Abstract
Two new strains of endophytic fungi were isolated from the bark of Moringa peregrina and identified as Aspergillus caespitosus LK12 and Phoma sp. LK13. These endophytes were identified through amplifying polymerase chain reaction (PCR) and sequencing the 18S internal transcribed spacer of DNA extracted from both endophytes. Pure cultures of endophytic fungi were subjected to extract and isolate gibberellins (GAs). Deuterated standards of [17,17-2H2]-GA1, [17,17-2H2]-GA3, [17, 17-2H2]-GA4 and [17, 17-2H2]-GA7 were used to quantify the endophytic fungal GAs. The analysis revealed that both the endophytes are producing bioactive GAs in various quantities (ng mL−1). A. caespitosus LK12 was producing GA1 (54.51 ± 1.23), GA4 (26.5 ± 0.65), and GA7 (2.87 ± 1.23) while Phoma sp. LK13 was secreting GA1 (4.8 ± 0.12), GA3 (8.65 ± 0.21), GA4 (23.7 ± 0.98), and GA7 (22.7 ± 0.73). The culture filtrate (CF) of A. caespitosus and Phoma sp. significantly increased the shoot length of GAs-deficient mutant waito-c and normal Dongjin-beyo rice seedlings as compared to control. Application of such growth-promoting and GAs-producing endophytes can ameliorate poorly growing crop plants.
Introduction
Endophytic fungi, residing in the host tissues without causing symptoms of disease, have been regarded a pivotal source of bioactive secondary metabolites (Zhou et al. Citation2014). Various classes of biologically active molecules including flavonoids, peptides, alkaloids, steroids, phenolics, terpenoids, lignans, and volatile organic compounds with many of them biologically active have been found to be produced by endophytes (Schulz & Boyle Citation2005; Hamilton & Bauerle Citation2012). In fact, a previous study by Schulz et al. (Citation2002) showed that about 51% of biologically active metabolites originate from endophytes compared to only 38% of novel substances originating from other soil microflora. In the past two decades, many novel bioactive compounds with antimicrobial, insecticidal, cytotoxic, and anticancer properties have been successfully isolated and characterized from endophytic fungi. The role of endophytic fungi in the production of plant growth-promoting substances (gibberellins [GAs]) is unexplored whilst identifying such microorganisms can extend greater benefits in improving agricultural productivity and human food security.
GAs are ubiquitous plant hormones that elicit various metabolic functions required for the growth of a plant, its seed germination, stem elongation, sex expression, flowering, formation of fruits, and senescence (Lee et al. Citation1998). Till now, more than 136 GAs have been identified (Bomke et al. Citation2008) which are named based on their discovery (http://www.plant-hormones.info/gibberellin_nomenclature.htm). Many fungal species, for example Gibberella fujikuroi, Sphaceloma manihoticola, Phaeosphaeria sp., Neurospora crassa, have been found to produce GAs (Rademacher Citation1994). Among fungi, endophytes are not only associated with plants but also in most cases they live inside the plant, and this can either be the aerial parts of the plant or the root. This symbiosis is without causing any disease symptoms to the host (Schulz & Boyle Citation2005; Hyde & Soytong Citation2008; Kumar et al. Citation2012; Waqas et al. Citation2014). Endophytes have been mostly reported to improve plant growth (Schulz & Boyle Citation2005). They can influence key aspects of plant physiology and increase host plant fitness against biotic and abiotic stresses (Khan et al. Citation2011a, Citation2011b). It has also been recently shown that strains of Penicillium commune (Choi et al. Citation2005), Phaeosphaeria sp. L487 (Kawaide Citation2006), Chrysosporium pseudomerdarium (Hamayun et al. Citation2009), Aspergillus fumigatus (Khan et al. Citation2011a), and Penicillium funiculosum (Khan et al. Citation2011b) can produce GAs. Previously some strains of Cephalotheca sulfurea and Phoma sp. have been reported to secret GAs, suggesting that these might have an active GAs biosynthesis pathway (Hamayun et al. Citation2010, Citation2012).
