Abstract
In the present study, different types of gibberellins (GAs) in the culture filtrate (CF) of Photorhabdus temperata M1021 were quantified. The analysis of CF helped in profiling various bioactive GAs: GA1, GA3, GA4, and GA7. Several physiologically inactive GAs: GA9, GA12, and GA20 were detected as well. Siderophore production was also investigated by growing P. temperata M1021 on chrome azurol-S blue agar plates. Furthermore, the strain was inoculated into ‘Waito-C’ (Oryza sativa L.) rice plants, which significantly (P < 0.05) increased plant growth attributes such as plant length, chlorophyll content, and fresh and dry biomass compared with those in controls. In a separate experiment, canola (Brassica napus L.) seeds treated with CF of M1021 were significantly (P < 0.05) accelerated germination rate as well as biomass production. Findings of the present study suggest that the strain M1021 contributes an important role in the plant growth by synthesizing a wide array of bioactive metabolites.
Introduction
Photorhabdus species are members of Enterobacteriaceae that invade and kill insects and live in symbiotic association with soil-dwelling entomopathogenic heterorhabditis nematodes (Akhurst Citation1980; Jang et al. Citation2012). Bacteria from the genus of Photorhabdus are currently used in many parts of the world as substitutes for chemical pesticide (agrochemical) application to control agricultural pests (Grewal et al. Citation2001). However, thorough investigation of the effects of Photorhabdus spp. on plant growth and development is essential before its development as a biocontrol agent. Genome analysis of Photorhabdus spp. has revealed many genes responsible for the production of metabolites other than proteinous metabolites (Duchaud et al. Citation2003; Park et al. Citation2013), and biochemical analyses of culture extracts have confirmed the existence of secondary metabolites such as antimicrobial and insecticidal compounds (Seo et al. Citation2012), siderophores (Watson et al. Citation2005), and phytohormones indole-3-acetic acid (Ullah et al. Citation2013).
Photorhabdus spp. produce siderophores for entomopathogenic and antimicrobial activities (Watson et al. Citation2010) however, in the rhizosphere, Siderophores chelate iron (Fe3+) and accelerate its mobility in the rhizosphere, making it available to plants (Römheld & Marschner Citation1981). Indirectly siderophores chelate Fe3+, making it inaccessible to pathogens and hence irradiate them from the surround environment and facilitate the plants to flourish (Joo et al. Citation2009; Glick Citation2012). Gibberellins (GAs) are plant hormones that initiate various metabolic functions, i.e. stem elongation, flowering, fruit formation, seed germination, and senescence (Taghavi et al. Citation2010; Kang et al. Citation2012). Besides the plants, GAs have been detected in fungi, for example, Aspergillus caespitosus LK12 and Phoma sp. LK13 (Khan et al. Citation2014), Paecilomyces formosus LHL10 (Khan et al. Citation2012), Aspergillus fumigatus sp. LH02 (Khan et al. Citation2011), Penicillium funiculosum LHL06 (Khan & Lee Citation2013). Similarly, bacterial genus e.g. Bacillus pumilus, Bacillus cereus, Bacillus macroides (Joo et al. Citation2004), Acinetobacter calcoaceticus SE370 (Kang et al. Citation2009), Promicromonospora sp. SE188 (Kang et al. Citation2012), Burkholderia cepacia SE4 (Kang et al. Citation2010), Pseudomonas spp. (Goswami et al. Citation2013) have been successfully analyzed for the GAs production. Entomopathogenic bacteria have been recently reported to produce plant growth regulators (Raddadi et al. Citation2008), that not only improve plant growth and development but also enhance the fitness of the host against a diverse array of environmental stresses (Joo et al. Citation2009; Kang et al. Citation2012). Bacteria produce GAs in culture extract that can be detected using gas chromatography coupled with mass spectrometry (GC/MS) with selected ion monitoring mode (SIM), which has been used extensively for the analysis of metabolites because of its greater separation efficiency that resolves very complex biological mixtures (Schneider et al. Citation1985).
Achieving the dual impacts of plant growth promotion and pest control with an entomopathogenic microbe is an ideal strategy for combatting various environmental and crop production threats. Exploring and assaying such beneficial microbes and their relationship to crop plants are new directions in agricultural biotechnology (Berg Citation2009). The present study used GC/MS–SIM to analyze plant growth-promoting hormones (GAs) in the culture extract of P. temperata M1021 and assessed the roles of these hormones in plant health and growth promotion. These analyses are initial steps in exploring the potential to exploit P. temperata as a bio-fertilizer, plant strengthener, phyto-stimulator, and biopesticide. Further study is needed to explore the many other potentially beneficial applications of P. temperata in agriculture.
