Abstract
Leaf curl disease caused by Cotton Leaf Curl Burewala virus (CLCuBuV) has been recognized as serious threat to cotton in Indian subcontinent. However, information about cotton–CLCuBuV interaction is still limited. In this study, the level of phenolic compounds, total soluble proteins, and malondialdehyde (MDA) and the activities of phenylalanine ammonia-lyase (PAL), peroxidase (POX), catalase (CAT), proteases, superoxide dismutase (SOD), and polyphenol oxidase (PPO) were studied in leaves of two susceptible (CIM-496 & NIAB-111) and two resistant (Ravi and Co Tiep Khac) cotton genotypes. Disease symptoms were mild in the resistant genotypes but were severe in highly susceptible genotypes. The results showed that phenolic compounds, proteins, PAL, POX, CAT, proteases, SOD, PPO, and MDA play an active role in disease resistance against CLCuBuV. The amount of total phenols, proteases, MDA, and PPO was significantly higher in leaves of CLCuBuV-inoculated plants of both resistant genotypes as in non-inoculated plants, and decreased in CLCuBuV-inoculated plants of both susceptible genotypes over their healthy plants. POX, protein content, SOD, and PAL activities showed lower values in resistant genotypes, while they decreased significantly in susceptible genotypes as compared to the noninoculated plants except PAL, which showed non-significant decrease. CAT was found to be increased in both susceptible and resistant genotypes with maximum percent increase in resistant genotype Ravi, as compared to non-inoculated plants. The results showed significantly higher concentrations of total phenols and higher activity of protease, MDA, SOD, and PPO in resistant genotype Ravi after infection with CLCuBuV, suggesting that there is a correlation between constitutive induced levels of these enzymes and plant resistance that could be considered as biochemical markers for studying plant-virus compatible and incompatible interactions.
Introduction
Leaf curl is one of the most common and destructive diseases of the upland cotton in Indian subcontinent and in Africa. Its causative agent has been characterized from Pakistan, India, and Sudan (Azhar et al. Citation2010) and disease was found to be caused by a complex of monpartite begomoviruses and a small symptom modulating, single stranded satellite DNA β component transmitted by the whitefly Bemisia tabaci (Akhtar et al. Citation2013). The begomovirus–betasatellite complexes that cause leaf curl disease in cotton in Asia and Africa are distinct. During the 1990s, the complex in Pakistan and India was shown to consist of multiple begomovirus species (often occurring as multiple infections more than one virus per plant), supporting a disease-specific betasatellite (Cotton leaf curl Multan betasatellite [CLCuMuB]) as well as an alphasatellite (Tahir et al. Citation2011). In 2001, a begomovirus strain that overcame existing resistance was reported in Pakistan (Akhtar et al. Citation2002; Mansoor et al. Citation2003). Recent sequence analysis of DNA showed that only a single begomovirus type is prevalent in Pakistan, in contrast to the situation before its appearance (Tahir et al. Citation2011). This virus is a recombinant that consists of sequences derived from cotton leaf curl Multan virus (CLCuMuV) and cotton leaf curl Kokhran virus (CLCuKV). This newly emerging recombinant is presently designated as cotton leaf curl Burewala virus (CLCuBuV). Only a single type of DNA β satellite, a recombinant mostly originating from CLCuMuB DNA β but with some sequence from a satellite isolated from tomato, is found to be associated with the disease but apparently lacking an alphasatellite (Amrao et al. Citation2010; Tahir et al. Citation2011). Recently, CLCuBuV has been reported from India dominating in many fields (Kumar et al. Citation2011; Rajagopalan et al. Citation2012).
CLCuBuV-infected plants are usually stunted and bushy. Younger leaves of infected plants can show downward cupping, followed by either upward or downward curling of leaf margins, swelling and darkening of veins, which frequently develop into cup-shaped leaf-like out-growths called ‘enations’. Leaves from infected plants become thickened and more brittle than those from healthy plants. Severely infected leaves can show rolling and a reduction in size followed by malformation of leaf petioles, branches, and the main stem (Akhtar et al. Citation2008). The use of resistant varieties is the safest, economical, and effective option to manage, but unfortunately introgressed host plant resistance was rapidly overcome by the resistance-breaking strain CLCuBuV during 2001 and all the available cultivated genotypes from Gossypium hirsutum at this time are susceptible (Akhtar et al. Citation2010). However, interestingly, the native cotton G. arboreum also known as ‘Desi cotton’ and G. herbaceum appears not to be infected with cotton leaf curl disease (CLCuD) till the first inception of disease (Akhtar et al. Citation2010, Citation2013). The fact shows that the area under G. arboreum and G. herbaceum is limited as compared to G. hirsutum, which have high yield and excellent fiber traits (Chandra & Sreenivasan Citation2011; Akhtar et al. Citation2013). However, utilization of disease resistant/susceptible attributes from G. arboreum and G. herbaceum to G. hirsutum genotypes is the demand of time after the emergence and re-emergence of CLCuD in Indian subcontinent (Akhtar et al. Citation2013).
The development of more CLCuD-resistant cotton is necessary to alleviate future threats to cotton in Indian subcontinent. Disease resistance in plants is associated with activation of a wide array of defense responses that slow down or halt infection at certain stages of the host–pathogen interaction. Plants have evolved various preexisting physical and chemical barriers, as well as inducible defense responses that interfere with pathogen establishment (Jones & Dangl Citation2006; Zhao et al. Citation2008; Vanitha et al. Citation2009). However, this requires comprehensive studies and understanding of the adaptive mechanisms and responses to CLCuBuV infection that allows survival of cotton plant. Because viruses cannot be cultured in vitro, our knowledge about their physiology, biochemistry, and molecular biology is limited. Moreover, we did not find any comprehensive previous report regarding biochemical alterations in cotton plants infected by the CLCuBuV. In the present study, therefore, investigation was conducted on the quantitative estimations of phenolic compounds, protein content, phenylalanine ammonia-lyase (PAL), peroxidase (POX), catalase (CAT), proteases, superoxide dismutase (SOD), polyphenol oxidase (PPO) activities, and malondialdehyde (MDA) contents, indicating their role in CLCuBuV-inoculated and noninoculated plants of resistant and susceptible cotton genotypes.
