Abstract
The symbiotic association between plant and arbuscular mycorrhizal fungi is an evolutionary conserved association that resulted from co-evolutionary events where both partners benefit from each other. Different plant genes, hormones, and miRNAs regulate this symbiotic association at different levels. Of those, the most important signaling molecules that play critical roles in symbiotic association are plant-derived strigolactones and fungal-derived lipochito-oligosaccharides. These molecules regulate the symbiotic association at the initial stage of symbiosis. Subsequent signaling events of these two molecules activate downstream signaling cascades to develop a proper symbiotic relationship between plants and fungi.
| Abbreviations | ||
| SL | = | strigolactone |
| CCD | = | carotenoid cleavage dioxygenase |
| ABA | = | abscisic acid |
| NCED | = | 9-cis-epoxycarotenoid cleavage dioxygenase |
| TB | = | teosinte branched |
| AM | = | arbuscular mycorrhiza |
| DMI | = | does not make infection |
| CCaMK | = | calcium and calmodulin-like protein kinase |
| STR | = | symbiosis-induced transporter |
| SYM | = | symbiosis |
| HRGP | = | hydroxyproline-rich glycoprotein |
| AGP | = | ADP-glucose pyrophosphorylase |
| NUP | = | nucleoporin |
| PT | = | phosphate transporter |
| MST | = | monosaccharide transporter |
| AMT | = | ammonium transporter |
| SCWC | = | stochastic calculus of wrapped compartments |
| IBA | = | indole-3-butyric acid |
| IAA | = | indole-3-acetic acid |
| DGT | = | diageotropic |
| BSH | = | bushy |
| GA | = | gibberellic acid |
| JA | = | jasmonic acid |
| SA | = | salicylic acid |
| LNP | = | lectin nucleotide phosphohydrolase |
| CPK | = | calcium-dependent protein kinase |
| miRNA | = | microRNA |
| HAP | = | haem peroxidase |
Introduction
A major event in the life of the planet occurred around 400–500 myr ago, when plants colonized the land habitat (Selosse & Le Tacon Citation1998). The roots of those plants were colonized by an ancient group of fungi known as Glomeromycota or arbuscular mycorrhizal fungi (AMF; Bonfante & Genre Citation2008; Tisserant et al. Citation2013). The AMF are considered as obligated symbionts (Colard et al. Citation2011; Tisserant et al. Citation2012). Evidence to support this scenario was primarily based on phylogenetic information in fossil records (Remy et al. Citation1994). Humphreys et al. (Citation2010) demonstrated that a mutualistic relationship between the AMF and a member of the most ancient clades of land plants promotes carbon uptake, growth, and sexual reproduction in the plant (Humphreys et al. Citation2010). They are important for plant growth, nutrition, protection from pathogens, diversity, nutrient cycling, and sustainable ecosystem (Sanders & Croll Citation2010). Different tool kits of mycorrhizal symbiosis are conserved in basal land plant (Delaux et al. Citation2013). A key goal in mycorrhizal research is to understand the molecular basis of the establishment, regulation, and functioning of the symbiosis. The symbiotic association induces important physiological changes in both partners that led to the reciprocal benefit from each other, and these events resulted from several modifications in plant and fungal partners (Herre et al. Citation1999; Perotto et al. Citation2014). However, there is lack of sufficient knowledge in the genomic, metabolomic, and proteomic events of the fungal side that needs to focus more on understanding the AMF symbiosis event efficiently.
The AMF symbiosis starts with the germination of fungal spores. The establishment of symbiosis includes hyphal branching, appressorium development, colonization, formation of intracellular arbuscules, and concomitant production of extraradical mycelium (Akiyama et al. Citation2005). At the molecular level, signals are exchanged between two partners, leading to a stage-specific pattern of gene expression and hormonal signaling. The corresponding gene products could be responsible for the morphological and physiological changes necessary for the integration of two partners into one association (Harrier Citation2001; Gutjahr & Parniske Citation2013).
Research on the mycorrhizal symbiosis is driven by its fundamental importance for plant growth and development (Miransari Citation2010; Franzini et al. Citation2013; Willmann et al. Citation2013). In spite of great importance being given to mycorrhizal symbiosis, much of basic biology of fungal partner is still lacking (Sanders & Croll Citation2010). The basic information on AMF, such as ploidy, meiosis, recombination events, and segregation, and other events that play a major role in symbiosis needs to be understood thoroughly. Even some findings are more particular to specific fungal phylum and conventional model of evolution, and Mendelian genetics are difficult to apply in studying inheritance, segregation, gene expression, and genomics of the fungal partner (Bever et al. Citation2008; Oehl et al. Citation2011).