Materials and methods
Culture preparation and maintenance
The present study was carried out with P. temperata strain M1021, identified, and characterized from soil entomopathogenic nematodes collected from the South Korean locations reported in our previous study (Jang et al. Citation2012). Whilst the draft genome sequencing of the strain has also been accomplished and the sequence is available in GenBank Whole-Genome Shotgun (WGS) database under the accession no. AUXQ00000000 (Park et al. Citation2013). During the present study the agricultural aspects of the strain were analyzed to determine its effect on plant growth. During the present study, the strain M1021 was routinely cultured in Luria-Bertani (LB) broth (0.5% yeast extract, 1% NaCl, 1% tryptone) and incubated at 28 ± 2°C for 48 h to prepare a pre-culture with an initial optical density of 0.6 at 600 nm. The strain was cultured on LB agar plant and in broth for both short- and long-term storage and subsequent use in experiments.
Siderophore production assay with chrome azurol-S (CAS) blue agar
All laboratory wares were treated with 6 M HCl to eliminate iron and rinsed thoroughly with autoclaved distilled water (DW). The CAS assay was modified for M1021 to test its capability for siderophore production. One litter CAS blue agar was prepared according to the procedure of Schwyn and Neilands (Citation1987). Solution A consisted of 60.5 mg CAS dissolved in 50 ml double distilled water and mixed with 10 ml ferric (Fe3+) solution (1 mM FeCl3 6H20, 10 mM HCl). The solution was added to 72.9 mg hexadecyltrimethylammonium dissolved in 40 ml water with gentle stirring. The dark blue solution was autoclaved at 121°C for 15 min. Solution B was prepared with 750 ml water, 15 g agar, 30.24 g PIPES, and 12 g of a solution of 50% (w/w) NaOH to raise the pH to 6.8. Solution B was autoclaved. The two solutions were mixed and agitated carefully to avoid foam formation. The CAS blue agar was poured into Petri plates and allowed to solidify and room temperature. The M1021 culture was grown on the LB agar plates for 3 days at 28 ± 2°C and equal size colonies were transferred to the CAS blue agar plates. The plates were incubated at 28 ± 2°C for 12 days and diameter of the colonies was measure at regular interval of time. Uninoculated CAS blue agar plates were used as control.
Quantification of GAs from the extract of M1021
To characterize the GAs secreted by M1021, we cultivated M1020 in LB broth for 7 days at 28 ± 2°C in a shaking incubator at 200 ± 20 rpm. The bacterial culture was then centrifuged at 10,000× g for 10 min at 4°C, and the supernatant was filtered through a 0.45-µm cellulose acetate filter. The culture filtrate (CF) was used to extract and purify GAs as described by Lee et al. (Citation1998). The filtrate was acidified to pH 2.8 ± 0.2 with 1 N HCl and extracted three times with a 2× volume of ethyl acetate (EtOAc). Then, the organic layers were combined and dried under vacuum at 45°C in a rotary evaporator. Nearly dry residues were resuspended in 60% methanol (MeOH), and the pH was adjusted to 8 with 2 N NH4OH. Before further analysis, deuterated GA internal standards ([17, 17-2H2] GA1,2,…n) were added to the CF. Quantification of GAs was performed according to the method of Lee et al. (Citation1998).
The extract was then passed through a Davisil C18 open column (90–130 µm; Alltech, Deerfield, IL, USA) and eluted with 40 ml 60% MeOH, and the eluent was saturated by evaporating excess solvent at 45°C under vacuum. The sample was then dried onto diatomaceous earth and loaded onto a SiO2 partitioning column (deactivated with 20% water) to separate the GAs as a group from more polar impurities. GAs were eluted with 80 ml 95:5 (v/v) EtOAc:hexane saturated with formic acid. The eluent was dried at 45°C in vacuum and resuspended in 5 ml EtOAc, and formic acid residues were neutralized via dropwise addition of a 2 N NaOH solution. Then, the solution was partitioned three times against 5 ml 0.1 M phosphate buffer (pH 8.0). Exactly 1 g polyvinylpolypyrrolidone was slurried for 1 h with the combined aqueous phases, and the pH was reduced to 2.5 with 6 N HCl. The extract was partitioned again with a 1× volume of EtOAc. The partitioning was repeated three times, combined fractions were dried under vacuum, and the residues were resuspended in 2 ml of 100% MeOH.
The sample was prepared for high-performance liquid chromatography (HPLC) using a C18 column (Waters Corp., Milford, MA, USA) and eluted at 1 ml min−1 with the following gradient: 0–5 min, isocratic 28% MeOH in 1% aqueous acetic acid; 5–35 min, linear gradient from 28% to 86% MeOH; 35–36 min, 86% to 100% MeOH; 36–40 min, isocratic 100% MeOH. Forty-eight fractions of 1.0 ml each were collected. The fractions were then evaporated using a Savant Automatic Environmental Speed-Vac (AES 2000, Madrid, Spain). Methyl esters of the fractions were prepared by dissolving the residue in 1 ml MeOH and adding 1 ml ethereal diazomethane. The excess diazomethane was removed under a stream of dry, oxygen-free nitrogen gas. The methylated samples were redissolved in EtOAc before analysis with a GC/MS–SIM system (6890N Network GC System and 5973 Network Mass Selective Detector; Agilent Technologies, Palo Alto, CA, USA).