Materials and methods
Plants material
Four cotton genotypes, namely, Ravi (G. arboreum), Co Tiep Khac (G. herbaceum), CIM-496 (G. hirsutum), and NIAB-111 (G. hirsutum) with known CLCuD-resistance and susceptibility levels were grown in glasshouse (Akhtar et al. Citation2010, Citation2013).
Source of viral inoculum
The inoculum of CLCuBuV was obtained from artificially inoculated plants of a highly susceptible cotton variety CIM-496 that were maintained in a glasshouse.
Virus transmission and identification
Five, six-week-old plants of each test genotypes of G. arboreum, G. herbaceum, and G. hirsutum were inoculated by graft inoculation with CLCuBuV, according to the method described by Akhtar et al. (Citation2013). Data were recorded on the percentage of successful grafts, percentage of disease transmission, latent period (average time required for first symptom appearance after grafting), and type of symptoms developed 90 days post inoculation. A same second set was left uninoculated to serve as control. The presence of CLCuBuV was confirmed through PCR using cotton begomovirus component and β-specific primers (Akhtar et al. Citation2013).
Estimation of total phenols
One-gram leaf samples from CLCuBuV-inoculated noninoculated plants of each test genotypes were collected after 30 days of inoculation, cut into small pieces, and then put into the smearing methanol until the green color is extracted. Leaf tissues were homogenized after decanting the methanol in the Polytrone (Pt1600E) and then again boiled in methanol for further five minutes and filtered through Whatman No. 1 filter paper. Residual material was washed with 80 percent acidified (0.1% HCL conc.) methanol. Using rotavapour (BUCHI R-114), methanol was evaporated and the aqueous layer was collected to adjust final volume as ml/g of weight with distilled water. Aqueous portion of extract was then washed with n-hexane to remove green color. The concentration of phenolic compounds in the leaves of CLCuBuV-inoculated and non-inoculated plants was determined by using the folin ciocalteau reagent modified method of Bray and Thorpe (Citation1954). 0.2 ml of above prepared leaf extract was taken in test tubes and add 4ml of 4% Na2CO3 solution, then 0.2 ml of Folin Ciocalteu reagent was added after 3 minutes with constant shaking on vortex mixer and placed. Absorption was measured in Double Beam Spectrophotometer (Hitachi u-2800) at 750 nm after 30 minutes. Chlorogenic acid was used as standard, and the total phenolic concentration was calculated as mg/g fresh weight of cotton leaves.
Total soluble protein content
Fully emerged leaves (0.5 g) were ground in cold extraction buffer. Samples were centrifuged at 15,000×g for 10 min at 4°C, and the supernatant was separated. Total soluble protein concentration was measured by dye binding assay, as described by Bradford (Citation1976).
Phenylalanine ammonia-lyase (PAL)
PAL activity was assayed following the method of Ngadze et al. (Citation2012). Leaf tissue samples were cut into small pieces of about 5 mm long from each of the treatment, and 2.5 g of homogenate was mixed with 5 ml of a buffer containing 50 mM Tris, 15 mM 2-mercaptoethanol, and 5% polyvinylpolypyrrolidone (PVPP; Sigma) and filtered through 4 layers of muslin cloth. The filtrate was centrifuged at 13,000 rpm for 5 min at 4°C. One ml of supernatant was mixed with 2 ml of 0.05 M borate buffer (pH 8.8) and 1 ml of 0.02 M L-phenylalanine. The samples were incubated at 30°C for 1 h. The reaction was stopped by adding 0.2 ml of 6 M trichloroacetic acid (TCA), and PAL activity was measured after 2 h of sectioning. One activity unit was defined as a change in absorbance of 0.01 at 290 nm h−1 g−1 protein.
Peroxidase (POD)
For the estimation of POD activity, leaves were homogenized in a medium composed of 50 mM potassium phosphate buffer (pH 7.0), 0.1 mM EDTA, and 1 mM dithiothreitol (DTT). Activity of POD was measured using the method of Chance and Maehly (Citation1955), with some modification. For measurement of POD, activity assay solution (3mL) contained 50 mM phosphate buffer (pH 7.0), 20 mM guaiacol, 40 mM H2O2, and 0.1 mL enzyme extract. The reaction was initiated by adding the enzyme extract. Increase in absorbance of the reaction solution at 470 nm was recorded after every 20s. One unit POD activity was defined as an absorbance change of 0.01 unit min−1.
Catalase (CAT)
Leaves from both CLCuBuV-inoculated and noninoculated plants were thoroughly mixed in a medium composed of 50 mM potassium phosphate buffer, pH 7.0, and 1 mM DTT (for the estimation of CAT) for measurement of CAT activity. Assay solution (3 mL) contained 50 mM phosphate buffer (pH 7.0), 5.9 mM H2O, and 0.1 mL enzyme extract. Reduction in absorbance of the reaction solution at 240 nm was recorded after every 20 s. An absorbance change of 0.01 unit's min−1 was defined as one unit CAT activity. On fresh weight basis, enzyme activity was expressed (Hameed et al. Citation2011).
Protease
For the estimation of protease activity, leaf samples (0.5 g) from CLCuBuV-inoculated and noninoculated plants were extracted in 50 mM potassium phosphate buffer (pH 7.8) (Hameed et al. Citation2011). Protease activity was determined by the casein digestion assay described by Drapeau (Citation1974). A series of tubes were equilibrated with 2.0 ml of 1% casein at 37°C for 5 min. To all the tubes, 100 µl of protease extracts was added and mixed well. A reagent blank was also included. Exactly 10 min after adding sample, reaction was stopped by adding 2.0 ml TCA solution and mixed well. Tubes were then allowed to stand for 10 min, and then reaction solution was filtered to remove the precipitate formed during reaction. The absorbance of filtrate was measured at 280 nm. By this method, one unit is that amount of enzyme, which releases acid soluble fragments equivalent to 0.001 A280 per min at 37°C and pH 7.8. Enzyme activity was expressed on protein basis (Hameed et al. Citation2011, Citation2013).
Superoxide dismutase (SOD)
For the estimation of SOD activity, leaves were homogenized in a medium composed of 50 mM potassium phosphate buffer (pH 7.0), 0.1 mM EDTA and 1 mM DTT as described by Dixit et al. (Citation2001). The activity of SOD was assayed by measuring its ability to inhibit the photochemical reduction of nitroblue tetrazolium (NBT) following the method of Giannopolitis and Ries (Citation1977). One unit of SOD activity was defined as the amount of enzyme, which caused 50% inhibition of photochemical reduction of NBT.