Moringa peregrina Fiori (Moringaceae), a deciduous tree, is an economically important plant of the Arabian and North African region (Hegazy et al. Citation2008; Zaghloul et al. Citation2011). Besides its ecological importance, it is mainly used in folk medicine and eaten as a vegetable (Marwah et al. Citation2007; Hegazy et al. Citation2008; Goma & Picó Citation2011; Zaghloul et al. Citation2011). It is effective for abdominal pain, burns, constipation, and headache while its bark has been used to remove freckles (Miier & Morris Citation1989). Its associated microflora has been poorly known. Looking at the importance of its bark and the potential of the tree to withstand extreme environmental conditions, in the present study, we aimed to assess the endophytes present in the bark.
Materials and methods
Fungal endophyte isolation
The bark of M. peregrina Fiori (Moringaceae), growing in the wild mountains of Jabal Al-Akhdar (23°04′ 22.00 N″; 57°40′ 07.00 E″), Sultanate of Oman, was detached from the tree. The climatic condition of the region is arid and M. peregrina trees are growing in extreme water-deficient conditions (Ψ = −4.23 hPa). About 20 samples of bark randomly collected from different position of the stem, i.e. above the soil surface and below the twigs of four trees, were stored in a sterilized polythene zip-bag during transportation. The samples were thoroughly washed in running tape water. To isolate endophytic microbes, the method of Arnold et al. (Citation2007) was adopted. Briefly, the 20 bark samples were further sliced into 60 pieces (0.5 cm) and surface sterilized with 2.5% sodium hypochlorite (30 min in a shaking incubator at 120 rpm) and washed with autoclaved double distilled water (DDW) to remove contaminants and microorganisms. The bark pieces were carefully placed in petri-plates containing Hagem media (0.5% glucose, 0.05% KH2PO4, 0.05% MgSO4·7H2O, 0.05% NH4Cl, 0.1% FeCl3, 80 ppm streptomycin, and 1.5% agar, pH 5.6 ± 0.2). The sterilized bark parts were also imprinted on separate Hagem plates to ensure the effectiveness of the surface sterilization (Arnold et al. Citation2007). The newly emerged fungal spots from the bark were isolated and grown on potato dextrose agar (PDA) medium under sterile conditions. A total of eight different fungal strains were isolated and grown on PDA media. Rate of prevalence was calculated as the average % frequency of isolates per bark samples of stem. These strains were inoculated in Czapek broth (50 ml; 1% glucose, 1% peptone, 0.05% KCl, 0.05% MgSO4·7H2O, and 0.001% FeSO4·7H2O; pH 7.3 ± 0.2) and grown for 7 days (shaking incubator − 120 rpm; temperature 30°C) to separate the liquid culture medium and the fungal mycelia (centrifugation 2500×g at 4°C for 15 min). The culture medium (culture filtrate [CF], 50 ml) and mycelium were immediately shifted to −70°C freezer and then freeze-dried (Virtis Freeze Dryer, Gardiner, NY, USA) for 4–7 days. The lyophilized CF was diluted with 1 ml of autoclaved DDW, while the mycelia were used for genomic DNA extraction.
GAs screening using mutant rice
Presence or absence of GAs in the pure culture of endophytes was analyzed by performing screening bioassay on (1) waito-c and (2) Oryza sativa L. cv. Dongjin-byeo. Waito-c is a GAs biosynthesis dy mutant with dwarf phenotype while Dongjin-beyo has active GAs biosynthesis pathway with a normal growth pattern. Rice seeds were surface sterilized with 2.5% sodium hypochlorite for 30 min, rinsed with autoclaved DDW and incubated for 24 h with 20 ppm uniconazole (except Dongjin-byeo) to obtain equally germinated seeds. The mutant was treated with uniconazole to further inhibit the GAs pathways. For providing axenic conditions, the pre-germinated waito-c and Dongjin-byeo seeds were transferred to autoclaved small plastic pots containing 0.8% water: agar medium (Khan et al. Citation2011a, Citation2011b; Redman et al. Citation2011). After attaining two leaves stage, the CF (20 µl) harvested from endophytes was applied at the apex of rice seedlings. After 7 days, the rice growth was recorded and compared with CF of medium and G. fujikuroi KCCM12329-treated rice plants.