For each GA, 1 µl of sample was injected into the GC/MS instrument. Full-scan mode (the first trial), three major ions of the supplemented [17-2H2] GA internal standards, and the bacterial GAs were monitored simultaneously. The bacterial CF GAs were calculated from the peak area ratios. The retention time was determined using hydrocarbon standards to calculate Kovats retention indices (). The data were reported in nanograms.
Table 1. GC/MS–SIM analysis of HPLC fractions of pure culture filtrate of M1021.
Bioassay on ‘Waito-C’
Culture extract of M1021 was tested in a bioassay with ‘Waito-C’ (Oryza sativa L.) rice plants to monitor effects on plant health such as plant morphology and physiology. Seeds were surface sterilized via soaking in 75% ethanol for 2 min and then disinfected (sodium hypochlorite:water:0.05% Triton X-100 in a v/v ratio of 3:2:2) for 1 min. Seeds were washed thoroughly, soaked in autoclaved DW, stored in a desiccator at 4°C for 72 h, and germinated on autoclaved filter paper. Two ‘Waito-C’ seedlings were transplanted per pot (50 ml) filled with 30 ml of 0.8% (w/v) water agar medium. The plants were grown in a controlled environment in a growth chamber with 16 h, 30°C day (light intensity, 20,000 lx) and 8 h, 20°C night regimens and a relative humidity of approximately 70%. The inoculum was prepared by inoculating 3 ml of LB broth with a single colony of M1021 and incubated at 28 ± 2°C for 48 h at 200 rpm. The culture broth was used to seed 100 ml LB broth, which was further seeded into 1 L LB broth. The culture broth was then incubated at 28 ± 2°C for 7 days in a shaking incubator at 200 rpm. Seven-day-old culture was used as inoculum, and ‘Waito-C’ seedlings were treated with 5 ml bacterial suspension at the two-leaf stage. Growth attributes such as plant length, fresh biomass, dry biomass, and chlorophyll content were recorded after 21 days of treatment and compared with those of control seedlings treated with LB or DW. The experiment consisted of 10 plants per treatment, and three replications were performed.
Seed germination test on Canola
An experiment was designed to analyze the effects of M1021 extract on seed germination and dormancy and biomass production. Canola (Brassica napus L.) seeds were used for the assay. Seeds were surface sterilized via treatment with 70% ethanol for 1 min followed by 2% NaOCl treatment for 30 s. M1021 was grown in LB broth for 7 days at 28 ± 2°C in a shaking incubator at 200 ± 20 rpm. The culture broth was partitioned into pellets and supernatant via centrifugation at 10,000× g for 10 min at 4°C, and the supernatant was filtered through a 0.2-µm cellulose acetate filter. Up to 50 ml supernatant was lyophilized at −70°C in a freeze dryer. The lyophilized supernatant was mixed with 2 ml autoclaved deionized DW, and 1 ml of this solution was applied to the canola seeds. The seeds were then transferred to sterilized filter papers and soaked with 1 ml autoclaved deionized DW in a Petri dish.
The germination test lasted for 1 week at 28°C and 70% humidity and monitored the effects of the treatment on seed germination as well as the biomass of the canola plant seedlings. Canola seeds treated with LB media and DW were used as positive and negative controls, respectively. The experiment consisted of 10 seeds per treatment, and 3 replications were performed.
Statistical analysis
Data were analyzed statistically for standard deviation using EXCEL software (Microsoft). Mean values were compared with Duncan's multiple range test at a P value of 0.05 (analysis of variance; SAS release 9.1; SAS, Cary, NC, USA).
Results and discussion
GAs quantification culture extract
M1021 CF was subjected to HPLC and GC/MS–SIM analysis for the quantification of GAs. GC/MS–SIM analysis detected GA ion signals in correlation with [2H2] GA standards (see ) and revealed the presence of GA1, GA3, GA4, GA7, GA9, GA12, and GA20. These GAs were identified and quantified by comparing their mass spectra and Kovats retention indices with those available from a spectral library (Supplemental data). The bioactive GA1, GA3, GA4, and GA7 were the most abundant GAs and were present in concentrations of 25 ng, 8.16 ng, 1.32 ng, and 0.4 ng ml−1 of CF, respectively. However, physiologically inactive GAs such as GA9, GA12, and GA20 were also present in concentrations of 30.39 ng, 4.45 ng and 10 ng ml−1 of CF, respectively ().
