-range linkage disequilibrium in a durum wheat elite collection . Mol Breed. , 15 : 271 – 289 .
- Maccaferri , M , Ratti , C , Rubies-Autonell , C , Vallega , V , Demontis , A , Stefanelli , S , Tuberosa , R and Sanguineti , MC . 2011 . Resistance to soil-borne cereal mosaic virus in durum wheat is controlled by a major QTL on chromosome arm 2BS and minor loci . Theor Appl Genet. , 123 : 527 – 544 .
- Mantovani , P , Maccaferri , M , Sanguineti , MC , Tuberosa , R , Catizone , I , Wenzl , P , Thomson , B , Carling , J , Eric , H De Ambrogio , E . 2008 . An integrated DArT-SSR linkage map of durum wheat . Mol Breed. , 22 : 629 – 648 .
- Marone , D , Del Olmo , AI , Laidò , G , Sillero , JC , Emeran , AAM , Russo , MA , Ferragonio , P , Giovanniello , V , Mazzucotelli , E De Leonardis , AM . 2009 . Genetic analysis of durable resistance against leaf rust in durum wheat . Mol Breed. , 24 : 25 – 39 .
- Maule , AJ , Caranta , C and Boulton , MI . 2007 . Sources of natural resistance to plant viruses: status and prospects . Mol Plant Pathol. , 8 : 223 – 231 .
- Merkle , OG and Smith , EL . 1983 . Inheritance of resistance to soil borne mosaic in wheat . Crop Sci. , 23 : 1085 – 1086 .
- Modawi , RS , Heyne , EG , Brunetta , D and Willis , WG . 1982 . Genetic studies of field reaction to wheat soil-borne mosaic virus . Plant Dis. , 66 : 1183 – 1184 .
- Nachit , MM , Elouafi , I , Pagnotta , MA , El Saleh , A , Iacono , E , Labhilili , M , Asbati , A , Azrak , M , Hazzam , H Benscher , D . 2001 . Molecular linkage map for an interspecific recombinant inbred population of durum wheat (Triticum turgidum L. var. durum) . Theor Appl Genet. , 102 : 177 – 186 .
- Narasimhamoorthy , B , Gill , BS , Fritz , AK , Nelson , JC and Brown-Guedira , GL . 2006 . Advanced backcross QTL analysis of a hard winter wheat×synthetic wheat population . Theor Appl Genet. , 112 : 787 – 796 .
- Peng , JH , Nora , L and Capitan , V . 2005 . Characterization of EST-derived microsatellites in the wheat genome and development of eSSR markers . Funct Integra Genomics. , 5 : 80 – 96 .
- Rao , AS and Brakke , MK . 1969 . Relation of soil-borne wheat mosaic virus and its fungal vector, Polymyxa graminis . Phytopathol. , 59 : 581 – 587 .
- Ratti , C , Pisi , A , Vallega , V and Rubies-Autonell , C . 2005 . Molecular characterization of Italian soil-borne cereal mosaic virus isolates . Parasitica. , 61 : 11 – 16 .
- Ratti , C , Rubies-Autonell , C , Maccaferri , M , Stefanelli , S , Sanguineti , MC and Vallega , V . 2006 . Reaction of 111 cultivars of Triticum durum Desf. from some of the world's main genetic pools to soil-borne cereal mosaic virus . J Plant Dis Protect. , 113 : 145 – 149 .
- Ratti , C , Rubies-Autonell , C , Maccaferri , M , Corneti , S , Stefanelli , S , Sanguineti , MC , Demontis , A , Massi , A and Vallega , V . 2009 . Inheritance of resistance to cereal soil-borne mosaic virus in a durum wheat population of lines derived from the cross ‘Meridiano/Claudio’: results of a two-year study . Annu Wheat Newsl. , 55 : 106 – 108 .
- Röder , MS , Korzun , V , Wendehake , K , Plaschke , J , Tixier , MH , Leroy , P and Ganal , MW . 1998 . A microsatellite map of wheat . Genetics. , 149 : 2007 – 2023 .
- Rubies-Autonell , C and Vallega , V . 1987 . Observations on a mixed soil-borne wheat mosaic virus and wheat spindle streak mosaic virus infection in durum wheat (Triticum durum Desf.) . J Phytopathol. , 119 : 111 – 121 .
- Rubies-Autonell C, Ratti C, Pisi A, Vallega V. 2006 . Behaviour of durum and common wheat cultivars in regard to soil-borne cereal mosaic virus (SBCMV): results of fifteen trials . Proceedings of 12th Congress of the Mediterranean Phytopathological Union; June 2006; Rhodes , Greece , p. 100 – 102 .