Polyphenol oxidase (PPO)
PPO activity for CLCuBuV-inoculated and noninoculated plants was measured according to the method described by Ngadze et al. (Citation2012). Leaf tissue samples were cut into small pieces of about 5 mm long from each of the treatment and were ground in liquid nitrogen using a mortar and pestle, and 2.5 g of homogenate was mixed with 5 ml of 0.05 M sodium phosphate buffer (pH 6.0) containing 5% polyvinylpolypyrrolidone (PVPP, wt/vol). The homogenate was filtered through 4 layers of muslin cloth, and the filtrate was centrifuged at 13,000 rpm for 5 min at 4°C. One ml of supernatant was transferred to a new tube and mixed with 2.9 ml of 0.05 M sodium phosphate buffer and 1 ml of 0.1 M catechol (Sigma). The mixture was aliquoted into three portions for measurement of PPO activity. The absorbance at 546 nm was measured for 4 min at 20-s intervals, and the values per minute were calculated and the results were presented as U µl−1 min−1 (Ngadze et al. Citation2012).
Malondialdehyde (MDA)
Levels of lipid POD in the leaf tissue of CLCuBuV-inoculated and noninoculated plants were calculated in terms of MDA (MDA, a product of lipid peroxidation) content determined by the thiobarbituric acid (TBA) reaction. A 0.25 g leaf sample was homogenized in 5 mL 0.1% TCA. The homogenate was centrifuged at 10,000×g for 5 min. In 1 mL aliquot of the supernatant, 4 mL of 20% TCA containing 0.5% TBA was added. The mixture was heated at 95°C for 30 min and then rapidly cooled in an ice-bath. After centrifuging at 10,000×g for 10 min, the absorbance of the supernatant at 532 nm was read and the value for the nonspecific assimilation at 600 nm was subtracted. The MDA content was considered by using extinction coefficient of 155 mM−1 cm−1 (Hameed et al. Citation2011).
Statistical analysis
The data collected from all experiments was analyzed separately for each parameter and subjected to two-way analysis of variance (ANOVA) using XL-STAT 2012. The means were compared for significance using Tukey's HSD test and the values presented are mean of three replicates ± standard error (SE).
Results and discussion
Disease response
CLCuBuV-inoculated plants of all tested genotypes showed a wide range of symptoms depending on their genetic makeup. Susceptible genotypes from G. hirsutum (CIM-496 and NIAB-111) developed disease symptoms 10 days post inoculation and showed sever symptoms like leaf curling & rolling, swelling and darkening of veins, and leaf-like out-growths called ‘enations’ within 20–25 days post inoculation. All the graft inoculated plants of resistant genotypes from G. arboreum (Ravi) and G. herbaceum (Co Tiep Khac) initiated disease at 25 and 24 days post inoculation, respectively. CLCuD symptoms on these genotypes started as slight vein swelling/darkening and ‘enations’ on the veins on the undersides of leaves. These symptoms were developed on few leaves of the grafted plants of G. arboreum (Ravi) and G. herbaceum (Co Tiep Khac). No increase in the disease severity value was observed till the end of the experiment (90 days post inoculation). CLCuBuV was readily detected in both symptomatic and symptom-less leaves of these plants through PCR. The lower disease severity in the leaves of CLCuBuV-inoculated plants of genotypes Ravi (G. arboreum) and Co Tiep Khac (G. herbaceum) compared with CIM-496 and NIAB-111 established the clear-cut classes of resistance and susceptibility, confirming findings of Akhtar et al. (Citation2013) who observed similar trends and ranked them as resistant and susceptible to CLCuBuV.
Total phenols
Phenolic compounds may contribute to enhance the mechanical strength of host cell walls by the synthesis of lignin and suberin that are involved in the formation of physical barriers that can block the spread of pathogens (Ngadze et al. Citation2012; Singh et al. Citation2014). In the present study, the amount of total phenols was significantly higher in leaves of noninoculated and inoculated plants of both resistant genotypes (Ravi and Co Tiep Khac), while it was significantly lower in susceptible genotypes (CIM-496 and NIAB-111; ). The maximum amount of total phenols in noninoculated plants was observed in resistant cultivar Ravi followed by Co Tiep Khac, NIAB-111, and CIM-496. This high level of phenolic content in these plants has been correlated with increased resistance to CLCuBuV as the accumulation of total phenols is usually found to be higher in resistant genotypes compared to susceptible ones (Velazhahan & Vidhyasekaran Citation1994; Pradeep & Jambhale Citation2002; Ghosal et al. Citation2004; Meena et al. Citation2008; Singh et al. Citation2010).
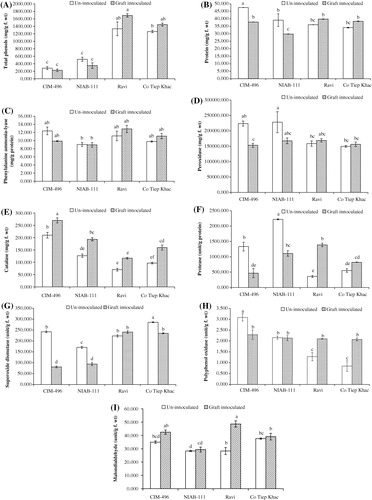
Altering the level of phenolic compounds in plants has been demonstrated to change disease susceptibility (Yao et al. Citation1995). In our study, the amount of total phenols was found to be decreased in inoculated plants of susceptible genotypes CIM-496 (20.17%) and NIAB-111 (32.02%) while increased in inoculated plants of resistant genotypes Ravi (21.11%) and Co Tiep Khac (12.86%), as compared to their noninoculated plants. However, the decrease or increase in both cases was statistically nonsignificant. A possible explanation for decrease in phenolic compounds after CLCuBuV inoculation in susceptible genotypes is that many plant pathogens actively suppress the expression of plant defense reactions during successful infection (Markakis et al. Citation2010). These results are in line with the results reported on other plant-pathogenic fungal bacterial and viral interactions, which showed that certain common phenols and phenolic substances are toxic to pathogens, which have long been considered as important defense-related compounds whose levels are naturally high in the resistant varieties of many crops (Gogoi et al. Citation2001) and accumulates in plants after infection, especially in resistant varieties (Agrios Citation1997).