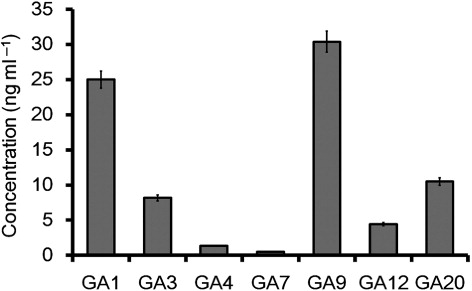
Previous reports have provided detailed information about various bioactive metabolites such as GAs produced by bacterial species (Joo et al. Citation2009; Kang et al. Citation2012). In the present study, we detected different bioactive and inactive GAs in the CF of M1021, and these results are strongly supported by earlier reports suggesting that entomopathogenic bacteria produce plant hormones. Raddadi et al. (Citation2008) have reported that several strains of Bacillus thuringiensis produce various growth-promoting metabolites including phytohormones. Similarly, Ullah et al. (Citation2013) have reported that P. temperata produces plant growth hormones. These reports suggest that entomopathogenic bacteria can interfere with plant hormones in complex ways. Furthermore, many species of bacteria have been demonstrated to produce various GAs but unlike the plants, there is no known role for GAs, rather they seem to be secondary metabolites that are not directly involved in the normal growth and development of the bacteria. However, the exogenous GAs produced by various bacterial strains are actively involved in the plant growth and promotion (Waqas et al. Citation2014). For example, Kang et al. (Citation2009) have reported that a strain of A. calcoaceticus SE370 secretes 10 GAs into its growth environment, including the bioactive GA1, GA3, and GA4. Similarly, Joo et al. (Citation2004, Citation2005) have identified GAs in B. cereus MJ-1, B. macroides CJ-29, and B. pumilus CJ-69. Several studies of Azospirillum sp. have characterized GAs using capillary GC/MS. GA analysis using HPLC coupled with GC/MS–SIM gives more reliable results than those obtained using thin-layer chromatography, bioassays, or HPLC with ultraviolet detection, which have poor resolution and decreased reliability (Hamayun et al. Citation2011; Waqas et al. Citation2013; Khan et al. Citation2014). GC/MS–SIM is an established method for identifying targeted novel secondary metabolites, and our results were confirmed when we detected no GAs in the bacterial-free culture broth treated as same using negative control. The repetition of the experiment and correlation with deuterated GA standards further confirmed our findings.
Siderophore production assay
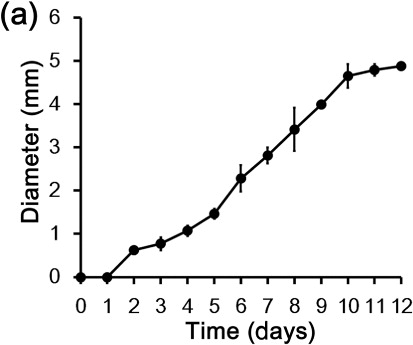
To evaluate siderophore production in M1021, we stabbed M1021 colonies over CAS blue agar plates and incubated the plates at 28 ± 2°C. The plates were observed for 12 days for bacterial siderophore production. The color of the CAS blue agar changed to pink and then orange and the diameters of the haloes were dilated with the elapse of time during the incubation (). Orange haloes around the M1021 colonies indicated the presence of chelators such as siderophores produced by bacterial colonies in the surrounding media, and increases in the diameter of the orange zones with time suggested the augmentation of siderophores in the medium. Watson et al. (Citation2005) have also investigated siderophore production in P. temperata on agar plates and reported similar results, confirming that Photorhabdus bacteria produce siderophores. Previous reports reinforce the results of the present study and suggest that iron association imparts the blue color in the CAS media, which changes from blue to orange in the presence of iron chelators because the iron is detached from the dye. Iron uptake in plants is reportedly enhanced by siderophores produced by several microbes (Miethke & Marahiel Citation2007; Pérez-Miranda et al. Citation2007). Photorhabdus spp. have been investigated by Watson et al. (Citation2005) and Ciche et al. (Citation2003) for the siderophore production at genetic level and reported exbD and ngrA genes (PPTase), encoding a component of TonB protein complex and 4′-phosphopantetheinyl transferase, respectively. These genes are responsible for the siderophores production in the bacterium. TonB complex and (PPTase) enable the Photorhabdus spp. to flourish in conditions where iron is not freely available in the environment (medium). According to Watson et al. (Citation2010), siderophore production is an entomopathogenic characteristic of the Photorhabdus spp. through which they chelate the Fe3+ from the hemolymph, leaving the insect deprived of Fe3+, resulting death of the host. Previously, Römheld and Marschner (Citation1981) and Zocchi et al. (Citation2007) have suggested that Fe3+ chelating potential and chemical stability of the siderophores can be exploited for the plant growth promotion, as siderophores chelate iron, making it unavailable to pathogens and thereby suppressing the growth of microbial pathogens in specific niches surrounding plants and improving plant health and growth indirectly. Others have hypothesized that siderophores chelate Fe+3, significantly increasing its mobility in the soil and making it more accessible to plants (Römheld & Marschner Citation1981; Dobbelaere et al. Citation2003). Overall, the above results support the inference that in addition to being plant growth hormones, siderophores produced by M1021 could be factors that promote plant health and growth. However further study is required to identify the exact pathway and type(s) of the specific siderophore(s) secreted by strain M1021 in the medium.