In this section we discussed current advances in functional genomics, nutrient uptake, hormonal signaling, calcium signaling events, and the role of miRNAs in mycorrhizal symbiosis. The AMF are obligated symbionts of the plants that grow toward the root of the plant and subsequently penetrate through it. Upon penetrating the root, it passes through the first layer of the cells and subsequently spread toward the root cortex (Sanders & Croll Citation2010). The fungi penetrate the cell wall of cortical cells and invaginate the host cell membrane and form a branched structure known as arbuscule (Gianinazzi-Pearson Citation1996; Veiga et al. Citation2013; Rich et al. Citation2014). The arbuscules are involved in the bidirectional exchange of nutrients between the host plant and the fungus (Smith & Smith Citation2011; Smith & Smith Citation2012). The hyphae of fungus grow out of the root into the soil and act as an extension of the root system that helps in nutrient uptake (Karandashov & Bucher Citation2005). In contrast to nutrient uptake, fungi take carbohydrates from the plant partner by using its arbuscules (Schuszler et al. Citation2006). The extraradical hyphae colonizes the adjacent root and forms a belowground hyphal network thus connecting many different plants within the ecosystem (Bonfante & Requena Citation2011).
Symbiotic events
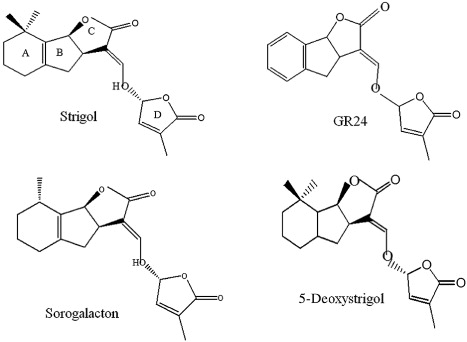
Strigolactones (SLs) are plant root exudates and are popularly known as a germination stimulant of AMF spore (Akiyama et al. Citation2005; Besserer et al. Citation2006; Bhattacharya et al. Citation2009; Prandi et al. Citation2011; Quain et al. Citation2014; Waldie et al. Citation2014). These SLs are now considered as a novel class of plant hormones that inhibits shoot branching (Scaffidi et al. Citation2014; Waldie et al. Citation2014). All the SLs contain a tricyclic lactone ring (ABC part) that connects via an enol ether bridge to a butenolide group (D ring; ; Yoneyama et al. Citation2009; Akiyama et al. Citation2010; Xie et al. Citation2013). These compounds have one or two methyl groups on the A ring and one or more hydroxyl or acetyl-oxyl group on the A/B ring (Xie et al. Citation2007). The 5-deoxystrigol is considered as the most common precursor of various SLs and their derivatives (; Besserer et al. Citation2006; Rath et al. Citation2006).
It is proposed that SLs originate from the carotenoid biosynthetic pathway (Matusova et al. Citation2005). The carotenoid cleavage dioxygenase 7 and 8 (CCD7 and CCD8) play significant roles in SL biosynthesis (; Domagalska & Leyser Citation2011; Delaux et al. Citation2013). Matusova et al. (Citation2005) suggested that plant hormone abscisic acid derived from the carotenoid biosynthetic pathway is involved in the regulation of SLs production (Matusova et al. Citation2005; López-Ráez et al. Citation2010). The ABA-deficient mutant in maize (viviparous14, vp14) and tomato (notabilis), with a null mutation in the gene encoding the 9-cis-epoxycarotenoid cleavage dioxygenase (NCEDs) induced a low level of germination in Striga hermonthica and Phelipanche ramosa seeds (Tan et al. Citation1997; Seo & Koshiba Citation2002; Lefebvre et al. Citation2006; Bouwmeester et al. Citation2007). A reduction in SLs in the root exudates of tomato mutant notabilis was confirmed by LC-MS/MS analysis (López-Ráez et al. Citation2010). But it is not clear, whether the reduction in SL production was due to mutation in NCEDs or due to reduced levels of ABA.