Total soluble protein content
Involvement of protein components in plant diseases resistance has been documented in many plant pathogenic interactions (Tornero et al. Citation2002; Carvalho et al. Citation2006). In noninoculated plants, total soluble protein content was significantly higher in leaves of susceptible genotype CIM-496 but was similar in leaves of susceptible genotype NIAB-111 and both resistant genotypes Ravi and Co Tiep Khac (). Usually, infected plants show a high protein content, which could be due to both the activation of the host defense mechanisms and the pathogen attack mechanisms (Agrios Citation1997). However, protein content was found to be decreased significantly in CLCuBuV-inoculated plants of both susceptible genotypes (CIM-496, 20.39% and NIAB-111, 23.64%), while it was found to change nonsignificantly in leaves of inoculated plants of both resistant genotypes Ravi (9.41%) and Co Tiep Khac (11.04%), as compared to their noninoculated plants. A possible explanation for significant decrease in total soluble protein contents in susceptible genotypes after CLCuBuV inoculation may be due to the high level of susceptibility of these genotypes. Similar results have been reported in maize, tomato, grapevine, and apple infected with mollicutes (Favali et al. Citation2001; Bertamini et al. Citation2002a, Citation2002b; Musetti Citation2010).
Phenylalanine ammonia-lyase activity
PAL is the primary enzyme in the phenylpropanoid metabolism and plays a significant role in the synthesis of several defense-related secondary compounds such as phenols and lignin (Hemm et al. Citation2004; Tahsili et al. Citation2014). Inhibition of PAL affects subsequent biosynthetic pathways of phenolic compounds (Jayaraj et al. Citation2010). In present study, PAL activity was higher in leaves of noninoculated plants of susceptible genotype CIM-496, followed by resistant genotypes Ravi and Co Tiep Khac and susceptible genotype NIAB-111 (). PAL activity was found to change nonsignificantly in inoculated plants of both susceptible genotypes CIM-496 (20.22%) and NIAB-111 (1.18%) and in leaves of inoculated plants of both resistant genotypes Ravi (13.48%) and Co Tiep Khac (12.01%), as compared to their noninoculated plants. Similar results were reported by Ogawa et al. (Citation2006) in tobacco leaves inoculated with tobacco mosaic virus. Several studies also indicated that the activation of PAL and subsequent increase in phenolic content in plants is a general response associated with disease resistance. It is known that the presence of phenolic compounds in plants and their synthesis in response to infection is associated with resistance and studies have indicated that increased accumulation of phenolics is due to increase in PAL activity – offered protection against diseases (Jayaraj et al. Citation2010).
Peroxidase activity
POX is one of the first enzymes responding and providing fast defense against plant pathogens (Sulman et al. Citation2001). POXs are involved in the lignification, suberification, polymerization of hydroxy-proline-rich glycoproteins, regulation of cell wall elongation, wound healing, and resistance against pathogens in plants (Hammond-Kosack & Jones Citation1996; Yoshida et al. Citation2003; Maksimov et al. Citation2014). POX activity was significantly higher in noninoculated plants of both susceptible genotypes, as compared to resistant genotypes (). The highest POX activity in noninoculated plants was observed in NIAB-111 followed by CIM-496, Ravi, and Co Tiep Khac. POX activity decreased significantly in CLCuBuV-inoculated plants of susceptible genotypes CIM-496 (31.54 %) and NIAB-111 (26.497 %) compared to the noninoculated plants, while it changed nonsignificantly in CLCuBuV-inoculated leaves of both resistant genotypes, i.e. Ravi (6.23 %) and Co Tiep Khac (4.43 %) as compared to their noninoculated plants. Infection with plant pathogens led to an induction in POX activity in plant tissues and a greater increase was recorded in resistant plants compared to the susceptible ones (Mydlarz & Harvell Citation2006). Similar increase in POX activity as observed in resistant genotypes in this study was observed in tomato and bell pepper infected with tobacco mosaic virus and tomato mosaic tobamovirus (Madhusudhan et al. Citation2009); cucumber mosaic virus and zucchini yellow mosaic virus-infected Cucumis sativus and Cucurbita pepo plants (Riedle-Bauer Citation2000); tobacco mosaic virus infected tobacco plants (Kiraly et al. Citation2002); tomato yellow leaf curl virus infected tomato plants (Dieng et al. Citation2011) and a number of resistant interactions involving several plant patho-systems.
Catalase activity
CAT is an oxygen-scavenging enzyme that plays the role of specific peroxidative, protecting cells from the toxic effects of substrates (H2O2) during development, which are otherwise lethal (Choodamani et al. Citation2009; Patel et al. Citation2011; Hameed & Iqbal Citation2014). In our study, CAT activity was significantly higher in noninoculated plants of susceptible genotype CIM-496 as compared to susceptible NIAB-111, resistant genotypes Ravi and Co Tiep Khac (). CAT activity was found to increase in leaves of CLCuBuV-inoculated plants of all tested genotypes. Maximum increase in CAT activity after CLCuBuV inoculation was noted in resistant genotype Ravi (40%), followed by Co Tiep Khac (39.58%), NIAB-111 (34.48%), and CIM-496 (22.22%), as compared to their noninoculated plants. Similar increases in foliar CAT activity were also observed in Algerian-susceptible but not in Algerian-Resistant barley (Hordeum vulgare L.) leaves inoculated with Blumeria graminis (Vanacker et al. Citation1998).
Protease activity
Proteases are involved in protein maturation, degradation, and protein rebuilding in response to different external stimuli and in removal of abnormal, misfolded proteins (Grudkowska & Zagdańska Citation2004). In the present study, protease activity was significantly higher in noninoculated plants of both susceptible genotype (CIM-496 and NIAB-111), as compared to non-inoculated plants of both resistant genotypes Ravi and CoTiep Khac (). Protease activity was found to be decreased significantly in leaves of CLCuBuV-inoculated plants of both susceptible genotypes CIM-496 (64.96%) and NIAB-111 (50.08%), while it was found to be increased significantly in leaves of CLCuBuV-inoculated plants of resistant genotype Ravi (74.04%) but nonsignificantly in resistant genotype Co Tiep Khac (332.86%), as compared to the noninoculated plants. Present observations showed that protease activity was altered in all genotypes and have significant role in resistance against CLCuBuV. There may be a possibility that proteases were relatively more vulnerable to pathogenesis than other proteins and genotypes accumulating more proteases before disease infection had higher disease severity. In contrast, there was much correlation between postinfectional protease activity and disease severity indicating the involvement of these enzymes in pathogenesis. Postinfectional protease activity level was generally higher when compared with preinfectional level in both resistant genotypes. Proteolytic processing is emerging as a common mechanism in regulating the defense response in plants against pathogens (Hameed et al. Citation2010).