Plant growth promotion by P. temperata M1021
The entomopathogenic role of M1021 is well established and has been thoroughly studied, but its genome also contains genes responsible for the production of beneficial metabolites such as plant growth hormones (Park et al. Citation2013). The role of M1021 in plant growth regulation has been poorly investigated. Plant bioassays carried out in the present study revealed the positive effects of M1021 on the health and growth of rice plants and showed that microbial treatment enhanced the growth attributes of plants. ‘Waito-C’ (dwarf plants that are mutant for reduced GA production) were selected for the plant growth assay. Inoculation of M1021 into ‘Waito-C’ plants significantly enhanced their growth attributes: plant length, the fresh and dry biomass of the plants, and chlorophyll content were significantly increased compared to those in control plants treated with LB media and DW.
M1021 application increased shoot length by 66.58% and 40.87% compared with that associated with DW and LB treatment, respectively (). The dry weights of the shoots and roots of M1021-treated plants also increased significantly. M1021-treated plants were 48.97% and 37.13% heavier than DW- and LB-treated plants, respectively. Similarly, chlorophyll content of the ‘Waito-C’ plants was measured in response to M1021 treatment. The results showed that the bacterial treatment increased chlorophyll content by 14.46% compared with that in controls. The present study demonstrated that the bioactive GAs and siderophores produced by M1021, may be potential factors of growth promoting rice plants. This bacterium produced seven different GAs including four physiologically bioactive GA1 (25 ng ml−1), GA3 (8.16 ng ml−1), GA4 (1.36 ng ml−1), and GA7 (0.5 ng ml−1). The results suggest that the bioactive GAs can be a vital factor, contributing in plant growth promotion even at very low concentrations. However, concentrations of the exogenous bioactive GAs higher than 1000 ng ml−1 inhibit the growth promotion in plants (Inada & Shimmen Citation2000). Previously, Kang et al. (Citation2009) and Joo et al. (Citation2009) observed that inoculation of rice seedlings with rhizobacteria e.g. Promicromonospora sp. SE188, Burkholderia sp. KCTC 11096BP and A. calcoaceticus (Rhizobium strains) increased the seedling vigor, root length, shoot length, and yield of rice plants whilst the rhizobacteria was also producing different types of bioactive GAs. The secretion of bioactive metabolites into growth medium by microbes indicates great advantages for agricultural cash crops such as wheat, rice, Brassica juncea, and barley (Khan et al. Citation2013). Earlier reports have disclosed that bioactive cultures can activate endogenous GA biosynthesis pathways in treated plants (Kang et al. Citation2012). GAs act as signaling molecules which degrade the DELLA protein complex and allow the phytochrome interacting factors to bind the gene promoters and regulate gene expression (Davière et al. Citation2008). Siderophore production is another characteristic of the M1021 through which bacteria chelate Fe3+, making it unavailable to pathogens as well as increase it mobility in rhizosphere and thereby, improve plant growth attributes (Römheld & Marschner Citation1981; Zocchi et al. Citation2007).
Table 2. Effects of Photorhabdus temperata M1021 culture extract on growth attributes of ‘Waito-C’ rice seedlings.
Dwarf rice (‘Waito-C’) was used in this experiment, as it is a GA biosynthesis mutant line with a passive dy gene that synthesizes bioactive GAs via a C13 hydroxylation pathway (Ikeda et al. Citation2001). ‘Waito-C’ rice seeds have been treated with uniconazole for additional suppression of this GA biosynthesis pathway (Ikeda et al. Citation2001). The use of ‘Waito-C’ rice can detect very small amounts of bioactive metabolites present in a culture (Hamayun et al. Citation2010; Khan et al. Citation2011). Agar and water were used as the growing media for the rice. Therefore, the rice seeds received no nutrients during the bioassay so that the sole effect of bacterial CF could be accurately measured (Khan et al. Citation2011).
Seed germination and biomass augmentation
Photorhabdus bacteria produce a variety of secondary metabolites during their life cycles that could be extremely valuable in agriculture. We treated canola seeds with a culture extract of M1021 to investigate its role in the induction of germination. Treatment with the culture extract significantly (P < 0.05) induced the germination of canola seeds compared with controls. The results indicated that culture extract stimulated seed germination to 40% after 1 day of treatment, whereas no germination was detected in the LB- or DW-treated control seeds (). The germination rate accelerated to 70% on the second day compared with a rate of 36.6% in the controls. Furthermore, treatment on the third and fourth days induced germination up to 76.6% and 83.3%, respectively, whereas seed germination remained at 36.6% and increased to 43.3% in both controls on the third and fourth days, respectively. The experiment was concluded on fifth day, on which bacterial treatment resulted in 93.3% seed germination compared with 43.3% and 50% germination in the LB and DW controls, respectively. In addition, the results indicated that the fresh biomass of the canola seedlings was significantly (P < 0.05) increased by culture extract treatment compared with that in the controls (LB and DW treatment; ).