Endophyte identification by polymerase chain reaction (PCR)
An efficient method was developed for the isolation of genomic DNA from endophytic fungi, because usual Cetyltrimethylammonium bromide (CTAB) extraction method and mycelial grinding were causing DNA shearing. Rich mycelial culture was obtained by growing fungus in Czapek culture broth (supplemented with 1% glucose and peptone) for 7 days on rotary shaking incubator (120 rpm and 28°C) and lyophilized for 24 hrs. A 0.5 g of lyophilized sample was broken carefully in 2 ml eppendorf, with the blunt end spatula or with a glass rod. Double volume of lysis buffer (20 mM Tris–HCL, pH 8.0; 10 mM EDTA; 1% SDS) containing 1% of 2-mercaptoethanol was added. The mixture was vortexed briefly (30 sec) to obtain homogeneity and left to incubate for 2 hr in water bath set at 55°C. Pre-heated 4% CTAB extraction buffer (250 µl/ml) was added to lysed cells mixture and incubated further at 65°C for 1 hr. Chloroform extraction followed by iso-propanol precipitation yielded condensed strand of nucleic acid, which was cleaned from RNA using 10 µl of RNase A for 2 hr of incubation at 37°C. The isolated DNA was suspended in 50 µl of autoclaved DDW and tested for purity (Hamayun et al. Citation2009).
The fungal isolate was identified through sequence analysis of the internal transcribed region (ITS) of 18S rDNA, using universal primers ITS1 (5′-TCC GTA GGT GAA CCT GCG G-3′) and ITS4 (5′-TCC TCC GCT TAT TGA TAT GC-3′) (Taylor & Bruns Citation1999). A 25 µl of PCR mixture contained 2.5 µl of dNTPs and Ex-Taq buffer, 2 µl of each primer, 0.5 µl of DNA sample, and 0.2 µl of Ex-Taq polymerase. The remaining volume was adjusted with 15.3 µl of autoclaved DDW. For the amplification of ITS1 and ITS4 regions of 18S rDNA, the reaction cycle consisted of initial denaturation (95°C) for 2 min, 35 cycles of denaturation (95°C) for 30 s, annealing (55°C) for 60 s, extension (72°C) for 30 s and a final extension time for 5 min (72°C). The resultant products were gene cleaned using a Nucleogen gene clean kit, ligated in T-vector using Takara Perfect T-cloning kit, and then inserted into DHa Escherichia coli competent cells (RBC) by overnight incubation (37°C). Transformed cells were selected, grown overnight (37°C) in LB broth, and their plasmids were extracted using SolGent Plasmid mini-prep kit, which were later sequenced.
The BLAST search program (http://www.ncbi.nlm.nih.gov/BLAST) was used to compare the sequence homology of nucleotide of 18S ITS region of fungi. The closely related sequences obtained were aligned through CLUSTAL W using MEGA version 5.1 software (Tamura et al. Citation2011), and the maximum parsimony (MP) tree was constructed using the same software. The bootstrap replications (1K) were used as a statistical support for the nodes in the phylogenetic tree.