Superoxide dismutase activity
SOD is one of the most important scavenging enzymes and catalyzes the dismutation of superoxide radicals to activity oxygen species hydrogen peroxide (Ehsani-Moghaddam et al. Citation2006; Hameed & Iqbal Citation2014). SOD activity was significantly higher in leaves of noninoculated plants of resistant genotypes Co Tiep Khac, followed by susceptible CIM-496, resistant Ravi and Co Tiep Khac (). SOD activity was found to decrease significantly in CLCuBuV-inoculated plants of both susceptible genotypes CIM-496 (66.69%), NIAB-111 (45.05%) and a resistant genotype Co Tiep Khac (17.67%), while it changed nonsignificantly in leaves of inoculated plants of resistant genotypes Ravi (7.53%), as compared to their noninoculated plants. SOD activity was also shown to increase in strawberry leaves infected by Mycosphaerella fragariae and the increase was greater for the resistant cultivars than for the susceptible ones (Ehsani-Moghaddam et al. Citation2006). Some fungal pathogens could benefit from an increase in ROS levels generated in the host cells during the defense against their infection as a facilitating factor to maximize their tissue colonization and nutrient uptake (Govrin & Levine Citation2000). Thus, significantly higher SOD activity in resistant cultivar Ravi could be a strategy of the plant to restrict CLCuBuV colonization, because the excess ROS can be removed (Govrin & Levine Citation2000; Ehsani-Moghaddam et al. Citation2006).
Polyphenol oxidase activity
PPO is important in the initial stage of plant defense where membrane damage causes release of phenols such as chlorogenic acid. PPO catalyzes the oxidation of phenolics to free radicals that can react with biological molecules, thus creating an unfavorable environment for pathogen development (Jockusch Citation1966; Mohamed et al. Citation2012). PPO activity was significantly higher in noninoculated plants of both susceptible genotypes (CIM-496 and NIAB-111), as compared to noninoculated plants of both resistant genotypes Ravi and Co Tiep Khac (). The maximum amount of PPO activity in leaves of noninoculated plants was observed in susceptible genotype CIM-496, followed by susceptible NIAB-111, resistant Ravi, and Co Tiep Khac. The PPO activity was found to be decreased significantly in CLCuBuV-inoculated plants of susceptible genotype CIM-496 (25.82 %), slightly decreased in inoculated plants of susceptible genotype NIAB-111 (1.11 %), and significantly increased in resistant genotypes Ravi (78.31 %) and Co Tiep Khac (59.33 %), as compared to the noninoculated plants. Many studies have shown that PPO is induced in response to infection by different pathogens (Constabel et al. Citation2000; Stewart et al. Citation2001; Vanitha et al. Citation2009). Systemic induction of PPO expression in resistant genotypes under present study in response to CLCuBuV might provide an additional line of defense to protect plants against further attack by pathogen and insects, as previously reported in other plant–pathogen interactions (Thipyapong et al. Citation1995; Stout et al. Citation1999). In addition, it was also interesting to note that resistant cotton genotypes (Ravi and Co Tiep Khac) that showed enhanced PPO activity after CLCuBuV inoculation also recorded a higher concentration of total phenols after CLCuBuV inoculation. It appears that these resistant genotypes produce more secondary metabolites involved in plant defense mechanisms than the susceptible genotypes tested. Total soluble phenols together with PPO appear to play a role in resistance to CLCuD, since these compounds were present in considerably higher levels in leaves of CLCuBuV-inoculated cotton plants of resistant genotypes. This is supported by the work of Li and Steffens (Citation2002), Thipathi and Verma (Citation1975), Kumar et al. (Citation1991), and Ngadze et al. (Citation2012).
Malondialdehyde content
MDA is considered a general indicator of lipid peroxidation (Bharwana et al. Citation2014; Hameed & Iqbal Citation2014). The MDA produced during lipid peroxidation is an indicator of cellular damage at the cell membrane by pathogenic infection (Dallagnol et al. Citation2011; Aly et al. Citation2012). The MDA content was statistically similar in noninoculated plants of both susceptible and resistant genotypes, while it was found to be increased in CLCuBuV-inoculated plants of all tested genotypes (). The extent of the cellular damage caused by the oxidative stress related to the plant response against the pathogen infection can be estimated by the products of the peroxidation of membrane lipids (Dallagnol et al. Citation2011; Debona et al. Citation2012). In the present study, MDA levels were found to be significantly increased in inoculated leaves of resistant genotype Ravi (41.83%) but were not changed significantly in inoculated plants of other resistant and susceptible genotypes, indicating that the lipid peroxidation occurred concomitantly with the appearance of mild vein thickening symptoms during the incompatible host–pathogen interaction in resistant genotype Ravi. El-Zahabi et al. (Citation1995) also reported the same results and provided evidence that Erysiphe gramins f. sp. hordei leads to significant changes in the antioxidant metabolism of barley. They suggested that in the compatible interaction, the fungus grows rapidly in the leaf tissues, affecting numerous cells. Thus, the stronger induction of antioxidative enzymes may be explained by the fact that the fungus elicits a stress reaction from more cells than in an incompatible reaction. However, no increase of lipid peroxidation was observed in the compatible interaction (with substantially decreasing ascorbic acid levels), and in the incompatible interaction, in which fewer cells were stressed, the rate of lipid peroxidation was intensively elevated. This suggests that the induced antioxidative processes are probably not only stressed reactions, but those they also can contribute to the suppression of lipid peroxidation and necrotic symptom expression (El-Zahabi et al. Citation1995).
Conclusion
Early and elevated levels of expression of various defense enzymes are an important feature of plant resistance to pathogens (Vanitha & Umesha Citation2008). Our findings indicate that phenolic compounds, protein, PAL, POX, CAT, proteases, SOD, PPO, and MDA play an active role in disease resistance against CLCuBuV. However, total phenolics, protease, MDA, SOD, and PPO were found to differ significantly in resistant and susceptible varieties after infection with CLCuBuV. Thus, it can be argued that there is a correlation between constitutively induced levels of total phenolics with these enzymes and plant resistance and that these enzymes could be considered as biochemical markers for studying plant–virus interactions in compatible and incompatible interactions. The preinfectional and postinfectional presence of high level of phenolics and postinfectional significant increase in proteases in resistant cultivar Ravi may have further role in disease resistance; thus, additional studies are needed to characterize the phenolic compounds and proteases involved.