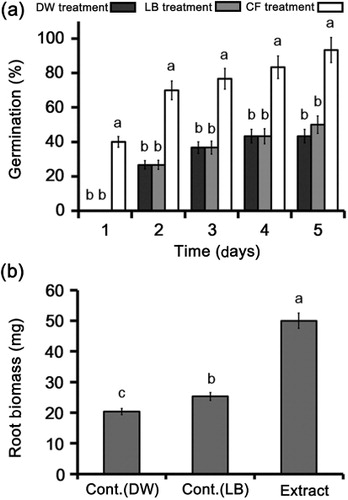
The seed germination capacity of M1021 might be attributed to its capacity to produce GAs (Glick Citation2012). GAs are ubiquitous plant hormones that initiate various metabolic functions during physiological activities such as plant growth, cellular totipotency, and seed germination (Großelindemann et al. Citation1992; Hedden & Kamiya Citation1997). Early studies of seed germination support the results of the present study and suggest that GAs accumulation in the environment stimulates massive cell proliferation, enhancing seed germination (Grosselindemann et al. Citation1991). Furthermore, according to Groot and Karssen (Citation1987), various bioactive GAs are involved in the mobilization of storage carbohydrates and hence weaken the endosperm cells surrounding the radicle tip in the seed, causing early germination and biomass production in seedling. All of this evidence indicates that the bioactive GAs produced by M1021 potentially contribute to plant growth and seed germination.
Conclusion
Bacteria of the genus Photorhabdus are used as biocontrol agents against a wide range of insect pests, and their antibacterial and antifungal characteristics have been well documented. However, to the best of our knowledge, no study involved examination of the capability of these bacteria to promote plant growth. To the best of our knowledge this study is the first in which we have analyzed the potential of P. temperata M1021 as a polyvalent biocontrol agent, bio-stimulator, and bio-fertilizer. The results suggest that M1021 produces several physiologically active and inactive GAs and has the capacity to produce siderophores that could be growth-promoting factors in ‘Waito-C’ rice plants. In short, we conclude that M1021 could be a factor for plant health and growth and that broader field trails to test its use as a bio-fertilizer for increased crop production in eco-friendly farming systems are justified.
Supplemental data
Supplemental data for this article can be accessed at http://dx.doi.org/10.1080/17429145.2014.942956.
Supplementary_Material.doc
Download MS Word (44.5 KB)Funding
This research was supported by the Basic Science Research Program through the National Research Foundation of Korea funded by the Ministry of Education [grant number NRF-2013R1A1A2010298].
Additional information
Funding
References
- Akhurst RJ. 1980. Morphological and functional dimorphism in Xenorhabdus spp., bacteria symbiotically associated with the insect pathogenic nematodes Neoaplectana and Heterorhabditis. J Gen Microbiol. 121:303–309.
- Berg G. 2009. Plant–microbe interactions promoting plant growth and health: perspectives for controlled use of microorganisms in agriculture. Appl Microbiol Biotechnol. 84:11–18. 10.1007/s00253-009-2092-7
- Ciche TA, Blackburn M, Carney JR, Ensign JC. 2003. Photobactin: a catechol siderophore produced by Photorhabdus luminescens, an entomopathogen mutually associated with Heterorhabditis bacteriophora NC1 nematodes. Appl Environ Microbiol. 69:4706–4713.
- Davière JM, De-Lucas M, Prat S. 2008. Transcriptional factor interaction: a central step in DELLA function. Curr Opin Genet Dev. 18:295–303. 10.1016/j.gde.2008.05.004
- Dobbelaere S, Vanderleyden J, Okon Y. 2003. Plant growth-promoting effects of diazotrophs in the rhizosphere. Crit Rev Plant Sci. 22:107–149. 10.1080/713610853
- Duchaud E, Rusniok C, Frangeul L, Buchrieser C, Givaudan A, Taourit S, Bocs S, Boursaux-Eude C, Chandler M, Charles J-F, et al. 2003. The genome sequence of the entomopathogenic bacterium Photorhabdus luminescens. Nat Biotech. 21:1307–1313. 10.1038/nbt886
