Abstract
The hydroperoxide lyase (HPL) pathway for six carbon (C6) volatiles and the allene oxide synthase (AOS) pathway for jasmonates (JAs) share the first part of the pathway. To avoid competition, a separate localization of HPL and AOS might be important. A fusion protein comprising Arabidopsis HPL and green fluorescent protein was transported into chloroplasts, where AOS was located. Arabidopsis harboring β-glucuronidase (GUS) gene downstream of Arabidopsis HPL promoter (pAtHPL::GUS) showed different GUS activity in floral organs compared with that from pAtAOS::GUS. With pAtHPL::GUS, wounding enhanced GUS activity at the periphery of cotyledons; while with pAtAOS::GUS, GUS activity was high in the vasculature. The distribution of the ability to form C6 volatiles correlated with the profile of HPL promoter activity; however, this ability unchanged after wounding. Inconsistency between the AOS promoter activity and JA levels was also evident. Thus, an additional factor should also control the ability to form C6 volatiles and JAs.
1. Introduction
Plants have evolved complex signaling pathways to ensure effective responses to biotic and abiotic challenges, as well as to developmental stimuli. The oxylipin pathway is one such pathway, which, upon activation by environmental and developmental inputs, induces the synthesis of a diverse group of bioactive compounds known as oxylipins (Feussner & Wasternack Citation2002; Howe & Schilmiller Citation2002). Oxylipin biosynthesis is initiated by lipoxygenases, leading to the oxygenation of polyunsaturated fatty acids, mainly linoleic and α-linolenic acids, to yield their 9- or 13-hydroperoxides (9-/13-hydroperoxyoctadecadienoic acid, and 9-/13-hydroperoxyoctadecatrienoic acid [9-/13-HPOT]; Chehab et al. Citation2006). These hydroperoxides are metabolized by a group of cytochrome P450 enzymes present in different branch pathways, to generate the oxylipins (Feussner & Wasternack Citation2002). Among the oxylipin branch pathways, the hydroperoxide lyase (HPL) and the allene oxide synthase (AOS) are considered the two major plant stress response pathways. AOS and HPL are related cytochrome P450s, designated CYP74A and 74B, respectively (Song & Brash Citation1991; Shibata et al. Citation1995a, Citation1995b; Nelson Citation1999), which metabolize a common fatty acid hydroperoxide substrate (13-HPOT) to different classes of bioactive oxylipins.
13-HPL (CYP74B) cleaves 13-HPOT at the C12-C13 bond to produce two carbonyl compounds: (Z)-3-hexenal and 12-oxo-(Z)-9-dodecenoic acid (Grechkin and Hamberg Citation2004). A nicotinamide adenine dinucleotide phosphate dependent reductase reduces (Z)-3-hexenal to (Z)-3-hexen-1-ol (Matsui et al. Citation2012), which can be further converted to (Z)-3-hexen-1-yl acetate by acetyl-coenzyme A: (Z)-3-hexen-1-ol acetyltransferase (D'Auria et al. Citation2007). In some plants, (Z)-3-hexenal is converted to (E)-2-hexenal spontaneously or enzymatically (Noordermeer et al. Citation1999; Matsui Citation2006; D'Auria et al. Citation2007). These C6-aldehydes, alcohols, and their corresponding esters of the HPL pathway are collectively known as green leaf volatiles (GLVs; Matsui Citation2006; Nyambura et al. Citation2011). Insecticidal, fungicidal, and bactericidal activities have been reported for (Z)-3-hexenal and its related aldehydes (Hamilton-Kemp et al. Citation1992; Croft et al. Citation1993; Hammond et al. Citation2000; Vancanneyt et al. Citation2001; Nakamura and Hatanaka Citation2002; Hubert et al. Citation2008; Kishimoto et al. Citation2008). They also function as airborne infochemicals in specific plant–herbivore, plant–carnivore, and plant–plant relationships (Arimura et al. Citation2009; Sugimoto et al. Citation2014).
AOS (CYP74A) transforms 13-HPOT to epoxy octadecatrienoic acid (EOT), which is converted spontaneously into α- and γ-ketols, and 12-oxophytodienoic acid (OPDA). In the presence of allene oxide cyclase, EOT is specifically converted to OPDA, and then, to jasmonic acid (JA) after several enzymatic reaction steps (Froehlich et al. Citation2001; Mosblech et al. Citation2009). JA and related cyclopentanone products of the AOS pathway (jasmonates, JAs) are essential signals in the defense against mechanical wounding and attacks by herbivores and necrotrophic pathogens; they are also involved in developmental processes (Creelman & Mullet Citation1997; Staswick & Lehman Citation1999; Brioudes et al. Citation2009; Glauser et al. Citation2009; Hause et al. Citation2009; Erb et al. Citation2012).
JAs from the AOS pathway and GLVs from the HPL pathway exert distinct bioactivities and functions; therefore, their formation should be finely tuned. Given that HPL and AOS show similar substrate specificities (Taurino et al. Citation2013), it is assumed each pathway is fine-tuned to avoid competition between HPL and AOS for the same substrate (13-HPOT).
The temporal expression profiles of AOS and HPL show a partial overlap. Wound-inducible increases in HPL and AOS transcript levels have been documented in Arabidopsis (Bate et al. Citation1998; Laudert & Weiler Citation1998; Matsui et al. Citation1999; Kubigsteltig et al. Citation1999; Howe et al. Citation2000). Exogenous application of methyl jasmonate (MeJA) increases both HPL and AOS transcript levels (Avdiushko et al. Citation1995; Kohlmann et al. Citation1999; Matsui et al. Citation1999; Sivasankar et al. Citation2000; Ziegler et al. Citation2001).
The subcellular localization of AOS and HPL should also be considered in understanding to what degree these two enzymes would compete. AT_Chloro (http://www.grenoble.prabi.fr/at_chloro/), a database dedicated to the chloroplast proteome from Arabidopsis, shows that Arabidopsis thaliana AOS (AtAOS) is targeted to the chloroplast envelope. Additionally, a link between AtAOS accumulation and chloroplast rhomboid proteases, both of which reside in the chloroplast envelope, has been reported (Knopf et al. Citation2012). Most 13-HPLs (CYP74B) examined to date have an N-terminal extension that is predicted to be a chloroplast transit peptide, according to a prediction method such as ChloroP (http://www.cbs.dtu.dk/services/ChloroP/). Froehlich et al. (Citation2001) showed that tomato HPL and AOS are targeted to the outer and inner membranes of the chloroplast envelope, respectively. Rice HPL3 (OsHPL3), which has the shortest extension of its N-terminal among the three HPLs, was transported to chloroplasts when a fusion protein of the transit peptide of OsHPL3 with green fluorescent protein (GFP) was expressed in Arabidopsis leaves (Savchenko et al. Citation2014). However, localization of HPL in Arabidopsis has not been reported. There is no entry for HPL in AT_Chloro because the proteome analysis was carried out with Col-0, a natural hpl loss-of-function mutant (Duan et al. Citation2005).