Quantification of GAs from bioactive endophyte
To characterize GAs secreted in the pure culture, endophytes were cultivated in Czapek broth for 7 days at 30°C (120 rpm) in 500 ml Erlenmeyer flask (150 ml liquid medium) as described previously (Waqas et al. Citation2014). The culture medium (CF) was used to extract and purify GAs as described by Lee et al. (Citation1998) and Hamayun et al. (Citation2009). Briefly, the CF (pH 2.5) was partitioned with ethyl acetate (EtOAc) and the organic layer was vacuum dried. It was added with 60% methanol (MeOH) while the pH was increased to 8.0 ± 0.3 by adding 2N NH4OH. Before column chromatography, deuterated GAs internal standards ([17, 17-2H2] GA1, GA3, GA4, and GA7 were added to the CF. The quantification of GAs was performed according to the method of Lee et al. (Citation1998). The extracts were passed through a Davisil C18 column (90–130 µm; Alltech, Deerfield, IL, USA). The eluent was reduced to near dryness at 40°C in vacuum. The sample was then dried onto Celite and loaded onto SiO2 partitioning column (deactivated with 20% water) to separate the GAs as a group from more polar impurities. GAs were eluted with 80 ml of 95:5 (v/v) EtOAc: hexane saturated with formic acid. This solution was dried at 40°C in vacuum, re-dissolved in 4 ml of EtOAc, and partitioned three times against 4 ml of 0.1 M phosphate buffer (pH 8.0). Drop-wise addition of 2 N NaOH was required during the first partitioning to neutralize residual formic acid. One gram polyvinylpolypyrrolidone (PVPP) was added to the combined aqueous phases, and this mixture was slurried for 1 h. The pH was reduced to 2.5 with 6 N HCl. The extract was partitioned three times against equal volumes of EtOAc. The combined EtOAc fraction was dried in vacuum, and the residue was dissolved in 3 ml of 100% MeOH. This solution was dried on a Savant Automatic Environmental Speedvac (AES 2000, Madrid, Spain). The extract of fungal CF was subjected to high performance liquid chromatography (HPLC) using a 3.9×300 m Bondapak C18 column (Waters Corp., Milford, MA, USA) and eluted at 1.0 ml/min with the following gradient: 0–5 min, isocratic 28% MeOH in 1% aqueous acetic acid; 5–35 min, linear gradient from 28% to 86% MeOH; 35–36 min, 86–100% MeOH; 36–40 min, isocratic 100% MeOH. Forty-eight fractions of 1.0 ml each were collected (Supplementary information 1Footnote1). The fractions were then prepared for gas chromatography/mass spectrometry (GC/MS) with selected ion monitoring (SIM) system (6890N Network GC System, and 5973 Network Mass Selective Detector; Agilent Technologies, Palo Alto, CA, USA). For each GAs, 1 µl of sample was injected in GC/MS (Supplementary information 1). Full-scan mode (the first trial) and three major ions of the supplemented [17-2H2] GAs internal standards and the fungal GAs were monitored simultaneously. The fungal CF GAs (GA1, GA3, GA4, and GA7) were calculated from the peak area ratios. The retention time was determined using hydrocarbon standards to calculate the Kovats retention index (KRI) value (Supplementary information 1). The data were calculated in nano-grams per milliliter (for fungal CF) while the analyses were repeated three times.
Statistical analysis
The data were analyzed statistically for standard deviation and error by using GraphPad Prism (version 5.0; CA USA). The mean values were compared using Duncan's multiple range tests at P < .05 (SAS, Cary, NC, USA).
Results and discussion
Endophytes isolation
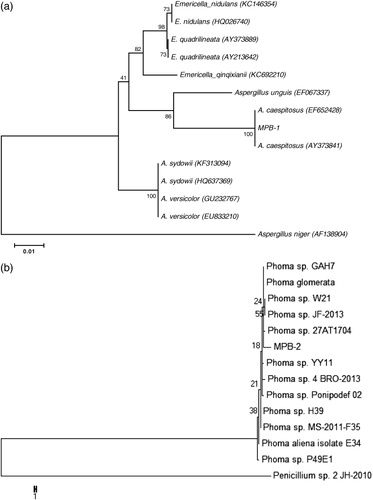
We isolated 11 fungal endophytes from 20 bark explants, which were further partitioned into 60 pieces (0.5 cm) randomly. These endophytes were grouped into eight different morpho-types on the basis of colony shape, thickness, color of aerial hyphae, colony reverse color, growth rate and pattern, margin characteristics, surface texture, and growth depth into medium (Tehler Citation1995). The rate of prevalence was 18.61%. Among eight endophytes (), MPB-1 and MPB-2 endophytes were found to be more abundant in bark samples.
Table 1. Effect of cultural filtrates of isolated endophytes on growth of GA mutant Waito-C and wild-type rice seedlings.