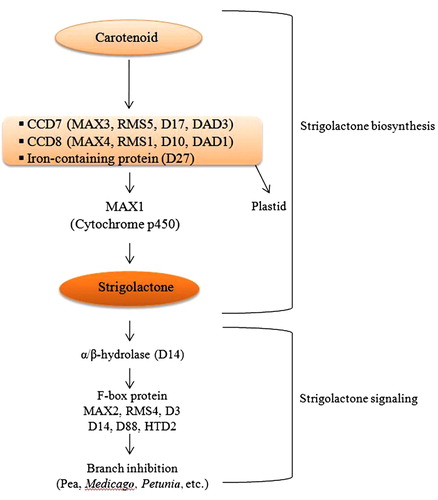
The biosynthesis of SL is usually boosted under phosphate-limiting conditions (López-ráez & Bouwmeester Citation2008; Yoneyama et al. Citation2010; Czarnecki et al. Citation2013; Yamada et al. Citation2014). A pair of GRAS-type transcription factors, nodulation signaling pathway (SNP) 1 and 2, play important roles in the regulation of SL biosynthesis under low phosphate condition (Liu et al. Citation2011). Both of these factors are indispensable for nodule formation in legumes. The GRAS and SNP genes are required for appropriate regulation of DWARF27 gene that encodes β-carotene isomerase (Alder et al. Citation2012; Waters et al. Citation2012). The β-carotene isomerase catalyzes the first committed step in SL biosynthesis (Alder et al. Citation2012). It is reported that F-box protein D3/MAX2/RMS4 interacts with α/β-hydrolase family receptor D14/DAD2/HTD2 to perceive SL signaling (Kagiyama et al. Citation2013; Nakamura et al. Citation2013). Once SL binds to D14/DAD2/HTD2, it undergoes conformational changes promoting its interaction with D3/MAX2/RMS4 that leads to the activation of E3-ligase-mediated protein degradation, permitting the SL responses (Gutjahr & Parniske Citation2013; Kagiyama et al. Citation2013; Nakamura et al. Citation2013). The F-box protein D3/RMS4 is also required for AM colonization in rice and pea (Yoshida et al. Citation2012; Foo, Yoneyama, et al. Citation2013; Gutjahr Citation2014). Early reports suggest the role of D27 and D14 genes in SL biosynthetic pathway as well as in the early phase of symbiotic association (Liu et al. Citation2011; Delaux et al. Citation2013). The Arabidopsis BRANCHED1 (BRC1), encoding a TCP transcription factor, is closely related to TEOSINTE BRANCHED1 (TB1) of maize that acts downstream of auxins and SLs (Dun et al. Citation2009).
Successful symbiosis with AM fungi relies on the fine-tuning and appropriate control of host gene expression and its physiological responses (Bhattacharya et al. Citation2009; Bonfante & Requena Citation2011; Chabaud et al. Citation2011). A molecular dialog is established between the host plant and the AM fungus that prepares both the partners for subsequent root colonization (Balzergue et al. Citation2013; Geurts & Vleeshouwers Citation2012). The communication and signal exchange start prior to the initial cell-to-cell contact between the two symbionts. The plant root exudes SLs that carry out the stimulatory effect on fungus and lead to its proper growth and development. The fungal hyphae in turn produce diffusible molecules known as ‘Myc factors’ that are perceived by plant roots (analogous to the rhizobial Nod factors; Kosuta et al. Citation2003; Akiyama et al. Citation2005; Balzergue et al. Citation2013; Delaux & Guillaume Citation2013; Gutjahr & Paszkowski Citation2013). The perception of Myc factor by the host cell triggers rapid and transient elevation in the intracellular calcium ion, alteration in the cellular architecture, and transcriptional reprogramming in the root (Delaux & Guillaume Citation2013; Gutjahr & Paszkowski Citation2013; Bucher et al. Citation2014). Besides the role of Myc factor, AM fungi also secrete lipochitooligosaccharides that stimulate the formation of AM symbiosis (Maillet et al. Citation2011; Herrbach et al. Citation2014).