- Glick BR. 2012. Plant growth-promoting bacteria: mechanisms and applications. Scientifica. 2012:15. 10.6064/2012/963401
- Goswami D, Vaghela H, Parmar S, Dhandhukia P, Thakker JN. 2013. Plant growth promoting potentials of Pseudomonas spp. strain OG isolated from marine water. J Plant Interact. 8:281–290. 10.1080/17429145.2013.768360
- Grewal PS, Nardo EABD, Aguillera MM. 2001. Entomopathogenic nematodes: potential for exploration and use in South America. Neotropical Entomol. 30:191–205. 10.1590/S1519-566X2001000200001
- Großelindemann E, Lewis M, Hedden P, Graebe J. 1992. Gibberellin biosynthesis from gibberellin A12-aldehyde in a cell-free system from germinating barley (Hordeum vulgare L., cv. Himalaya) embryos. Planta. 188:252–257. 10.1007/BF00216821
- Groot SPC, Karssen CM. 1987. Gibberellins regulate seed germination in tomato by endosperm weakening: a study with gibberellin-deficient mutants. Planta. 171:525–531. 10.1007/BF00392302
- Grosselindemann E, Graebe JE, Stockl D, Hedden P. 1991. ent-Kaurene biosynthesis in germinating barley (Hordeum vulgare L., cv Himalaya) caryopses and its relation to alpha-amylase production. Plant Physiol. 96:1099–1104. 10.1104/pp.96.4.1099
- Hamayun M, Khan SA, Khan AL, Afzal M, Lee I-J. 2011. Endophytic Cephalotheca sulfurea AGH07 reprograms soybean to higher growth. J Plant Interact. 7:301–306. 10.1080/17429145.2011.642013
- Hamayun M, Khan SA, Khan AL, Rehman G, Kim YH, Iqbal I, Hussain J, Sohn EY, Lee IJ. 2010. Gibberellin production and plant growth promotion from pure cultures of Cladosporium sp. MH-6 isolated from cucumber (Cucumis sativus L.). Mycologia. 102:989–995. 10.3852/09-261
- Hedden P, Kamiya Y. 1997. Gibberellin biosynthesis: enzymes, genes and their regulation. Annu Rev Plant Physiol Plant Mol Biol. 48:431–460.
- Ikeda A, Ueguchi-Tanaka M, Sonoda Y, Kitano H, Koshioka M, Futsuhara Y, Matsuoka M, Yamaguchi J. 2001. Slender rice, a constitutive gibberellin response mutant, is caused by a null mutation of the SLR1 gene, an ortholog of the height-regulating gene GAI/RGA/RHT/D8. Plant Cell. 13:999–1010. 10.1105/tpc.13.5.999
- Inada S, Shimmen T. 2000. Regulation of elongation growth by gibberellin in root Segments of Lemnaminor. Plant Cell Physiol. 41: 932–939. 10.1093/pcp/pcd018
- Jang EK, Ullah I, Lim JH, Lee IJ, Kim JG, Shin JH. 2012. Physiological and molecular characterization of a newly identified entomopathogenic bacteria, Photorhabdus temperata M1021. J Microbiol Biotechnol. 22:1605–1612. 10.4014/jmb.1203.03068
- Joo GJ, Kang SM, Hamayun M, Kim SK, Na CI, Shin DH, Lee IJ. 2009. Burkholderia sp. KCTC 11096BP as a newly isolated gibberellin producing bacterium. J Microbiol. 47:167–171. 10.1007/s12275-008-0273-1
- Joo GJ, Kim YM, Kim JT, Rhee IK, Kim JH, Lee IJ. 2005. Gibberellins-producing rhizobacteria increase endogenous gibberellins content and promote growth of red peppers. J Microbiol. 43:510–515.
- Joo GJ, Kim YM, Lee IJ, Song KS, Rhee IK. 2004. Growth promotion of red pepper plug seedlings and the production of gibberellins by Bacillus cereus, Bacillus macroides and Bacillus pumilus. Biotechnol Lett. 26:487–491. 10.1023/B:BILE.0000019555.87121.34
- Kang S-M, Joo G-J, Hamayun M, Na C-I, Shin D-H, Kim H, Hong J-K, Lee I-J. 2009. Gibberellin production and phosphate solubilization by newly isolated strain of Acinetobacter calcoaceticus and its effect on plant growth. Biotechnol Lett. 31:277–281. 10.1007/s10529-008-9867-2
- Kang SM, Hamayun M, Joo GJ, Khan AL, Kim YH, Kim SK, Jeong HJ, Lee IJ. 2010. Effect of Burkholderia sp. KCTC 11096BP on some physiochemical attributes of cucumber. Eur J Soil Biol. 46:264–268.