Bioactivity of endophytes
The endophytes were screened for their role in promoting the growth of GAs-deficient mutant waito-c and wild-type Dongjin-beyo rice seedlings. The activation of suppressed GAs biosynthesis in waito-c would reveal the presence of bioactive secondary metabolites while the effects were rectified using normal rice (Khan et al. Citation2011a, Citation2011b; Waqas et al. Citation2014). The screening results showed that CF application of endophytic strains MPB-1 and MPB-2 significantly increased the mutant and normal rice growth (). The shoot length, chlorophyll contents, and shoot fresh weight of waito-c and normal rice seedlings were significantly enhanced by the CF application of MPB-1 and MPB-2 as compared to the CF of G. fujikuroi and DDW applied to rice. The growth-promoting effects of MPB-1 and MPB-2 were not significantly different from each other ().
Dwarf rice (waito-c) is a GAs biosynthesis mutant line having passive dy gene which synthesizes bioactive GAs through C13 hydroxylation pathway (Ikeda et al. Citation2001). The waito-c rice seeds were treated with uniconazole to further suppress the GAs biosynthesis pathway (Ikeda et al. Citation2001). The use of waito-c rice can help detecting very small amounts of GAs if present in the sample (Hamayun et al. Citation2010). Agar and water, on the other hand, were used as growing medium for rice. During the bioassay, the rice seeds were fully grown on agar water media till the harvest in order to accurately measure the sole effect of fungal CF (Khan et al. Citation2011a, Citation2011b). Fusarium fujikuroi produces a wide array of diterpenoids including a group of phytohormones mainly GAs (Albermann et al. Citation2013). We compared the effect of CF of wild-type F. fujikuroi with that of our endophytes and found a similar growth-promoting behavior (Hamayun et al. Citation2010; Khan et al. Citation2011a, Citation2011b, Citation2013). The results of bioassay showed that MPB-1 and MPB-2 have significantly higher growth stimulatory effects; therefore, the CF of the strain might contain biologically active secondary metabolites.
GAs production by endophytes
The CF of MPB-1 and MPB-2 was subjected to chromatography and GC/MS SIM analysis for the quantification of GAs. For comparison, the wild type of G. fujikuroi was also subjected to GA analysis (Bomke et al. Citation2008). The results showed that the CF of both the bioactive endophytes contains GAs. The results of GC/MS SIM analysis evidenced the present of GA ion signals in correlation with [2H2] GA standards. The GA produced by both the endophytes has been given in . Total four different GAs (GA1, GA3, GA4, and GA7) in various amount (ng mL−1) were detected in the CF of MPB-1 (GA1 – 54.51 ± 1.23; GA4 – 26.5 ± 0.65; GA7 – 2.87 ± 1.23) and MPB-2 (GA1 – 4.8 ± 0.12; GA3 – 8.65 ± 0.21; GA4 – 23.7 ± 0.98; GA7 – 22.7 ± 0.73) (Supplementary information 1). The detection of bioactive GAs gives us the insight of GA production biosynthesis pathway in our isolated endophytes. The presence of GA4 in CF of both endophytes shows the subsequent conversion of GA14 and GA4 on desaturation then yields GA3 and GA7. While GA1 production is different from them, which is the minor product of 13-hydroxylation of GA4 and does not convert into GA3 (Albermann et al. Citation2013; Waqas et al. Citation2014).
Table 2. GAs quantification of the CFs of fungal endophytes.