Cellular and tissue distribution of expression of HPL and AOS should be taken into account to evaluate their competition as well as their distinct physiological functions. We addressed this issue using reporter assays with GFP and β-glucuronidase (GUS). We also examined the metabolic levels of GLVs and JAs. First, we examined subcellular and tissue localizations of HPL and C6-aldehydes. Thereafter, we examined responses of HPL promoter and AOS promoter against mechanical wounding with the GUS reporter system. We also examined distribution of GLVs and JAs in leaf tissues upon mechanical wounding. The differences in the spatial and temporal expression patterns of HPL and AOS observed in this study provide an additional insight into how these two genes are regulated to avoid substrate competition during oxylipin synthesis.
2. Materials and methods
2.1. Plant material and growth conditions
Wild-type ecotypes (Col-0 and No-0) of Arabidopsis thaliana and transgenic lines pAtAOS::GUS (C24; Kubigsteltig et al. Citation1999; provided by Dr. Ines Kubigsteltig, Ruhr-Universität, Bochum, Germany), and pVSP::GUS (Col-gl1; Xie et al. Citation1998; provided by Prof. John G. Turner, University of East Anglia, UK) were grown sterilely on B5 medium plates at 22°C with light from fluorescent lights (14 h light/10 h dark). Seedlings grown for 8 days were used for GUS staining. For the analysis of C6-volatiles and phytohormones in dissected leaves, 4-week-old Arabidopsis wild-type ecotype Nössen-0 (No-0) grown in soil (Vermiculite and Metro-mix) at 22°C and 70% relative humidity with light from fluorescent lights (14 h light/10 h dark) was used.
2.2. Construction of the reporter systems: pAtHPL::GFP and pAtHPL::GUS
Genomic DNA was purified from Col-0 leaves, and the DNA encoding the N-terminal half (from Met1 to Asn275) of AtHPL (At4g15440) was polymerase chain reaction (PCR) amplified with primers (5′-GTCGACATGTTGTTGAGAACGATGGCG-3′ and 5′-CCATGGGTTCTCGTCGATGAAAT-3′). The resultant amplicon was inserted into SalI– and NcoI–digested 35Ω-sGFP (S65T; Chiu et al. Citation1996; provided by Dr. Y. Niwa, University of Shizuoka, Shizuoka, Japan). The plasmid was propagated in Escherichia coli and then transiently introduced into Arabidopsis leaflet using particle bombardment.
To prepare the GUS construct, the promoter region (−2120 to +33 nucleotides relative to the translation start codon) of AtHPL (At4g15440) was PCR amplified with primers (5′-CCCAAGCTTCACATTGCTCTGAACTGAATCGCCTAG-3′ and 5′-CGGGATCCGCGGGGAAGTCGCCGCCATCGTTC-3′), and the resultant amplicon was inserted into BamHI- and HindIII-digested pBI101.3. The gene was introduced into Col-0 Arabidopsis following the floral dip method using Agrobacterium tumefaciens (LBA4404) as a transient host (Weigel & Glazerbrook Citation2002). Transgenic plants (pAtHPL::GUS) were selected by kanamycin resistance and PCR confirmation of the transgene. Homozygotes of the T5–T7 generations were used for the GUS monitoring. Homozygous pAtHPL::GUS plants were crossed with coi1 (Xie et al. Citation1998; provided by Prof. John G. Turner, University of East Anglia, UK) or dad1 (Ishiguro et al. Citation2001). Homozygotes of coi1 confirmed its male sterility among the progeny of coi1 heterozygotes. Homozygotes of dad1 were obtained by rescuing its male sterility by dipping young buds into 0.5 mM MeJA suspended with 0.05% (w/v) Tween 20.
2.3. GFP reporter assays
AtHPL::GFP was introduced into leaf epidermal cells of Arabidopsis (Col-0) by particle bombardment, as previously described (Tanaka et al. Citation2013), in which the helium pressure was 4 kgf cm−2 under a vacuum of 80 kPa. Following bombardment, the agar plate was filled with water to prevent desiccation. After incubation overnight at 22°C in the dark, leaves were viewed with a TCS SP5 confocal laser scanning microscopy (Leica Microsystems, Wetzlar, Germany) using an HCX PL APO CS 20.0 × 0.70 WATER UV objective lens. The GFP and chlorophylls were excited by the argon laser line (488 nm). The fluorescence of GFP was detected at 500–530 nm, whereas chlorophyll autofluorescence was detected at 700–730 nm. Images were processed with Photoshop CS6 (Adobe Systems, San Jose, CA, USA).
2.4. GUS activity assays
Histochemical staining of plant tissues for GUS activity was performed according to published protocols (Weigel & Glazerbrook Citation2002). Briefly, samples were placed in substrate solution (50 mM sodium phosphate pH 7.2, 2 mM potassium ferrocyanide, 2 mM potassium ferricyanide, and 0.2% Triton X-100 containing 1 mM 5-bromo-4-chloro-3-indoyl-β-D-glucoronide), vacuum infiltrated for 20 min, and then incubated at 37°C for 18 h. Subsequently, the samples were subjected to a series of 20, 35, and 50% ethanol, and then fixed in formalin–ethanol–acetic acid solution and finally transferred to 70% ethanol to remove the chlorophyll. Mechanical wounding was accomplished by applying pressure for 5 s using forceps on one side of the cotyledon of 8-day-old seedlings. The mid-vein was carefully left intact. Control samples were not wounded.
2.5. Determination of JAs
To determine the amounts of JAs in the mid-veins and the remaining leaf lamina, fully developed rosette leaves of 4-week-old A. thaliana ecotype No-0 plants were wounded by applying pressure with forceps three times on one side of the leaf lamina (corresponding to ca. 30% wound area on the treated side). Samples pooled from at least three different plants were taken from unwounded (control) and wounded tissue at different time points after wounding. The leaf was dissected into three parts using a sharp razor blade. Approximately 50 mg of tissue samples from the mid-vein and the opposite unwounded leaf lamina were collected within 30 s in disruptor tubes containing two glass beads (2 mm internal diameter [i.d.]) and two steel beads (3 mm i.d.), flash-frozen in liquid nitrogen, and stored at −80°C until use. One milliliter of ethyl acetate spiked with 20 ng of D2-JA (Sigma-Aldrich, St. Louis, MO, USA), used as the internal standard, was added to each sample, which was then completely homogenized on a MicroSmash homogenizer (MS-100R; Tomy Digital Biology Co. Tokyo, Japan). After centrifugation at 12,000 × g for 10 min at 4°C, supernatants were transferred to fresh 2-mL microtubes. Each pellet was re-extracted with 0.5 mL of ethyl acetate and centrifuged; supernatants were combined and then evaporated to dryness on a vacuum concentrator. The residue was resuspended in 0.5 mL of 70% methanol (v/v) and centrifuged to clarify the phases. The supernatants were pipetted into glass vials and then analyzed by liquid chromatography–tandem mass spectrometry (LC-MS/MS).