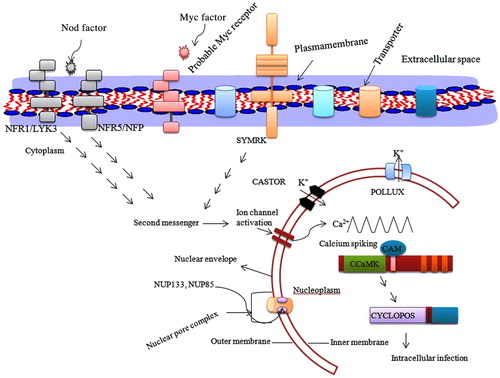
Cytological studies show that the nuclear division that occurs in the nuclei within the germination hyphae is necessary for the symbiotic hyphal growth (Requena et al. Citation2000). Earlier it was demonstrated that GmTOR2 gene from Glomus mossae, a homolog of Saccharomyces cerevisae, controls cell cycle arrest in response to nutrient starvation (Requena et al. Citation2000). Requena et al. (Citation2000) also suggested a possible role of GmGIN1 of G. mossae in the signaling event during spore germination before symbiosis. The genes encoding germin-like, nodulin 26-like, and four other proteins of unknown function are activated at the appressorium stage suggesting their role in the early stage of mycorrhizal colonization (Brechenmacher et al. Citation2004). Other genes encoding cell wall protein PsENOD12A and C1p serine protease are activated in pea root during appressoria formation (Cathy et al. Citation1999). A proline-rich protein which is encoded by ENOD11 activates the epidermal cells before, during pre-penetration apparatus formation (PPA), and at the late stage of mycorrhizal development, and even in arbuscule-containing cells. Three ion channel genes of Medicago truncatula DMI1 (does not make infection 1), DMI2, and DMI3 are required for the induction of pre-penetration apparatus in plants (Genre et al. Citation2005; Siciliano et al. Citation2007; Genre & Bonfante Citation2010). A novel IPD3 protein interacts with the DMI3 protein, involved in pre-penetration apparatus formation (Messinese et al. Citation2007). Using mutants of Lotus japonicus, five genes, namely CASTOR, POLLUX, NUP85, NUP133, and CYCLOPS, were identified from legumes required for the development of the mycorrhizal symbiosis in the plant roots (; Banba et al. Citation2008; Charpentier et al. Citation2008; Gutjahr et al. Citation2008; Yano et al. Citation2008; T. Hayashi et al. Citation2014). All these genes are common to Rhizobium-legume symbiosis as well as mycorrhizal symbiosis and are induced at early stage of signal transduction. The genes necessary for mycorrhizal formation in rice were also well investigated (Sanders & Croll Citation2010). The putative orthologs of CASTOR, POLLUX, CCaMK (DMI3), and CYCLOPS in rice and legumes are necessary for mycorrhiza formation (; Banba et al. Citation2008; Gutjahr et al. Citation2008). However, a number of other genes are also identified in rice that appear to be specific to mycorrhiza formation (Charpentier et al. Citation2008; Gutjahr et al. Citation2008; Yano et al. Citation2008). These genes are divided into two different groups, AM13 and AM11. These groups of genes are expressed at early stages of symbiosis before arbuscule formation. The genes AM10, 11, 14, 15, 18, 20, 24, 25, 26, 29, 31, 34, 39, 42, and PT11 are expressed at the later stage of development, and some are in arbusculated cells (Gutjahr et al. Citation2008). The functions encoded by most of these genes are currently unknown.
Gallou et al. (Citation2012) performed potato microarray analysis that led to finding of changes in transcript level at pre-, early, and late stages in potato root colonization by Glomus sp. MUCL41833 (Gallou et al. Citation2012). Data analysis revealed 526 up-regulated and 132 down-regulated genes during pre-stage, 272 up-regulated and 109 down-regulated during early stages, and 734 up-regulated and 122 down-regulated genes during the late stage of root colonization. The most important class of regulated genes that reported to play a significant role is associated with plant stress and in particular with the WRKY transcription factor genes. The WRKY transcription factor genes are regulated at the pre-stage in the root colonization process. The expression profiles demonstrated wide transcriptional variation in pre-, early, and late stage of root colonization. It is found that the WRKY transcription factor genes are involved in controlling the mechanism of arbuscular mycorrhizal establishment by regulating the plant defense genes.