GC/MS SIM is an established method to identify targeted novel secondary metabolites (Supplementary information 1) and the absence of the GA in the fungal-free culture broth supported our results. The repetition of the experiment and correlation with deuterated GA standards further confirmed our findings. Many fungal/endophytes species for example G. fujikuroi, S. manihoticola, Phaeosphaeria sp., N. crassa (Rademacher Citation1994), P. commune (Choi et al. Citation2005), Phaeosphaeria sp. L487 (Kawaide Citation2006), Penicillium citrinum, and P. funiculosum (Khan et al. Citation2011a) have been known to produce GAs. Previously A. fumigatus LHL02 (Khan et al. Citation2011c), Phoma sp. GAH7, and Phoma herbarum (Hamayun et al. Citation2009) were reported for GA production. The detection of these GAs suggests the existence of a GA gene cluster as reported for G. fujikuroi, Phaeospheria sp. L487, and Sphaceloma sp. (Bomke et al. Citation2008). Endophytes have been mostly reported for their behavior to stimulate plant growth. They can influence key aspects of plant physiology and host's protection against biotic and abiotic stresses (Schulz & Boyle Citation2005). Secretion of bioactive metabolites such as phytohormones by endophytes can extend greater benefits to the host plants even during extreme environmental conditions (Khan et al. Citation2011a, Citation2011b) while on the other hand, being eco-friendly, these can be extradited for commercial application and use at industrial levels.
Identification of endophytes
Fungal endophyte isolates were identified by PCR amplification and sequencing of ITS region. Phylogenetic analysis of the MPB-1 and MPB-2 was carried out through MEGA 5.1 using MP method. A consensus tree was made for MPB-1 from 13 (12 references and 1 clone) aligned ITS1 and ITS4 with 1000 bootstrap replications. The criteria for selection of strains to construct dendrogram were based on BLAST search showing maximum homology percentage, query coverage, and lowest E values with newly endophytes isolated in this study. Results of BLASTn search in National Center of Biotechnology Information (NCBI) revealed that MPB-1 has 100% sequence homology with Aspergillus caespitosus. In MP dendrogram, MPB-1 formed 100% bootstrap support with A. caespitosus (). The phylogenetic tree of MPB-2 formed 55% clade with Phoma sp. Therefore, on the basis of sequence similarity and phylogenetic analysis, fungal isolates MPB-1 and MPB-2 were identified as the new strains LK12 and LK13 of A. caespitosus and Phoma sp., respectively. Previously these fungi were isolated and identified from various sources; however, these are reported first time from the bark of Moringa. El-Nagerabi et al. (Citation2013) studied endophytic fungal population and isolated 80 different species including A. caespitosus and Phoma sorghina from the healthy leaves of Ziziphus spina-christi and Ziziphus hajanensis plants from the same study area over four years. They found that host-specific association with endophytes showed no seasonal variation. Thus, our study suggests the ecological importance of isolated endophytes, which could help the host plant to adopt the particular harsh arid and semi-arid natural habitat (Arnold & Lutzoni Citation2007). This confirms the ecological niche that many fungal endophytes play a central role in maintaining a suitable adoptive response of plant hosts.
Conclusion
GAs-producing endophytes can be an essential arsenal of plant, which can help the host and other crop plants to improve its growth. A similar effect was observed in the present study. Two strains of fungal endophytes belonging to A. caespitosus and Phoma sp. increased the growth of dwarf mutant and wild-type rice seedlings. Upon chromatographic and spectroscopic analysis of the CF, it was found that both the endophytes are producing physiologically active GAs. Application of such GAs-producing plant growth-promoting endophytes can not only improve plants growth but also extend great help in enforcing plant responses/resistance to biotic and abiotic stresses. However, similar properties need further analysis at tissue culture level application to host and field level application to economically important crops. Additionally, the presence of GAs suggests the existence of GA biosynthesis pathway, which could be further analyzed by the molecular techniques.
Supplementary_Material.doc
Download MS Word (47 KB)Acknowledgments
The authors would like to thank the financial support of The Oman Research Council through the open research grant (ORG/CBS/12/004), FURAP program, and National Research Foundation of Korea (2012R1A6A3A01040552 and 2011-0022027).