JA and jasmonoyl isoleucine (JA-Ile) were analyzed by LC-MS/MS (3200 Q-TRAP LC/MS/MS System; AB Sciex, Framingham, MA, USA) equipped with Prominence UFLC (Shimadzu, Kyoto, Japan). At a flow rate of 0.2 mL min−1, 2 µL of each sample was injected onto a Mightysil RP18 column (5 µm, 150 × 2 mm). A mobile phase composed of solvent A (water/acetonitrile/formic acid (90:10:0.1, v/v) and solvent B (acetonitrile/water/formic acid (95:5:0.1, v/v) was used in a gradient mode for separation. The solvent gradient used was 100% A to 100% B over 20 min, hold at 100% B for 5 min and then the solvent returned to 100% A for 15 min equilibration before the next injection. The mass spectrometry (MS) was used in negative ion mode and ions were detected using multiple reaction monitoring. The parent ions, daughter ions, and parameters used for their detection are listed in Supplemental Table S1. Quantification was made based on the internal standard added and standard curves.
2.6. Determination of GLVs
For the determination of C6 volatiles, fully developed rosette leaves of 4-week-old A. thaliana ecotype No-0 plant were mechanically wounded and dissected as described above. Approximately 50 mg of the dissected mid-veins and remaining leaf lamina were collected separately in a glass vial (22 mL; Perkin Elmer, Waltham, MA, USA) and immediately stored at −80°C until use. Samples were thawed for 10 min in a water bath set at 25°C, and then, 1 mL of saturated CaCl2 solution was added to halt any enzyme reactions. A solid-phase microextraction (SPME) fiber (50/30 µm DVB/Carboxen/PDMS Stable Flex; Supelco, Bellefonte, PA, USA) was exposed to the headspace of the vial for 30 min at 25°C. The fiber was inserted into the insertion port of a gas chromatography–MS apparatus (QP-2010 Plus; Shimadzu, Kyoto, Japan) equipped with a stabiliwax column (30 m × 0.25 mm i.d. × 0.25 µm film thickness; Restek, Bellefonte, PA, USA). The column temperature was programmed as follows: 40°C for 5 min, increasing by 5°C min−1 to 200°C for 2 min. The carrier gas (He) was delivered at a flow rate of 1.54 mL min−1. The glass insert was an SPME Sleeve (Supelco). The fiber was held in the injection port for 10 min at 240°C to fully remove any compounds from the matrix. Splitless injection with a sampling time of 1 min was used. The temperatures of the ion source and interface were 200°C and 240°C, respectively. The mass detector was operated in the electron impact mode, with an ionization energy of 70 eV. To identify each compound, retention indices and MS profiles of the corresponding authentic compounds were used. The quantities of GLVs were determined from the peak areas, based on a calibration curve constructed with known amounts of GLVs suspended in 1 mL of saturated CaCl2 solution.
3. Results
3.1. Determining the subcellular localization of AtHPL using a GFP reporter assay
A ChloroP prediction suggested that the N-terminal sequence consisting of 34 amino acids of AtHPL was a transit peptide for chloroplast targeting. We fused the complementary DNA (cDNA) sequences encoding the N-terminal half of AtHPL to the cDNA encoding GFP. The fusion protein was expressed transiently under the control of cauliflower mosaic virus 35S promoter in Arabidopsis leaves using the particle gun bombardment technique (Tanaka et al. Citation2013). The fluorescence signals of AtHPL::GFP fusions were found in chloroplasts, as identified by imaging of the autofluorescence of chlorophyll (). The image was almost identical to that obtained with GFP fused to the transit peptide derived from the Arabidopsis ribulose-1,5-biphosphate carboxylase small subunit (Chiu et al. Citation1996), while GFP without the additional peptide was located in the cytoplasm and nuclei (Chiu et al. Citation1996). These results showed that AtHPL is localized to the chloroplasts.
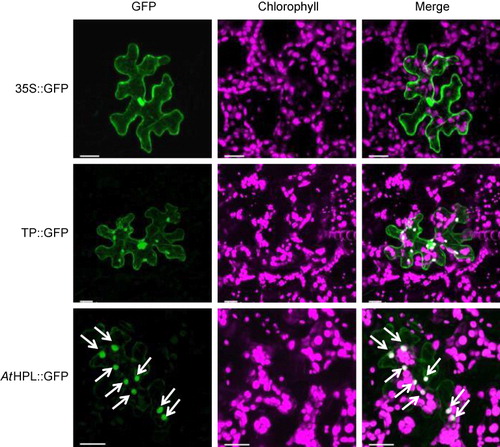
3.2. GUS reporter assay to determine the expression profile of AtHPL
We fused the promoter region (2152 bp of the 5′ upstream sequence) of the AtHPL gene (pAtHPL) to the GUS gene and transformed Arabidopsis (Col-0) with the fusion gene. When the transgenic plants were grown under normal growth conditions, we noticed significant GUS expression in the inflorescence (, , and ). This agrees with the results of northern blot analysis indicating that the transcript of AtHPL was abundant in inflorescences, flowers, and siliques (Bate et al. Citation1998; Matsui et al. Citation1999). High GUS activity was evident in carpels, filaments, peduncles, and sepals in open flowers; however, anthers, stigmas, and petals showed little activity. The GUS activity in the sepals of young buds was very high and decreased as the flowers matured (). The activity in siliques also decreased in their middle part during elongation.