- Kang S-M, Khan A, Hamayun M, Hussain J, Joo G-J, You Y-H, Kim J-G, Lee I-J. 2012. Gibberellin-producing Promicromonospora sp. SE188 improves Solanum lycopersicum plant growth and influences endogenous plant hormones. J Microbiol. 50:902–909. 10.1007/s12275-012-2273-4
- Khan AL, Hamayun M, Kang SM, Kim YH, Jung HY, Lee JH, Lee IJ. 2012. Endophytic fungal association via gibberellins and indole acetic acid can improve plant growth under abiotic stress: an example of Paecilomyces formosus LHL10. BMC Microbiol. 12:3–9. 10.1186/1471-2180-12-3
- Khan AL, Hamayun M, Kim Y-H, Kang S-M, Lee J-H, Lee I-J. 2011. Gibberellins producing endophytic Aspergillus fumigatus sp. LH02 influenced endogenous phytohormonal levels, isoflavonoids production and plant growth in salinity stress. Process Biochem. 46:440–447. 10.1016/j.procbio.2010.09.013
- Khan AL, Hussain J, Al-Harrasi A, Al-Rawahi A, Lee IJ. 2013. Endophytic fungi: resource for gibberellins and crop abiotic stress resistance. Crit Rev Biotechnol. 1–13. doi:10.3109/07388551.2013.800018
- Khan AL, Lee IJ. 2013. Endophytic Penicillium funiculosum LHL06 secretes gibberellin that reprograms glycine max L. growth during copper stress. BMC Plant Biol. 13:86. 10.1186/1471-2229-13-86
- Khan AL, Waqas M, Hussain J, Al-Harrasi A, Al-Rawahi A, Al-Hosni K, Kim MJ, Adnan M, Lee IJ. 2014. Endophytes Aspergillus caespitosus LK12 and Phoma sp. LK13 of Moringa peregrina produce gibberellins and improve rice plant growth. J Plant Interact. 9:731–737. 10.1080/17429145.2014.917384
- Lee I-J, Foster KR, Morgan PW. 1998. Photoperiod control of gibberellin levels and flowering in Sorghum. Plant Physiol. 116:1003–1011. 10.1104/pp.116.3.1003
- Miethke M, Marahiel MA. 2007. Siderophore-based iron acquisition and pathogen control. Microbiol Mol Biol Rev. 71:413–451. 10.1128/MMBR.00012-07
- Park G-S, Khan AR, Hong S-J, Jang E-K, Ullah I, Jung BK, Choi J, Yoo N-K, Park K-J, Shin J-H. 2013. Draft genome sequence of entomopathogenic bacterium Photorhabdus temperata strain m1021, isolated from nematodes. Genome Announc. 1:1–2.
- Pérez-Miranda S, Cabirol N, George-Téllez R, Zamudio-Rivera LS, Fernández FJ. 2007. O-CAS, a fast and universal method for siderophore detection. J Microbiol Methods. 70:127–131. 10.1016/j.mimet.2007.03.023
- Raddadi N, Cherif A, Boudabous A, Daffonchio D. 2008. Screening of plant growth promoting traits of Bacillus thuringiensis. Ann Microbiol. 58:47–52. 10.1007/BF03179444
- Römheld V, Marschner H. 1981. Iron deficiency stress induced morphological and physiological changes in root tips of sunflower. Physiol Plantarum. 53:354–360. 10.1111/j.1399-3054.1981.tb04512.x
- Schneider E, Kazakoff C, Wightman F. 1985. Gas chromatography-mass spectrometry evidence for several endogenous auxins in pea seedling organs. Planta. 165:232–241. 10.1007/BF00395046
- Schwyn B, Neilands JB. 1987. Universal chemical assay for the detection and determination of siderophores. Anal Biochem. 160:47–56. 10.1016/0003-2697(87)90612-9
- Seo S, Lee S, Hong Y, Kim Y. 2012. Phospholipase A2 inhibitors synthesized by two entomopathogenic bacteria, Xenorhabdus nematophila and Photorhabdus temperata subsp. temperata. Appl Environ Microbiol. 78:3816–3823. 10.1128/AEM.00301-12
- Taghavi S, van der Lelie D, Hoffman A, Zhang YB, Walla MD, Vangronsveld J, Newman L, Monchy S. 2010. Genome sequence of the plant growth promoting endophytic bacterium Enterobacter sp.638. PLoS Genet. 6:1–15. 10.1371/journal.pgen.1000943
- Ullah I, Khan A, Park G-S, Lim J-H, Waqas M, Lee I-J, Shin J-H. 2013. Analysis of phytohormones and phosphate solubilization in Photorhabdus spp. Food Sci Biotechnol. 22:25–31. 10.1007/s10068-013-0044-6
- Waqas M, Khan AL, Lee I-J. 2013. Bioactive chemical constituents produced by endophytes and effects on rice plant growth. J Plant Interact. 9:478–487.
- Waqas M, Khan AL, Lee I-J. 2014. Bioactive chemical constituents produced by endophytes and effects on rice plant growth. J Plant Interact. 9:478–487. 10.1080/17429145.2013.860562
- Watson RJ, Joyce SA, Spencer GV, Clarke DJ. 2005. The exbD gene of Photorhabdus temperata is required for full virulence in insects and symbiosis with the nematode Heterorhabditis. Mol Microbiol. 56:763–773. 10.1111/j.1365-2958.2005.04574.x
- Watson RJ, Millichap P, Joyce S, Reynolds S, Clarke D. 2010. The role of iron uptake in pathogenicity and symbiosis in Photorhabdus luminescens TT01. BMC Microbiol. 10:177. 10.1186/1471-2180-10-177
- Zocchi G, De Nisi P, Dell'Orto M, Espen L, Gallina PM. 2007. Iron deficiency differently affects metabolic responses in soybean roots. J Exp Bot. 58:993–1000. 10.1093/jxb/erl259