Acknowledgment
The authors are very thankful to Mr. Muhammad Tanvir Elahi, (RA), NIAB, for his useful assistance.
References
- Agrios GN. 1997. Plant pathology. 4th ed. San Diego (CA): Academic Press; 635 p.
- Akhtar KP, Haidar S, Khan MKR, Ahmad M, Sarwar N, Murtaza MA, Aslam M. 2010. Evaluation of Gossypium species for resistance to cotton leaf curl Burewala virus. Ann Appl Biol. 157:135–147. 10.1111/j.1744-7348.2010.00416.x
- Akhtar KP, Haq MA, Hussain M, Khan AI. 2002. Whitefly transmitted Geminiviruses and associated disorders in cotton: a review. Pak J Phytopathol. 14:140–150.
- Akhtar KP, Jamil FF, Haq MA, Khan IA. 2008. Comparison of resistance to cotton leaf curl disease (Multan/Burewala) among Gossypium hirsutum L. Varieties and breeding lines. J Phytopathol. 156:352–357. 10.1111/j.1439-0434.2007.01368.x
- Akhtar KP, Ullah R, Khan IA, Saeed M, Sarwar N, Mansoor S. 2013. First symptomatic evidence of infection of Gossypium arboreum with cotton leaf curl Burewala virus through grafting. Int J Agric Biol. 15:157–160.
- Aly AA, Mansour MTM, Mohamed HI, Abd-Elsalam KA. 2012. Examination of correlations between several biochemical components and powdery mildew resistance of flax cultivars. Plant Pathol J. 28:149–155. 10.5423/PPJ.2012.28.2.149
- Amrao L, Amin I, Shahid MS, Briddon, RW, Mansoor S. 2010. Cotton leaf curl disease in resistant cotton is associated with a single Begomovirus that lacks an intact transcriptional activator protein. Virus Res. 152:153–163. 10.1016/j.virusres.2010.06.019
- Azhar MT, Amin I, Anjum ZI, Arshad M, Briddon RW. 2010. Both malvaceous and non-malvaceous betasatellites are associated with two wild cotton species grown under field conditions in Pakistan. Virus Genes. 41:417–424. 10.1007/s11262-010-0521-4
- Bertamini M, Grando MS, Muthuchelian K, Nedunchezhian N. 2002a. Effect of phytoplasmal infection on photosystem II efficiency and thylakoid membrane protein changes in field grown apple (Malus pumila) leaves. Physiol Mol Plant Pathol. 61:349–356. 10.1006/pmpp.2003.0450
- Bertamini M, Nedunchezhian N, Tomasi F, Grando S. 2002b. Phytoplasma (Stolbur subgroup (Bois Noir-BN)) infection inhibits photosynthetic pigments, ribulose-1,5-biphosphate carboxylase and photosynthetic activities in field grown grapevine (Vitis vinifera L. cv. Chardonnay) leaves. Physiol Mol Plant Pathol. 61:357–366. 10.1006/pmpp.2003.0449
- Bharwana SA, Ali S, Farooq MA, Iqbal N, Hameed A, Abbas F, Ahmad MSA. 2014. Alleviation of lead toxicity by glycine betaine is related to elevated photosynthesis, antioxidant enzymes suppressed lead uptake and oxidative stress in cotton. Turk J Bot. 38:281–292. 10.3906/bot-1304-65
- Bradford MM. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Ann Biochem. 72:248–254. 10.1016/0003-2697(76)90527-3
- Bray HG, Thorpe WV. 1954. Analysis of phenolic compounds of interest in metabolism. Methods Biochem Anal. 1:27–52.
- Carvalho D, Anastacio Q, Luciana M. 2006. Proteins and isozymes electrophoresis in seeds of Desti (Leguminosae caesalpinioidea) artificially aged. Rev Arv. 30:19–21.
- Chance M, Maehly AC. 1955. Assay of catalases and peroxidases. Methods Enzymol. 2:764–817.
- Chandra M, Sreenivasan S. 2011. Studies on improved Gossypium arboreum cotton: Part I – fiber quality parameters. Indian J Fibre Textile Res. 36:24–34.
- Choodamani MS, Hariprasad P, Sateesh MK, Umesha S. 2009. Involvement of catalase in bacterial blight disease development of rice caused by Xanthomonas oryzae pv. Oryza. Int J Pest Manage. 55:121–127. 10.1080/09670870802601076
- Constabel CP, Yip L, Patton JJ, Christopher ME. 2000. Polyphenol oxidase from hybrid poplar, cloning and expression in response to wounding and herbivory. Plant Physiol. 124:285–295. 10.1104/pp.124.1.285
- Dallagnol LJ, Rodrigues FA, Martins SCV, Cavatte PC, DaMatta FM. 2011. Alterations on rice leaf physiology during infection by Bipolaris oryzae. Australas Plant Pathol. 40:360–365. 10.1007/s13313-011-0048-8
- Debona D, Rodrigues FÁ, Rios JA, Nascimento KJT. 2012. Biochemical changes in the leaves of wheat plants infected by Pyricularia oryzae. Phytopathology. 102:1121–1129. 10.1094/PHYTO-06-12-0125-R
- Dieng H, Satho T, Hassan AA, Aziz AT, Morales RE, Hamid SA, Miake F, Abubakar S. 2011. Peroxidase activity after viral infection and whitefly infestation in juvenile and mature leaves of Solanum lycopersicum. J Phytopathol. 159:707–712. 10.1111/j.1439-0434.2011.01830.x
- Dixit V, Pandey V, Shyam R. 2001. Differential antioxidative response to cadmium in roots and leaves of pea. J Exp Bot. 52:1101–1109. 10.1093/jexbot/52.358.1101
- Drapeau G. 1974. Protease from Staphylococcus aureus. In: Lorand L, editor. Method of enzymology, vol. 45b. New York: Academic Press; p. 469.