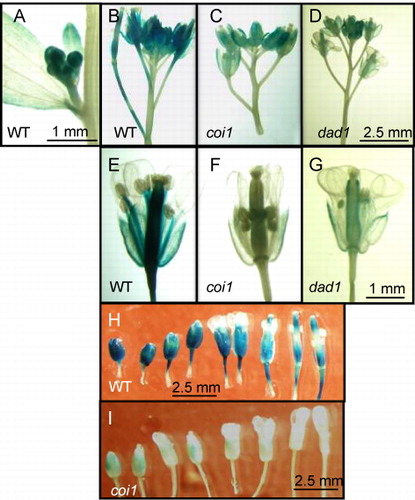
Mechanical wounding and MeJA treatment enhanced the expression of AtHPL (Matsui et al. Citation1999); therefore, it was assumed that expression of AtHPL was under the control of the JA-signaling pathway. COI1 is an F-box protein essential to transduce JA signaling through a specific binding of JA-Ile (Katsir et al. Citation2008; Mosblech et al. Citation2011; Wasternack & Hause Citation2013). DAD1 is a lipase essential to form JA in filaments and anthers (Ishiguro et al. Citation2001). When the GUS activity under the control of the AtHPL promoter was examined with a mutant lacking active COI1 or DAD1 (coi1 or dad1, respectively), the GUS activity was much lower than that found in the flowers of Col-0 (, , , , and ). However, there was still weak but substantial GUS activity with coi1 and dad1 at the rim parts of carpels and sepals. Thus, deficiency of active COI1 or DAD1 seemed to suppress the intensity of GUS activity, but did not affect the spatial profile of the activity.
3.3. The ability to form GLV in flowers
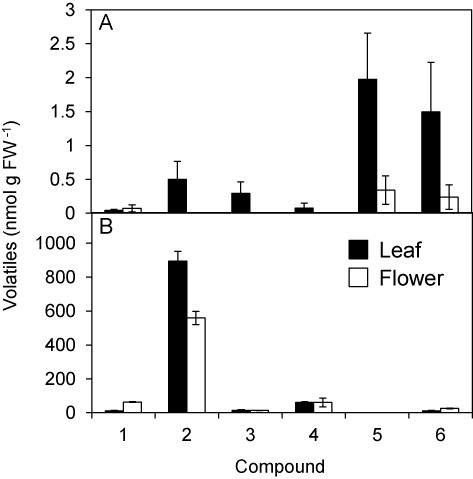
To clarify whether HPL is active in flowers, we examined GLV formation in flowers before and after freeze–thaw treatment to facilitate extensive tissue disruption and compared GLV formation with that in leaves (). The amounts of GLV formed and emitted from intact flowers were as low as 0.5 µmol g FW−1, and they were several folds lower than those emitted from intact leaves. The GLV compositions were similar between flowers and leaves: (Z)-3-hexen-1-yl acetate and (Z)-3-hexen-1-ol were the major GLVs emitted out from both organs when they were intact. When the flowers were disrupted through the freeze–thaw treatment, emission of GLVs, especially (Z)-3-hexenal, increased significantly. The composition of GLVs emitted from disrupted flowers was similar to that found in disrupted leaves (; Matsui et al. Citation2012).
3.4. Expression patterns of AtHPL gene after mechanical wounding
Classical studies showed that transcript levels of AtHPL and AtAOS increased after mechanical wounding (Bate et al. Citation1998; Kubigsteltig et al. Citation1999; Matsui et al. Citation1999). Here, we validated the previous results with higher spatial resolution using the GUS assay system. When the pHPL::GUS transgenic Arabidopsis was grown for 8 days aseptically in a plate with agar nutrients, weak expression of GUS was found throughout the cotyledons, with a tendency for higher expression at the rim parts (). At 30 min after mechanical wounding of one of the cotyledons, high GUS expression was observed at the rim parts. The high expression at the rim parts continued for approximately 24 h after mechanical wounding. The intensity and profile of GUS expression was similar in the other undamaged cotyledons. After 24 h, the GUS expression spread to the interior parts of both cotyledons. The emerging true leaves also showed intense GUS activity; however, the mid-vein of the leaves showed little activity.
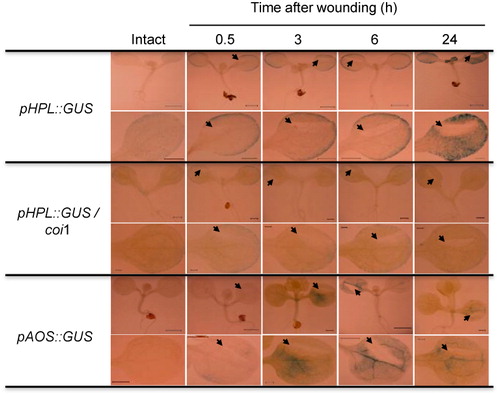
The intact 8-day-old cotyledon of pHPL::GUS/coi1 showed weaker GUS staining than that of pHPL::GUS/Col-0. Mechanical wounding increased the GUS activity at the rim part, but with an intensity that was much lower than that found with the reporter system with Col-0 background. The higher GUS activity persisted for least 24 h after wounding.
Expression of GUS under the control of the AtAOS promoter was also examined under the same conditions employed for pHPL::GUS to precisely compare the spatiotemporal expression profiles of these two genes. As reported previously (Kubigsteltig et al. Citation1999), GUS expression was barely detectable in intact Arabidopsis plants (). Significant GUS expression was observed at 3 h after mechanical wounding. GUS expression was relatively constant in the lamina, with intense expression in the veins. There was no specific GUS induction around the wound site, even though intense GUS expression was observed around the wound site when the same reporter system was examined with tobacco (Nicotiana tabacum) plants (Kubigsteltig et al. Citation1999). The expression profile was similar until 24 h after mechanical wounding. Slight GUS expression could be found at the base of the cotyledons, but expression in the other systemic cotyledons was not significant.
3.5. Oxylipin analysis in mid-vein and lamina of Arabidopsis leaves
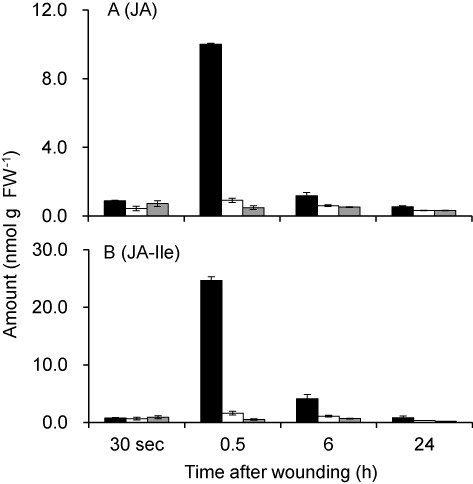
The GUS reporter assay shown above indicated that AtHPL and AtAOS showed distinct spatiotemporal expression patterns after mechanical wounding. The most remarkable difference was observed in the vascular tissues, where GUS activity was almost absent with pAtHPL::GUS, while the most intense GUS activity was observed with pAtAOS::GUS after mechanical wounding. To examine whether this spatial specificity correlates with spatial distribution of abilities to form GLVs and JAs, we mechanically wounded one side of 4-week-old Arabidopsis ecotype No-0 leaves using a scalpel without touching the mid-veins and harvested the middle part containing the mid-veins and the other side of the leaves for determination of JAs and GLVs ( and ).