- Rubies-Autonell C, Ratti C, Pisi A, Sarti A, Canestrale R, Vallega V. 2008 . Reaction of thirty-four durum wheat cultivars to soil-borne cereal mosaic virus in 2007 . In : Rush CM, Proc of the Seventh Symposium of the International Working Group on Plant Viruses with Fungal Vectors (IWGPVFV), Quedlingburg, Germany, August–September, 2008 . American Society of SB Technologists , Denver . p. 105 – 110 .
- Shaalan , M , Heyne , EG and Sill , WH . 1966 . Breeding wheat for resistance to soil-borne wheat mosaic virus, wheat streak-mosaic virus, leaf rust, stem rust, and bunt . Phytopathol. , 56 : 664 – 669 .
- Shirako , Y , Suzuki , N and French , RC . 2000 . Similarity and divergence among viruses in the genus Furovirus . Virol. , 270 : 201 – 208 .
- Somers , JD , Isaac , P and Edwards , K . 2004 . A high-density microsatellite consensus map for bread wheat (Triticum aestivum L.) . Theor Appl Genet. , 109 : 1105 – 1114 .
- Song , QJ , Shi , JR , Singh , S , Fickus , EW , Costa , JM , Lewis , J , Gill , BS , Ward , R and Cregan , PB . 2005 . Development and mapping of microsatellite (SSR) markers in wheat . Theor Appl Genet. , 110 : 550 – 560 .
- Sourdille , P , Cadalen , T , Guyomarc'h , H , Snape , JW , Perretant , MR , Charmet , G , Boeuf , C , Bernard , S and Bernard , M . 2003 . An update of the Courtot×Chinese spring intervarietal molecular marker linkage map for the QTL detection of agronomic traits in wheat . Theor Appl Genet. , 106 : 530 – 538 .
- Suprayogi , Y , Pozniak , CJ , Clarke , FR , Clarke , JM , Knox , RE and Singh , AK . 2009 . Identification and validation of quantitative trait loci for grain protein content in adapted Canadian durum wheat populations . Theor Appl Genet. , 119 : 437 – 448 .
- Torrance , L and Koenig , R . 2005 . “ Genus furovirus ” . In Virus taxonomy: eighth report of the international committee on taxonomy of viruses , Edited by: Fauquet , CM , Mayo , MA , Maniloff , J , Desselberger , U and Ball , LA . 1027 – 1032 . London : Elsevier Academic Press .
- Vallega , V and Rubies-Autonell , C . 1985 . Reactions of Italian Triticum durum cultivars to soil-borne wheat mosaic . Plant Dis. , 69 : 64 – 66 .
- Vallega , V , Rubies-Autonell , C , Turina , M , Ratti , C and Contoli , S . 1999 . Reactions to SBWMV of durum wheat cultivars grown in northern Italy during 1995–96 . Zeitschrift für Pflanzenkrankheiten und Pflanzenschutz. , 106 : 284 – 290 .
- Vallega , V , Rubies-Autonell , C , Ratti , C , Canestraie , R and Sarti , A . 2002 . Virus del mosaico comune del frumento: comportamento delle varietà di grano duro e tenero. Risultati di prove effettuate nel 1996–97. L . Informatore Agrario. , 58 : 69 – 73 .
- Vallega , V , Rubies-Autonell , C and Ratti , C . 2003 . Reaction of durum wheat cultivars to mixed SBWMV and WSSMV infection in central Italy . Phytopathol Mediterr. , 42 : 177 – 182 .
- Vallega , V , Rubies-Autonell , C and Ratti , C . 2006 . Resistance to accumulation of soil-borne cereal mosaic virus in eight cultivars of Triticum durum Desf . Parasitica. , 62 : 79 – 96 .
- Van Ooijen JW. 2004 . MapQTL 5, Software for the mapping of quantitative trait loci in experimental populations . Wageningen : Kyazma B.V.
- Van Ooijen JW, Voorips RE. 2004 . JoinMap Version 4.0, Software for the calculation of genetic linkage maps . Wageningen : Kyazma B.V .
- Verchot-Lubicz , J . 2005 . A new model for cell-to-cell movement of potexviruses . Mol Plant Microbe Interact. , 18 : 283 – 290 .
- Wenzl , P , Carling , J , Kudrna , D , Jaccoud , D , Huttner , E , Kleinhofs , A and Kilian , A . 2004 . Diversity arrays technology (DArT) for whole-genome profiling of barley . Proc Natl Acad Sci USA. , 101 : 9915 – 9920 .
- Xue , S , Zhang , Z , Lin , F , Kong , Z , Cao , Y , Li , C , Yi , H , Mei , M , Zhu , H Wu , J . 2008 . A high-density intervarietal map of the wheat genome enriched with markers derived from expressed sequence tags . Theor Appl Genet. , 117 : 181 – 189 .
- Zadoks , JC , Chang , TT and Konzak , CF . 1974 . A decimal code for the growth stages of cereals . Weed Res. , 14 : 415 – 421 .