- Ehsani-Moghaddam B, Charles MT, Carisse O, Khanizadeh S. 2006. Superoxide dismutase responses of strawberry cultivars to infection by Mycosphaerella fragariae. J Plant Physiol. 163:147–153. 10.1016/j.jplph.2005.04.025
- El-Zahabi HM, Gullner G, Királi Z. 1995. Effects of powdery mildew infection of barley on the ascorbate-glutathione cycle and other antioxidants in different host–pathogen interactions. Phytopathology. 85:1225–1230. 10.1094/Phyto-85-1225
- Favali MA, Sanità di Toppi L, Vestena C, Fossati F., Musetti R. 2001. Phytoplasmas associated with tomato stolbur disease. Acta Horticulturae. 551:93–99.
- Ghosal TK, Dutta S, Senapatiand SK, Deb DC. 2004. Role of phenol contents in legume seeds and its effects on the biology of Collosbrchus chinensis. Ann Plant Prot Sci.12:442–444.
- Giannopolitis CN, Ries SK. 1977. Superoxide dismutases occurrence in higher plants. Plant Physiol. 59:309–314. 10.1104/pp.59.2.309
- Gogoi R, Singh DV, Srivastava KD. 2001. Phenols as a biochemical basis of resistance in wheat against Karnal bunt. Plant Pathol. 50:470–476. 10.1046/j.1365-3059.2001.00583.x
- Govrin EM, Levine A. 2000. The hypersensitive response facilitates plant infection by the necrotrophic pathogen Botrytis cinerea. Curr Biol.10:751–757. 10.1016/S0960-9822(00)00560-1
- Grudkowska M, Zagdańska B. 2004. Multifunctional role of plant cysteine proteinases. Acta Biochimica Polonica. 51:609–624.
- Hameed A, Akhtar KP, Saleem MY, Asghar M. 2010. Correlative evidence for prooxidase involvement in disease resistance against Alternaria leaf blight of tomato. Acta Physiologiae Plantarum. 32:1171–1176. 10.1007/s11738-010-0512-z
- Hameed A, Bibi N, Akhter J, Iqbal N. 2011. Differential changes in antioxidants, proteases, and lipid peroxidation in flag leaves of wheat genotypes under different levels of water deficit conditions. Plant Physiol Biochem. 49:17–185. 10.1016/j.plaphy.2010.11.009
- Hameed A, Goher M, Iqbal, N. 2013. Drought induced programmed cell death and associated changes in antioxidants, proteases, and lipid peroxidation in wheat leaves. Biologia Plantarum. 57:370–374. 10.1007/s10535-012-0286-9
- Hameed A, Iqbal N. 2014. Chemo-priming with mannose, mannitol and H2O2 mitigate drought stress in wheat. Cereal Res Comm.
- Hammond-Kosack KE, Jones JDG. 1996. Resistance gene-dependent plant defense responses. Plant Cell. 8:1773–1791.
- Hemm MR, Rider SD, Ogas J, Murry DJ, Chapple C. 2004. Light induces phenylpropanoid metabolism in Arabidopsis roots. Plant J. 38:765–778. 10.1111/j.1365-313X.2004.02089.x
- Jayaraj J, Bhuvaneswari R, Rabindran R, Muthukrishnan S, Velazhahan R. 2010. Oxalic acid-induced resistance to Rhizoctonia solani in rice is associated with induction of phenolics, peroxidase and pathogenesis-related proteins. J Plant Interact. 5:147–157. 10.1080/17429140903291012
- Jockusch H. 1966. The role of host genes, temperature and polyphenol oxidase in the necrotization of TMV infected tobacco tissue. Phytopathol Z. 55:185–192. 10.1111/j.1439-0434.1966.tb02222.x
- Jones JDG, Dangl JL. 2006. The plant immune system. Nature. 444:323–329 10.1038/nature05286
- Kiraly Z, Barna B, Kecskes A, Fodor J. 2002. Down regulation of antioxidative capacity in a transgenic tobacco, which fails to develop acquired resistance to necrotization caused by tobacco mosaic virus. Free Radical Res. 36:981–991. 10.1080/1071576021000006581
- Kumar A, Pundhir VS, Gupka KC. 1991. The role of phenols in potato tuber resistance against soft rot by Erwinia carotovora ssp. Carotovora. Potato Res. 34:9–16. 10.1007/BF02358090
- Kumar A, Snehi SK, Raj SK, Kumar J, Khan JA. 2011. Association of cotton leaf curl Burewala virus and its satellite molecules with leaf distortion symptoms of cotton in India. New Dis Rep. 24:18. 10.5197/j.2044-0588.2011.024.018
- Li L, Steffens JC. 2002. Over expression of polyphenol oxidase in transgenic tomato plants results in enhanced bacterial disease resistance. Planta. 215:239–247. 10.1007/s00425-002-0750-4
- Madhusudhan KN, Srikanta BM, Shylaja MD, Prakash HS, Shetty HS. 2009. Changes in antioxidant enzymes, hydrogen peroxide, salicylic acid and oxidative stress in compatible and incompatible host tobamovirus interaction. J Plant Interact. 4:157–166. 10.1080/17429140802419516
- Maksimov I, Troshina N, Surina O, Cherepanova E. 2014. Salicylic acid increases the defense reaction against bunt and smut pathogens in wheat calli. J Plant Interact. 9:306–314.
- Mansoor S, Amin I, Iram S, Hussain M, Zafar Y. 2003. Breakdown of resistance in cotton to cotton leaf curl disease in Pakistan. Plant Pathol. 52:784. 10.1111/j.1365-3059.2003.00893.x
- Markakis EA, Tjamos SE, Antoniou PP, Roussos PA, Paplomatas EJ, Tjamos EC. 2010. Phenolic responses of resistant and susceptible olive cultivars induced by defoliating and nondefoliating Verticillium dahliae pathotypes. Plant Dis. 94:1156–1162. 10.1094/PDIS-94-9-1156
- Meena RK, Patni V, Arora DK. 2008. Study on phenolics and their oxidative enzyme in Capsicum annuum L. infected with Geminivirus. Asian J Exp Sci. 22:307–310.
- Mohamed H, EL-Hady AA, Mansour M, El-Rheem, El-Samawaty A. 2012. Association of oxidative stress components with resistance to flax powdery mildew. Trop Plant Pathol. 37:386–392. 10.1590/S1982-56762012000600002
- Musetti R. 2010. Biochemical changes in plants infected by Phytoplasmas. In: Weintraub PG, Jones P, editors. Phytoplasmas: genomes, plant hosts, and vectors. Wallingford: Printed and bound in the UK by MPG Books Group; 331 p.