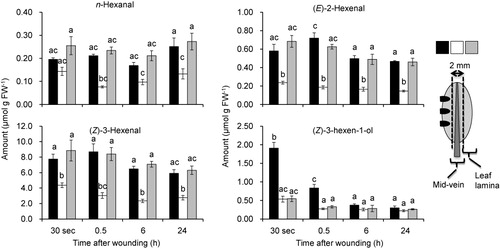
The amounts of GLVs from each part of leaves were determined by collecting volatiles emitted from the tissue sections after freeze–thaw disruption, using a SPME fiber for 30 min at 25°C (). Just after mechanical wounding (30 s), each leaf part showed substantial ability to form GLVs. With this procedure, n-hexanal, (E)-2-hexenal, (Z)-3-hexenal, and (Z)-3-hexen-1-ol were detected as major volatiles, and among them, (Z)-3-hexenal was the most abundant, as previously reported (Matsui et al. Citation2012). Among the sections, the lamina parts showed higher abilities to form GLVs than the mid-vein parts, except for (Z)-3-hexen-1-ol. Mechanical wounding resulted in little change in the GLV-forming ability, even though the GUS activity observed with the reporter system significantly increased after mechanical wounding ().
The amounts of oxylipins formed from the AOS pathway, namely, JA and JA-Ile, were also determined (). JA and JA-Ile remained at a low level just after mechanical wounding, but their amounts increased transiently at 30 min after wounding. The transient accumulation of JA and JA-Ile peaking at 30 min after wounding was found only in the directly wounded lamina. Both JA and JA-Ile decreased thereafter, and at 24 h after wounding, they returned to the base level.
4. Discussion
Among the oxylipin branch pathways, the AOS and HPL are involved in the two major plant stress response pathways. These two branches might compete for the same substrate, 13-HPOT (Taurino et al. Citation2013), because HPL and AOS show similar substrate specificity; however, the physiological functions associated with their end products are distinct. Their endocellular localization might be one way to avoid the competition. The chloroplast protein database dedicated to sub-plastidial localization (PPDB [Plant protein database] and AT_Chloro) indicated that AtAOS is targeted to the envelope and thylakoids of chloroplasts. On the other hand, HPLs show diverse localization patterns; in some plants, HPLs are localized to the lipid bodies (Mita et al. Citation2005), the outer envelope of chloroplasts (Froehlich et al. Citation2001), and the stroma (Bonaventure Citation2014); in some cases, no specific localization in a particular organelle is observed (Phillips & Galliard Citation1978; Shibata et al. Citation1995b; Noordermeer et al. Citation2000). TargetP prediction for AtHPL indicated a cytoplasmic localization, while ChloroP prediction indicated a 34-amino acid chloroplast transit peptide. PPDB suggested a location on the outer envelope; however, this has not been confirmed experimentally. Probably the fact that HPL in Col-0, the most commonly used Arabidopsis ecotype for proteomic analysis, has a 10-nucleotide deletion in its first exon resulting in an inactive truncated protein (Duan et al. Citation2005) hindered identification of HPL through proteomic analysis. In this study, in vivo GFP fusion assays showed that the N-terminal 34 amino acid (derived from No-0 ecotype) of AtHPL almost exclusively transported the fusion protein to the chloroplasts in the epidermal layer of Arabidopsis leaves. Even though it is still possible for the two CYP74 enzymes to be segregated at the level of sub-chloroplast membrane or even within the same membrane (Mita et al. Citation2005), the close localization of the two enzymes sharing the same substrate would cause disordered competition, especially when the enzymes form their products in the disrupted tissues for the rapid oxylipin burst (Matsui Citation2006; Glauser et al. Citation2008, Citation2009). These results prompted us to compare the spatiotemporal expression patterns of these two genes.
Based on the results obtained with pAtHPL::GUS reporter system, we found that the promoter of AtHPL was active in floral organs, especially in peduncles, carpels, filaments, and sepals. Kubigsteltig et al. (Citation1999) reported that the promoter activity of AtAOS was high at the pollen sacs and at the base of filaments. Apparently, the tissue specificity of AtHPL expression was largely distinct from that of AtAOS, except for the base of filaments; thus, the two enzymes could form their products from shared substrate (13-HPOT) mostly without competition. For example, pollen sacs must be important organs for JA formation, because JA is essential for pollen maturation and anther dehiscence (Ishiguro et al. Citation2001). There was little GUS activity in the pollen sacs of the pAtHPL::GUS plants, while the sacs were among most intensely stained organs in pAtAOS::GUS plants (Kubigsteltig et al. Citation1999). In the absence of HPL, AOS uses 13-HPOT exclusively, in a controlled manner, to adjust the best timing of pollen maturation.
As expected from the data obtained with transcript analyses, expression of GUS in pAtHPL::GUS was highly suppressed in an Arabidopsis mutant lacking the JA-signaling component (coi1) and the mutant lacking a lipase essential to JA formation in floral organs (dad1). DAD1 is specifically expressed in filaments and is involved in JA formation in floral organs, but is hardly involved in JA formation of Arabidopsis leaves, especially after wounding (Ellinger et al. Citation2010). Therefore, extensive suppression of GUS activity in coi1 and dad1 suggested that expression of AtHPL in peduncles, carpels and sepals was regulated by the JA (or JA-Ile) formed by DAD1 at the filaments, but not by JAs formed at the other organs. If this is the case, there should be a system to transport JA and/or JA-Ile from filaments to the other tissues of the floral organs. It is also possible that a secondary signal molecule, formed depending on DAD1 and COI1 in filaments, might be a mobile signal directly inducing AtHPL expression in the other parts of the floral organs.
GLVs formed in intact flowers were low, and Arabidopsis self-pollinates; therefore, HPL in Arabidopsis may not be directly involved in recruiting pollinators. Instead, based on the fact that GLVs are extensively formed only after disruption of flowers, HPL in floral organs might have an important role in defense against herbivores and necrotrophic pathogens, as reported with caryophyllene synthase in Arabidopsis flowers (Huang et al. Citation2012).