The mycorrhizal-induced plant genes play major roles in the development of symbiotic phase during mycorrhization process (Balestrini et al. Citation2007; Koegel et al. Citation2013). Nowadays, significant progress has been made in identification of related genes involved in symbiosis events (). Uses of ESTs, cDNA libraries, genomic libraries, and microarray analysis have the potential to screen differentially expressed transcripts. Three different M. truncatula-based lectine-like genes are induced in mycorrhizal roots during the colonization process (Wulf et al. Citation2003a). Expression of transcript corresponding to gene encoding lectin-like glycoprotein PsNLEC-1 is strongly expressed in root nodules of Pisum sativum (Kardailsky et al. Citation1994; Dahiya et al. Citation1997). Identification of numerous different lectin-like transcripts is also increased in mycorrhizal roots. This indicates their significant role during the symbiotic process. Toward understanding the molecular aspects of arbuscule development in AM fungi, Gomez et al. (Citation2009) used Affymetrix genechip Medicago genome array to document M. truncatula transcript profile associated with AM symbiosis and then developed laser micro-dissection (LM) of M. truncatula root cortical cells to enable the analysis of gene expression in individual cell types (Gomez et al. Citation2009). Experiments showed that within the arbuscule, genes associated with urea cycle, amino acid biosynthesis, and cellular autophagy are detected to be potentially much higher than the others. Analysis of gene expression in colonized cortical cells revealed the up-regulation of a lysine motif (LysM) receptor-like kinase (members of the GRAS transcription factor family) and a symbiotic-specific ammonium transporter (). The rice transcriptome analysis shows that 12 genes are exclusively expressed in mycorrhizal roots (Güimil et al. Citation2005). These genes are proposed as marker genes for AM symbiosis. Among them, one encodes for putative peroxidase OsAM1 (). Interestingly, 43% of the mycorrhiza-induced rice gene responds similarly to the infection by fungal pathogens, suggesting the conservation of the transcriptional activation pathway in mycorrhizal symbiosis (Güimil et al. Citation2005). In addition, about one third of the mycorrhizal responsive genes of rice are matched with homologous sequences of dicotyledonous plants that are up-regulated during AM symbiosis, suggesting a relatively conserved symbiotic mechanism in monocot and dicot plants (Güimil et al. Citation2005).
Table 1. Table showing list of genes and their roles during mycorrhizal symbiosis process.
Zhang et al. (Citation2010) have performed an experiment with M. truncatula mutant, stunted arbuscule (str), in which arbuscule development was impaired (Zhang et al. Citation2010). The AM symbiosis failed, which led to the identification of a second AM symbiosis-induced transporter STR2. The silencing of STR2 by RNA interference resulted in a stunted arbuscule phenotype. They also reported that STR1 and STR2 are co-expressed constitutively in the vascular tissue, and their expression was induced in cortical cells that contained arbuscules (Zhang et al. Citation2010). The STR1 heterodimerizes with STR2, and the resulting transporter was found in the peri-arbuscular membrane where its activity is necessary for arbuscule development and consequently functional AM symbiosis.
Upon development of hypopodium on the outer wall of root epidermal cells, both the plant and fungal partner enters into a novel developmental program that culminates fungal accommodation inside the root cell lumen that lead to fully functional symbiosis. It is reported that initiation of an accommodation program strongly correlates with the activation of SYM pathway (Gutjahr et al. Citation2008). As symbiosis occurs, it undergoes significant changes in the morphological organization of host cells to accommodate fungal partners. This includes the creation of new interface compartments within the host cell. The genes encoding both ADP-glucose pyrophosphorylase (AGP) and a hydroxyproline-rich glycoprotein (HRGP) are supposed to play important roles in this process. These genes are induced in mycorrhizal roots of M. truncatula and maize, respectively, and the transcripts are specifically localized in arbusculated cells (Balestrini et al. Citation1997; Van Burren et al. Citation1999). Many of the ESTs that are identified by Grunwald et al. (Citation2004) belong to the gene product involved in cell wall modification. One encodes an extensin-like glycoprotein that belongs a to large protein family characterized by hydroxy-proline rich motifs. Using a cDNA array, Liu et al. (Citation2003) reported the induction of MtCell gene during symbiosis. It suggested that MtCell is localized in the periarbuscular membrane and is involved in assembling the cellulose/hemicelluloses matrix at the interfaces. This Mtcell gene is associated with expanding tissue and cellulose synthesis.
An extracellular protein, expansin, with the possible role in cell wall loosening is up-regulated during intracellular colonization of AM fungi during early stage of mycorrhizal interactions. This indicates that these classes of proteins are involved in cell wall loosening and could be crucial in accommodating the AM fungus inside cortical cells. The mutational analysis of Mt-xht1 gene shows a systemic modification of cell wall structure to enable fungal penetration to the roots (Balestrini & Bonfante Citation2014). The cytoskeleton of host cell undergoes a massive change during the invasion of cortical cells and undergoes transient rearrangement, presumably to enable the development of arbuscular interfaces. This re-organization is induced by signals before mycorrhizal penetration to the host cells. The activation in α-tubulin promoter and accumulation of enhanced mRNA of β-tubulin gene occur during a cellular modification stage in arbuscule-containing cells (Zampieri et al. Citation2010). The β-tubulin gene also modulated during the symbiosis process (Rhody et al. Citation2003).