Notes
1. Supplemental Content may be viewed online at http://dx.doi.org/10.1080/17429145.2014.917384
References
- Albermann S, Elter T, Teubner A, Krischke W, Hirth T, Tudzynski B. 2013. Characterization of novel mutants with an altered gibberellin spectrum in comparison to different wild-type strains of Fusarium fujikuroi. Appl Microbiol Biot. 97:7779–7790. 10.1007/s00253-013-4917-7
- Arnold AE, Henk DA, Eells RL, Lutzoni F, Vilgalys R. 2007. Diversity and phylogenetic affinities of foliar fungal endophytes in loblolly pine inferred by culturing and environmental PCR. Mycologia. 99:185–206. 10.3852/mycologia.99.2.185
- Arnold AE, Lutzoni F. 2007. Diversity and host range of foliar fungal endophytes: are tropical leaves biodiversity hotspots? Ecology. 88:541–549. 10.1890/05-1459
- Bomke C, Rojas MC, Gong F, Hedden P, Tudzynski B. 2008. Isolation and characterization of the gibberellin biosynthetic gene cluster in Sphaceloma manihoticola. Appl Environ Microb. 74:5325–5339. 10.1128/AEM.00694-08
- Choi WY, Rim SO, Lee JH, Lee JM, Lee IJ, Cho KJ, Rhee IK, Kwon JB, Kim JG. 2005. Isolation of gibberellins producing fungi from the root of several Sesamum indicum plants. J Microbiol Biotechnol. 15:22–28.
- El-Nagerabi SAF, Elshafie AE, AlKhanjari SS. 2013. Endophytic fungi associated with Ziziphus species and new records from mountainous area of Oman. Biodiversitas. 14:10–16.
- Goma NH, Picó FX. 2011. Seed germination, seedling traits, and seed bank of the tree Moringa peregrina (Moringaceae) in a hyper-arid environment. Am J Bot. 98:1024–1030. 10.3732/ajb.1000051
- Hamayun M, Khan SA, Khan AL, Afzal M, Lee IJ. 2012. Endophytic Cephalotheca sulfurea AGH07 reprograms soybean to higher growth. J Plant Interact. 7:301–306. 10.1080/17429145.2011.642013
- Hamayun M, Khan SA, Khan AL, Rehman G, Sohn EY, Shah AA, Kim SK, Joo GJ, Lee IJ. 2009. Phoma herbarum as a new gibberellin-producing and plant growth promoting fungus. J Microbiol Biotechnol. 19:1244–1249.
- Hamayun M, Khan SA, Khan AL, Tang DS, Hussain J, Ahmad N, Anwar Y, Lee IJ. 2010. Growth promotion of cucumber by pure cultures of gibberellin-producing Phoma sp. GAH7. World J Microb Biot. 26:889–894. 10.1007/s11274-009-0248-3
- Hamilton CE, Bauerle TL. 2012. A new currency for mutualism? Fungal endophytes alter antioxidant activity in hosts responding to drought. Fungal Divers. 54:39–49. 10.1007/s13225-012-0156-y
- Hegazy AK, Hammouda O, Lovett-Doust J, Gomaa NH. 2008. Population dynamics of Moringa peregrina along altitudinal gradient in the northwestern sector of the Red Sea. J Arid Environ. 72:1537–1551. 10.1016/j.jaridenv.2008.03.001
- Hyde KD, Soytong K. 2008. The fungal endophyte dilemma. Fungal Div. 33:163–73.