Wound-inducible increases in HPL and AOS transcript levels have been documented in Arabidopsis (Bate et al. Citation1998; Laudert & Weiler Citation1998; Kubigsteltig et al. Citation1999; Matsui et al. Citation1999; Howe et al. Citation2000). Topological analysis using the GUS assay system showed that the spatiotemporal expression pattern of AtHPL after mechanical wounding was different from that of AtAOS. The present study showed that AtHPL is largely expressed in the mesophyll cells at the rim part of cotyledons, whereas AtAOS is preferentially expressed in the vasculature. The expression of AtAOS in the vascular tissues corresponded to the data reported by Kubigsteltig et al. (Citation1999). Thus, it can be concluded that the spatiotemporal expression of AtHPL and AtAOS after mechanical wounding is regulated differently.
A transient increase in the amounts of JA and JA-Ile after mechanical wounding was found only in wounded leaf lamina. By contrast, high GUS activity was observed with pAtHPL::GUS plants at the other side of lamina and even at the other systemic cotyledon after mechanical wounding. This suggested that local accumulation of JA/JA-Ile was not a prerequisite for induction of AtHPL expression. Taken together with the fact that the GUS activity of pAtHPL::GUS was suppressed extensively in coi1 background, we hypothesized that the expression of AtHPL is not directly regulated by JA/JA-Ile at the places where it was upregulated, but is regulated by an as-yet-unknown factor that is formed at the wounded site depending on COI1 after mechanical wounding.
GLVs, including (E)-2-hexenal, (Z)-3-hexenal, n-hexanal, as well as their corresponding alcohols or esters, are produced from mechanically wounded plant tissues (Hatanaka Citation1993; Matsui et al. Citation2012). After wounding, (Z)-3-hexenal as well as n-hexanal are the first products formed extensively in disrupted tissues, and they are converted into (Z)-3-hexen-1-ol and n-hexan-1-ol in the intact tissues adjacent to the wounds (Matsui et al. Citation2012). In the present study, the ability to form GLVs in the leaf lamina was shown to be higher than that in the mid-veins (). We estimated the amounts of GLVs after freeze–thaw disruption; therefore, C6-aldehydes were the most abundant GLVs. C6-aldehydes have been reported to have insecticidal, fungicidal, and bactericidal activities (Hamilton-Kemp et al. Citation1992; Croft et al. Citation1993; Hammond et al. Citation2000; Vancanneyt et al. Citation2001; Nakamura & Hatanaka Citation2002; Hubert et al. Citation2008; Kishimoto et al. Citation2008), and thus they play a protective role in plant defense. The distinct spatial expression of HPL and accumulation of GLVs at the rim part of the leaf lamina may contribute to the ad hoc defense in plants upon tissue disruption, especially that caused by chewing insects. In our experiment, however, there was no increase in the abilities to form GLVs in the rim part of leaf lamina, even though GUS activity increased after mechanical wounding. The supply of substrate (13-HPOT) might be a limiting factor to determine the ability. By contrast, there are several reports indicating that formation and/or emission of some GLVs, such as (Z)-3-hexen-1-ol or (Z)-3-hexen-1-yl acetate, are enhanced after herbivore damage and mechanical wounding (D’Auria et al. Citation2007; Chehab et al. Citation2008). Thus, it was conceivable that induction of the HPL gene after mechanical wounding has an important role in forming a subset of GLVs, C6-alcohol, and C6-acetate, which were largely involved in indirect defense or plant–plant interaction. To further test this hypothesis, quantification of each GLV emitted from each leaf section should be carried out. A technique to examine volatile emission from one leaf, or even from a small section of a leaf, should be developed in the future.
Supplemental data
Supplemental Table S1 for this article can be accessed at http://dx.doi.org/10.1080/17429145.2014.999836.
Supplementary_material.pptx
Download MS Power Point (49.1 KB)Acknowledgments
The authors thank Dr I. Kubigsteltig (Department of Plant Physiology, Ruhr-Universität Bochum, Germany) and Dr John G. Turner (School of Biological Sciences, University of East Anglia, UK) for providing the transgenic Arabidopsis pAtAOS::GUS and pVSP::GUS lines, respectively, used in this study.
Additional information
Funding
References
- Arimura G, Matsui K, Takabayashi J. 2009. Chemical and molecular ecology of herbivore-induced plant volatiles. Proximate factors and their ultimate functions. Plant Cell Physiol. 50:911–923.10.1093/pcp/pcp030
- Avdiushko S, Croft KPC, Brown GC, Jackson DM, Hamilton-Kemp TR, Hiderbrand D. 1995. Effect of volatile methyl jasmonate on the oxylipin pathway in tobacco, cucumber, and Arabidopsis. Plant Physiol. 109:1227–1230.10.1104/pp.10.4.1227
- Bate NJ, Sivasankar S, Moxon C, Riley JM, Thompson JE, Rothstein SJ. 1998. Molecular characterization of an Arabidopsis gene encoding hydroperoxide lyase, a cytochrome P-450 that is wound inducible. Plant Physiol. 117:1393–1400.10.1104/pp.117.4.1393
- Bonaventure G. 2014. Lipases and the biosynthesis of free oxylipins in plants. Plant Signaling Behav. 3:9.10.4161/psb.28429
- Brioudes F, Joly C, Szécsi J. 2009. Jasmonates control late development stages of petal growth in Arabidopsis thaliana. Plant J. 60:1070–1080.10.1111/j.1365-313X.2009.04023.x
- Chehab EW, Kaspi R, Savchenko T, Rowe H, Neger-Zakharov F, Kliebenstein D, Dehesh K. 2008. Distinct roles of jasmonates and aldehydes in plant-defense responses. PLoS One. 3:e1904.10.1371/journal.pne.0001904
- Chehab EW, Raman G, Walley JW, Perea JV, Banu G, Theng S, Dehesh K. 2006. Rice hydroperoxide lyases with unique expression patterns generate distinct aldehydes signatures in Arabidopsis. Plant Physiol. 141:121–134.10.1104/pp.106.078592
- Chiu W, Niwa Y, Zeng W, Hirano T, Kobayashi H, Sheen J. 1996. Engineered GFP as a vital reporter in plants. Curr Biol. 6:325–330.10.1016/S0960-9822(02)00483-9
- Creelman RA, Mullet JE. 1997. Biosynthesis and action of jasmonates in plants. Annu Rev Plant Physiol Mol Biol. 48:355–381.10.1146/annurev.arplant.48.1.355