- Mydlarz LD, Harvell CD. 2006. Peroxidase activity and inducibility in the see fan coral exposed to a fungal pathogen. Comp Biochem Physiol. 10:1016.
- Ngadze E, Icishahayo D, Coutinho TA, van der Waals JE. 2012. Role of polyphenol oxidase, peroxidase, phenylalanine ammonia lyase, chlorogenic acid, and total soluble phenols in resistance of potatoes to soft rot. Plant Dis. 96:186–192. 10.1094/PDIS-02-11-0149
- Ogawa D, Nakajima N, Seo S, Mitsuhara I, Kamada H, Oaci Y. 2006. The phenylalanine pathway is the main route of salicylic acid biosynthesis in tobacco mosaic virus-infected tobacco leaves. Plant Biotechnol. 23:395–398. 10.5511/plantbiotechnology.23.395
- Patel SJ, Subramanian RB, Jha YS. 2011. Biochemical and molecular studies of early blight disease in tomato. Phytoparasitica. 39:269–283. 10.1007/s12600-011-0156-6
- Pradeep T, Jambhale ND. 2002. Relationship between phenolics, polyphenol oxidase and peroxidase and resistance to powdery mildew in Zizhyphus. Indian Phytopathol. 55:195–196.
- Rajagopalan PA, Naik A, Katturi P, Kurulekar M, Kankanallu RS, Anandalakshmi R. 2012. Dominance of resistance-breaking cotton leaf curl Burewala virus (CLCuBuV) in northwestern India. Arch Virol. 157:855–868. 10.1007/s00705-012-1225-y
- Riedle-Bauer M. 2000. Role of reactive oxygen species and antioxidant enzymes in systemic virus infections of plants. Journal of Phytopathology. 148: 297–302. 10.1046/j.1439-0434.2000.00503.x
- Singh HP, Kaur S, Batish DR, Kohli RK. 2014. Ferulic acid impairs rhizogenesis and root growth, and alters associated biochemical changes in mung bean (Vigna radiata) hypocotyls. J Plant Interact. 9:267–274.
- Singh A, Singh KP, Rajwarand GS, Singh UP. 2010. Phenolic acid conteny-a criterion for selection of resistance apple cultivars against Psdosphaera leucotricha (EII. and Ev.) Salmon. J Arch Phytopathol Plant Prot. 43:1138–1143. 10.1080/03235400802343817
- Stewart RJ, Sawyer BJB, Bucheli CS, Robinson SP. 2001. Polyphenol oxidase is induced by chilling and wounding in pineapple. Australas J Plant Physiol. 28:181–191.
- Stout MJ, Fidantsef AL, Duffey SS, Bostock RM. 1999. Signal interactions in pathogen and insect attack: systemic plant-mediated interactions between pathogens and herbivores of the tomato, Lycopersicon esculentum. Physiol Mol Plant Pathol. 54:115–130. 10.1006/pmpp.1998.0193
- Sulman M, Fox G, Osman A, Inkerman A, Williams P, Michalowitz M. 2001. Relationship between total peroxidase activity and susceptibility to black point in mature grain of some barley cultivars. Proceeding of the 10th Australian Barley Technical Symposium. Canberra, ACT, Australia, 16–20 Sep. 2001. http://www.regional.org.au/au/abts/2001/t4/sulman.htm
- Tahir MN, Amin I, Briddon RW, Mansoor S. 2011. The merging of two dynasties—identification of an African cotton leaf curl disease-associated begomovirus with cotton in Pakistan. PLoS One. 6:e20366. 10.1371/journal.pone.0020366
- Tahsili J, Sharifi M, Safaie N, Esmaeilzadeh-Bahabadi S, Behmanesh M. 2014. Induction of lignans and phenolic compounds in cell culture of Linum album by culture filtrate of Fusarium graminearum. J Plant Interact. 9:412–417.
- Thipathi RK, Verma MN. 1975. Phenolic compounds and polyphenol oxidase activity in relation to resistance in potatoes against bacterial soft rot. Indian J Exp Biol. 13:414–416.
- Thipyapong P, Hunt MD, Steffens JC. 1995. Systemic wound induction of potato (Solanum tuberosum) polyphenol oxidase. Phytochemistry. 40:673–676. 10.1016/0031-9422(95)00359-F
- Tornero P, Chao R, Luthin W, Goff S, Dangl J. 2002. Large-scale structure, function, analysis, of Arabidopsis RPM1 disease resistance protein. Plant Cell. 14:435–450. 10.1105/tpc.010393
- Vanacker H, Carver TL. W, Foyer CH. 1998. Pathogen-induced changes in the antioxidant status of the apoplast in barley leaves. Plant Physiol. 117:1103–1114. 10.1104/pp.117.3.1103
- Vanitha SC, Niranjana SR, Umesha S. 2009. Role of phenylalanine ammonia lyase and polyphenol oxidase in host resistance to bacterial wilt of tomato. J Phytopathol. 157:552–557. 10.1111/j.1439-0434.2008.01526.x
- Vanitha SC, Umesha S. 2008. Variations in defense related enzyme activities in tomato during the infection with bacterial wilt pathogen. J Plant Interact. 3:245–253. 10.1080/17429140802032863
- Velazhahan R, Vidhyasekaran P. 1994. Role of phenolic compounds, peroxidase and polyphenol-oxidase in resistance of groundnut to rust. Acta Phytopathologica Entomologica Hungariaca. 29:23–29.
- Yao K, De Luca V, Brisson N. 1995. Creation of a metabolic sink for tryptophan alters the phenylpropanoid pathway and the susceptibility of potato to Phytophthora infestans. Plant Cell. 7:1787–1799.
- Yoshida K, Kaothien P, Matsui T, Kawaoka A, Shinmyo A. 2003. Molecular biology and application of plant peroxidase genes. Appl Microbiol Biotechnol. 60:665–670. 10.1007/s00253-002-1157-7
- Zhao CJ, Wang AR, Shi YJ, Wang LQ, Liu WD, Wang ZH, Lu GD. 2008. Identification of defense-related genes in rice responding to challenge by Rhizoctonia solani. Theor Appl Genet. 116:501–516. 10.1007/s00122-007-0686-y