Nutrient uptake
A major breakthrough in AMF symbiosis was achieved when the Pi transporter gene was characterized from Glomus versiforme extraradical hyphae involved in Pi uptake from soil (Harrison & van Buuren Citation1995). This Pi gene was induced at the transcriptional level in the presence of lower amount of Pi. Later the PT homolog GmosPT was reported from the Glmous mosseae that plays a similar role in Pi transport (Benedetto et al. Citation2005). The phosphate transporter genes encoding alkaline phosphatases were also described in Glomus intraradices and G. margarita (Tisserant et al. Citation1993; Aono et al. Citation2004). The transcript level of these genes are higher in the mycorrhizal root than in the germinating spore and external hyphae, suggesting their significant role in nutrient exchange with the host plant (Aono et al. Citation2004). The phosphate transporter MtPT4 gene from M. truncatula is located in the periarbuscular membrane, at the interface between arbuscules and the invaginated plant cell membrane, generally assumed as the site of nutrient exchange (Harrison et al. Citation2002). A gene Mtha1 encodes the plasma membrane H+ ATPase up-regulated in M. truncatula during AM symbiosis, and Mtha1 transcript get accumulated in arbuscule-containing cells (Wulf et al. Citation2003a; Krajinski et al. Citation2014). Karandashov and Bucher (Citation2005) demonstrated the role of phosphate transporter StPT3 gene (Karandashov & Bucher Citation2005). They reported that StPT3 is expressed in root cells harboring various mycorrhizal structures. The cell-to-cell contact between the partner is required to induce the phosphate transport system by Pi transporters (Güimil et al. Citation2005). The redundancy within the mycorrhiza-inducible Pi transporter pathways ensure that Pi transfer is evolutionary robust and relatively insensitive to mutation (Nagy et al. Citation2005). At present, it is confirmed that several Pi transporter genes were strongly induced during AM colonization process (). These genes are OsPT11 (Oryza sativa phosphate transporter11), LePT4 (Lycopersicon esculentum PT4), PtPT8 (Populus trichocarpa PT8), PtPT10 (Populus trichocarpa PT10), StPT4 (Solanum tuberosum PT4), StPT5 (Solanum tuberosum PT5), LePT4 (Lycopersicon esculentum PT4), PhPT4 (Petunia hybrid PT4), PhPT5 (Petunia hybrid PT5), LjPT3 (L. japonicus PT3), GmPT7 (Glycine max PT7), GmPT11 (Glycine max PT11), GmPT10 (Glycine max PT10), ZmPT6 (Zea maize PT6). The organic phosphate molecule delivers to the host plant via the mycorrhizal-induced phosphate transporter genes (Balestrini et al. Citation2007). The fungal partner activates part of the low Pi adaptation system of the plant partner by secreting phosphatases and improves the overall efficiency of Pi uptake. Many other plant Pi transporters and one fungal Pi transporter were identified in arbusculated cells (Loth-Pereda et al. Citation2011). The MtPT4 is essential for the acquisition of Pi delivery by AM fungi. The loss of MtPT4 function led to the premature death of arbuscules in M. truncatula plant, and fungus was unable to proliferate within the host root and thus symbiosis was terminated (Javot et al. Citation2007).