- Croft KPC, Juttner F, Slusarenko AJ. 1993. Volatile products of the lipoxygenase pathway evolved from Phaseolus vulgaris (L.) leaves inoculated with Pseudomonas syringae pv phaseolicola. Plant Physiol. 101:13–24.10.1104/pp.101.1.13
- D’Auria JC, Pichersky E, Schaub A, Hansel A, Gershenzon J. 2007. Characterization of BAHD acyltransferase responsible for producing the green leaf volatile (Z)-3-hexen-1-yl acetate in Arabidopsis thaliana. Plant J. 49:194–207.10.1111/j.1365-313X.2006.02946.x
- Duan H, Huang MY, Palacio K, Schuler MA. 2005. Variations in CYP74B2 (hydroperoxide lyase) gene expression differentially affect hexenal signaling in the Columbia and Landsberg erecta ecotypes of Arabidopsis. Plant Physiol. 139:1529–1544.10.1104/pp.105.067249
- Ellinger D, Stingl N, Kubigsteltig II, Bals T, Juenger M, Pollmann S, Berger S, Schuenemann D, Mueller MJ. 2010. Dongle and defective in anther dehiscence1 lipase are not essential for wound- and pathogen-induced jasmonate biosynthesis: redundant lipases contribute to jasmonate formation. Plant Physiol. 153:114–127.10.1104/pp.110.155093
- Erb M, Meldau S, Howe GA. 2012. Role of phytohormones in insect-specific plant reactions. Trends Plant Sci. 17:250–259.10.1016/j.tplants.2012.01.003
- Feussner I, Wasternack C. 2002. The lipoxygenase pathway. Annu Rev Plant Biol. 53:275–297.10.1146/annurev.arplant.53.100301.135248
- Froehlich JE, Itoh A, Howe GA. 2001. Tomato allene oxide synthase and fatty acid hydroperoxide lyase, two cytochrome P450s involved in oxylipin metabolism, are targeted to different membranes of chloroplast envelope. Plant Physiol. 125:306–317.10.1104/pp.125.1.306
- Glauser G, Dubugnon L, Mousavi SA, Rudaz S, Wolfender J-L, Farmer EE. 2009. Velocity estimates for signal propagation leading to systemic jasmonic acid accumulation in wounded Arabidopsis. J Biol Chem. 284:34506–34513.10.1074/jbc.M109.061432
- Glauser G, Grata E, Dubugnon L, Rudaz S, Farmer EE, Wolfender J-L. 2008. Spatial and temporal dynamics of jasmonate synthesis and accumulation in Arabidopsis in response to wounding. J Biol Chem. 283:16400–16407.10.1074/jbc.M801760200
- Grechkin AN, Hamberg M. 2004. The “heterolytic hydroperoxide lyase” is an isomerase producing a short-lived fatty acid hemiacetal. Biochim Biophys Acta. 1636:47–58.10.1016/j.bbalip.2003.12.003
- Hamilton-Kemp TR, McCracken CTJ, Loughrin JH, Andersen RA, Hildebrand DF. 1992. Effect of some natural volatile compounds on the pathogenic fungi Alternaria alternata and Botrytis cinerea. J Chem Ecol. 18:1083–1091.10.1007/BF00980064
- Hammond DG, Rangel S, Kubo I. 2000. Volatile aldehydes are promising broad-spectrum postharvest insecticides. J Agric Food Chem. 48:4410–4417.10.1021/jf000233+
- Hatanaka A. 1993. The biogeneration of green odor by green leaves. Phytochemistry. 34:1201–1218.10.1016/0031-9422(91)80003-J
- Hause B, Wasternack C, Strack D. 2009. Jasmonates in stress responses and development. Phytochemistry. 70:1483–1484.10.1016/j.phytochem.2009.07.004
- Howe GA, Lee GI, Itoh A, Li L, DeRocher AE. 2000. Cytochrome P450-dependent metabolism of oxylipins in tomato cloning and expression of allene oxide synthase and fatty acid hydroperoxide lyase. Plant Physiol. 123:711–724.10.1104/pp.123.2.711
- Howe GA, Schilmiller AL. 2002. Oxylipin metabolism in response to stress. Curr Opin Plant Biol. 50:230–236.10.1016/S1369-5266(02)00250-9
- Hubert J, Münzbergová Z, Santino A. 2008. Plant volatile aldehydes as natural insecticides against stored-product beetles. Pest Manag Sci. 64:57–64.10.1002/ps.1471
- Huang M, Sanchez-Moreiras AM, Abel C, Sohrabi R, Lee S, Gershenzon J, Tholl D. 2012. The major volatile organic compound emitted from Arabidopsis thaliana flowers, the sesquiterpene (E)-β-caryophyllene, is a defense against a bacterial pathogen. New Phytol. 193:997–1008.10.1111/j.1469-8137.2011.04001.x
- Ishiguro S, Kawai-Oda A, Ueda J, Nishida I, Okada K. 2001. The defective in anther dehiscence1 gene encodes a novel phospholipase A1 catalyzing the initial step of jasmonic acid biosynthesis, which synchronized pollen maturation, anther dehiscence, and flower opening in Arabidopsis. Plant Cell. 13:2191–2209.10.1105/tpc.13.10.2191
- Katsir L, Chung HS, Koo AJK, Howe GA. 2008. Jasmonate signaling: a conserved mechanism of hormone sensing. Curr Opin Plant Biol. 11:428–435.10.1016/j.pbi.2008.05.004
- Kishimoto K, Matsui K, Ozawa R, Takabayashi J. 2008. Direct fungicidal activities of C6-aldehydes are important constituents for defense responses in Arabidopsis against Botrytis cinerea. Phytochemistry. 69:2127–2132.10.1016/j.phytochem.2008.04.023
- Knopf RR, Feder A, Mayer K, Lin A, Rozenberg M, Schaller A, Adam Z. 2012. Rhomboid proteins in the chloroplast envelope affect the level of allene oxide synthase in Arabidopsis thaliana. Plant J. 72:559–571.10.1111/j.1365-313X.2012.05090.x
- Kohlmann M, Bachmann A, Weichert H, Kolbe A, Balkenhohl T, Wasternack C, Feussner I. 1999. Formation of lipoxygenase-pathway-derived aldehydes in barley leaves upon methyl jasmonate treatment. Eur J Biochem. 260:885–895.10.1046/j.1432-1327.1999.00231.x
- Kubigsteltig I, Laudert D, Weiler E. 1999. Structure and regulation of Arabidopsis thaliana allene oxide synthase gene. Planta. 208:463–471.10.1007/s004250050583
- Laudert D, Weiler EW. 1998. Allene oxide synthase: a major control point in Arabidopsis thaliana octadecanoid signalling. Plant J. 15:675–684.10.1046/j.1365-313x.1998.00245.x
- Matsui K. (2006). Green leaf volatiles. Hydroperoxide lyase pathway of oxylipin metabolism. Curr Opin Plant Biol. 9:274–280.10.1016/j.pbi.2006.03.002
- Matsui K, Sugimoto K, Mano J, Ozawa R, Takabayashi J. 2012. Differential metabolisms of green leaf volatiles in injured and intact parts of a wounded leaf meet distinct ecophysiological requirements. PLoS ONE. 7:e36433.10.1371/journal.pone.0036433
- Matsui K, Wilkinson J, Hiatt B, Knauf V, Kajiwara T. 1999. Molecular cloning and expression of Arabidopsis fatty acid hydroperoxide lyase, from tea leaves. Phytochemistry. 40:477–481.