- Ikeda A, Ueguchi-Tanaka M, Sonoda Y, Kitano H, Koshioka M, Futsuhara Y, Matsuoka M, Yamaguchi J. 2001. Slender rice, a constitutive gibberellin response mutant, is caused by a null mutation of the SLR1 gene, an ortholog of the height-regulating gene GAI/RGA/RHT/D8. Plant Cell. 13:999–1010. 10.1105/tpc.13.5.999
- Kawaide H. 2006. Biochemical and molecular analysis of gibberellins biosynthesis in fungi. Biosci Biotechnol Biochem. 70:583–590. 10.1271/bbb.70.583
- Khan AL, Hamayun M, Ahmad N, Hussain J, Kang SM, Kim YH, Adnan M, Tang DH, Waqas M, Radhakrishnan R, et al. 2011a. Salinity stress resistance offered by endophytic fungal interaction between Penicillium minioluteum LHL09 and Glycine max. L. J Microbiol Biotechnol. 21:893–902. 10.4014/jmb.1103.03012
- Khan AL, Hamayun M, Kim YH, Kang SM, Lee IJ. 2011b. Ameliorative symbiosis of endophyte (Penicillium funiculosum LHL06) under salt stress elevated plant growth of Glycine max L. Plant Physiol Biochem. 49:852–862. 10.1016/j.plaphy.2011.03.005
- Khan AL, Hamayun M, Kim YH, Kang SM, Lee JH, Lee IJ. 2011c. Gibberellins producing endophytic Aspergillus fumigatus sp. LH02 influenced endogenous phytohormonal levels, plant growth and isoflavone biosynthesis in soybean under salt stress. Process Biochem. 46:440–447. 10.1016/j.procbio.2010.09.013
- Khan AL, Hussain J, Al-Harrasi A, Al-Rawahi A, Lee IJ. 2013. Endophytic fungi: a source of gibberellins and crop resistance to abiotic stress. Crit Rev Biotechnol. doi:10.3109/07388551.2013.800018
- Kumar M, Sharma R, Jogawat A, Singh P, Dua M, Gill SS, Trivedi DK, Tuteja N, Verma AK, Oelmuller R, Johri AK. 2012. Piriformospora indica, a root endophytic fungus, enhances abiotic stress tolerance of the host plant. In: Tuteja N, Gill SS, Tiburcio AF, Tuteja R, editors. Improving crop resistance to abiotic stress. Verlag: Wiley-VCH; p. 231–234.
- Lee IJ, Foster K, Morgan PW. 1998. Photoperiod control of gibberellin levels and flowering in sorghum. Plant Physiol. 116:1003–1011. 10.1104/pp.116.3.1003
- Marwah RG, Fatope MO, Mahrooqi RA, Varma GB, Abadi HA, Al-Burtamani SKS. 2007. Antioxidant capacity of some edible and wound healing plants in Oman. Food Chem. 101:465–470.
- Miier AG, Morris M. 1989. Plants of Dhofar, the southern region of Oman. Traditional economic and medicinal uses. The office of the advisor for conservation of the environment. Sultanate of Oman: Diwan of Royal Court.
- Rademacher W. 1994. Gibberellin formation in microorganisms. Plant Growth Regul. 15:303–314. 10.1007/BF00029903
- Redman RS, Kim YO, Woodward CJDA, Greer C, Espino L. 2011. Increased fitness of rice plants to abiotic stress via habitat adapted symbiosis: a strategy for mitigating impacts of climate change. PLoS One. 6:e14823. 10.1371/journal.pone.0014823
- Schulz B, Boyle C. 2005. The endophytic continuum. Mycol Res. 109:661–686. 10.1017/S095375620500273X
- Schulz B, Boyle C, Draeger S, Rommert AK, Krohn K. 2002. Endophytic fungi: a source of biologically active secondary metabolites. Mycol Res. 106:996–1004. 10.1017/S0953756202006342
- Taylor DL, Bruns TD. 1999. Community structure of ectomycorrhizal fungi in a Pinus muricata forest: minimal overlap between the mature forest and resistant propagule communities. Mol Ecol. 8:1837–1850.
- Tehler A. 1995. Morphological data, molecular data, and total evidence in phylogenetic analysis. Can J Botany. 73:667–676. 10.1139/b95-309
- Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. 2011. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 28:2731–2739.
- Waqas M, Khan AL, Lee IJ. 2014. Bioactive chemical constituents produced by endophytes and effects on rice plant growth. J Plant Interact. 9:478–487.
- Zaghloul MS, Abdel-Wahab RH, Moustafa AA. 2011. Ecological assessment and phenotypic and fitness variation of Sinai's remnant populations of Moringa peregrina. Appl Ecol Env Res. 8:351–366.
- Zhou Z, Zhang C, Zhou W, Li W, Chu L, Yan J, Li H. 2014. Diversity and plant growth-promoting ability of endophytic fungi from the five flower plant species collected from Yunnan, Southwest China. J Plant Interact. 9:585–591.