- Mita G, Quarta A, Fasano P, De Paolis A, Di sansebastiano GP, Perrotta C, Iannacone R, Belfield E, Hughes R, Tsesmetzis N, et al. 2005. Molecular cloning and characterization of an almond 9-hydroperoxide lyase, a new CYP74 targeted to lipid bodies. J Exp Bot. 56:2321–2333.10.1093/jxb/eri225
- Mosblech A, Feussner I, Heilmann I. 2009. Oxylipins. Structurally diverse metabolites from fatty acid oxidation. Plant Physiol Biochem. 47:511–517.10.1016/j.plaphy.2008.12.011
- Mosblech A, Thurow C, Gatz C, Feussner I, Heilmann I. 2011. Jasmonic acid perception by COI1 involves inositol polyphosphates in Arabidopsis thaliana. Plant J. 65:949–957.10.1111/j.1365-313X.2011.04480.x
- Nakamura S, Hatanaka A. 2002. Green-leaf-derived C6 aroma compounds with potent antibacterial action that act on both gram-negative and gram-positive bacteria. J Agric Food Chem. 50:7639–7644.10.1021/jf025808c
- Nelson DR. 1999. Cytochrome P450 and the individuality of species. Arch Biochem Biophys. 369:1–10.10.1006/abbi.1999.1352
- Noordermeer MA, Van Dijken AJ, Smeekens SC, Veldink GA, Vliegenthart JF. 2000. Characterization of three cloned and expressed 13-hydroperoxide lyase isoenzymes from alfalfa with unusual N-terminal sequences and different enzyme kinetics. Eur J Biochem. 267:2473–2482.10.1046/j.1432-1327.2000.01283.x
- Noordermeer MA, Veldink GA, Vliegenthart JF. 1999. Alfalfa contains substantial 9-hydroperoxide lyase activity and 3Z:2E-enal isomerase. FEBS Lett. 443:201–204.10.1016/S0014-5793(98)01706-2
- Nyambura MC, Matsui K, Kumamaru T. 2011. Establishment of an efficient screening system to isolate rice mutants deficient in green leaf volatile formation. J Plant Interact. 6:185–186.10.1080/17429145.2010.544777
- Phillips DR, Galliard T. 1978. Flavor biogenesis: partial purification and properties of a fatty acid hydroperoxide cleaving enzyme from fruits of cucumber. Phytochemistry. 17:355–358.10.1016/S0031-9422(00)89315-2
- Savchenko T, Kolla VA, Wang CQ, Nasafi Z, Hicks DR, Phadungchob B, Chehab WE, Brandizzi F, Froehlich J, Dehesh K. 2014. Functional convergence of oxylipin and abscisic acid pathways controls stomatal closure in response to drought. Plant Physiol. 164:1151–1160.10.1104/pp.113.234310
- Shibata Y, Matsui K, Kajiwara T, Hatanaka A. 1995a. Fatty acid hydroperoxide lyase is a heme protein. Biochem Biophys Res Commun. 207:438–443.10.1006/bbrc.1995.1207
- Shibata Y, Matsui K, Kajiwara T, Hatanaka A. 1995b. Purification and properties of fatty acid hydroperoxide lyase from green bell pepper fruits. Plant Cell Physiol. 36 (1):147–156.
- Sivasankar S, Sheldrick B, Rothstein SJ. 2000. Expression of allene oxide synthase determines defense gene activation in tomato. Plant Physiol. 122:1335–1342.10.1104/pp.122.4.1335
- Song WC, Brash AR. 1991. Purification of an allene oxide synthase and identification of the enzyme as a cytochrome P-450. Science. 252:781–784.10.1126/science.1876834
- Staswick PE, Lehman CC. 1999. Jasmonic acid-signaled responses in plants. In: Agrawal AA, Tazun S, Bent E, editor. Induced plant defenses against pathogens and herbivores. St. Paul (MN): American Phytopathological Society Press; p. 117–136.
- Sugimoto K, Matsui K, Iijima Y, Akakabe Y, Muramoto S, Ozawa R, Uefune M, Sasaki R, Alamgir K.M, Akitake S. 2014. Intake and transformation to a glycoside of (Z)-3-hexenol from infested neighbors reveals a mode of plant odor reception and defense. Proc Natl Acad Sci USA. 111:7144–7149.10.1073/pnas.1320660111
- Taurino M, De Domenico D, Bonsegna S, Santino A. 2013. The hydroperoxide lyase branch of the oxylipin pathway and green leaf volatiles in plant/insect interaction. J Plant Biochem Physiol. 1:102.10.4172/2329-9029.1000102
- Tanaka Y, Nishimura K, Kawamukai M, Oshima A, Nakagawa T. 2013. Redundant function of two Arabidopsis COPII components, AtSec24B and AtSec24C, is essential for male and female gametogenesis. Planta. 238:561–575.10.1007/s00425-013-1913-1
- Vancanneyt G, Sanz C, Farmaki T, Paneque M, Ortego F, Castanera P, Sanchez-Serrano JJ. 2001. Hydroperoxide lyase depletion in transgenic potato plants leads to an increase in aphid performance. Proc Natl Acad Soc USA. 98:8139–8144.10.1073/pnas.141079498
- Wasternack C, Hause B. 2013. Jasmonates: biosynthesis, perception, signal transduction and action in plant stress response, growth and development. An update to the 2007 review in Annals of Botany. Ann Botany. 111:1021–1058.10.1093/aob/mct067
- Weigel D, Glazerbrook J. 2002. Arabidopsis: a laboratory manual. Cold Spring Harbor (NY): Cold Spring Harbor Laboratory Press.
- Xie DX, Feys BF, James S, Nieto-Rostro M, Turner JG. 1998. COI1: an Arabidopsis gene required for jasmonate-regulated defense and fertility. Science. 280:1091–1094.10.1126/science.280.5366.1091
- Ziegler J, Keinanen M, Baldwin IT. 2001. Herbivore-induced allene oxide synthase transcripts and jasmonic acid in Nicotiana attenuata. Phytochemistry. 58:729–738.10.1016/S0031-9422(01)00284-9