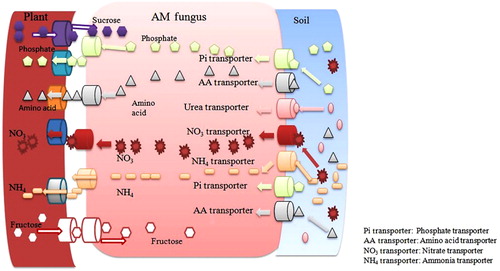
The monosaccharide transporter (MST2) from Glomus sp. with a broad substrate spectrum that functions at several symbiotic root location are demonstrated very well (Büttner & Sauer Citation2000; Schuszler et al. Citation2006; Helber et al. Citation2011; Slewinski Citation2011). The plant cell wall sugar can efficiently outcompete the Glc uptake capacity of MST2, suggesting that they can serve as the alternative carbon source (; Helber et al. Citation2011). The MST2 expression is closely correlated with that of mycorrhizal-specific phosphate transporter 4 (PT4). The reduction of MST2 expression using host-induced gene silencing resulted in impaired mycorrhiza formation, malformed arbuscules, and reduced PT4 expression. This finding highlighted the symbiotic role of MST2 and supported the hypothesis that the exchange of carbon for phosphate is tightly regulated (). During the symbiotic phase, C metabolisms of both partners are significantly modified at the level of gene expression (Balestrini & Lanfranco Citation2006). Ravnskov et al. (Citation2003) reported an up-regulation of the sucrose synthase gene which led to hypothesize that the sucrose synthase plays a major role in generating sink strength (Ravnskov et al. Citation2003). In 1996, Harrison reported a high expression of sugar transporter gene (Mtst1) in arbuscule-containing cells as well as the adjacent surrounding colonized area (Harrison Citation1996).
In addition to phosphate in mycorrhizal associations, the fungal partner assisted the plant partner by providing nitrogen (N) nutrient too (Hodge et al. Citation2001; Govindarajulu et al. Citation2005; Matsumura et al. Citation2013; Kranabetter Citation2014). Arbuscular mycorrhizal (AM) fungi have access to inorganic or organic forms of nitrogen, such as nitrate, ammonia, and urea. They easily translocate them from the extraradical surface to the intraradical mycelium via respective transporter molecule, where the nitrogen is transferred to the plant without any carbon skeleton () (Pérez-Tienda et al. Citation2011; Jin et al. Citation2012; Ellerbeck et al. Citation2013). However, the molecular form in which N is transferred, as well as the involved mechanism is still under debate. The NH4+ seems to be the preferred molecule (Guether et al. Citation2009). The transcript of LjAMT2 (Amt/Rh family) is up-regulated in transcriptome analysis of L. japonicus root upon colonization with G. margarita. This transcript is extensively expressed in mycorrhizal root, but not in the nodule (Guether et al. Citation2009). The ammonium transporter LjAMT2 transports NH3 instead of NH4+. The ammonium transporter binds to charged ammonium in the apoplastic interface compartment and releases the uncharged NH3 molecule into the cytoplasm. The role of ammonium transporter LjAMT2 is confirmed by stochastic calculus of wrapped compartment (SCWC) (Variant of stochastic calculus of looping sequence) in mycorrhizal symbiosis (Coppo et al. Citation2011). The X-ray structure study of ammonium transporter revealed the conduction mechanism of Amt/Rh (rhesus) protein that involved in the single- file diffusion of NH3 molecule (Zheng et al. Citation2004; Andrade et al. Citation2005; Lamoureux et al. Citation2010). However it is suggested that AmtB could be filled with water molecule and the presence of water molecule in the pore lumen indicate, Amt/Rh protein work as plain NH3 channel (Lamoureux et al. Citation2010). The Amt/Rh protein also plays variety of permeation mechanism including passive diffusion of NH3.
Hormonal regulation
The relationship between host root and AM fungi requires continuous exchange of different signals those help them for proper development of symbiosis. The phytohormones are important signaling molecules known to regulate many growth and developmental process in plants (Barker & Tagu Citation2000; Foo, Yoneyama, et al. Citation2013; Gutjahr Citation2014). In maize and M. truncatula, level of indole-3-butyric acid (IBA) get increases whereas, level of indole-3-acetic acid (IAA) remains unaltered in mycorrhizal symbiotic plant (Ludwig-Müller et al. Citation1997; Ludwig-Müller & Güther Citation2007). In Glycine max, IAA level present in higher concentration in AM root than in control plant (Meixner et al. Citation2005). Altered level of auxin and cytokinin was observed in Solanum lycopersicum harbored with mycorrhizal symbiont (Torelli et al. Citation2000; Shaul-keinan et al. Citation2002). It is speculated that auxin, more particularly IBA may facilitate colonization of host by increasing the number of lateral root during early growth phase of plant development (Kaldorf & Ludwig-Müller Citation2000; Ludwig-Müller & Güther Citation2007). The application of inhibitor against IBA, root growth and lateral root induction reduced significantly that led to reduced AM colonization (Kaldorf & Ludwig-Müller Citation2000). The transcript level of leghemoglobin get up-regulated in AM colonized root, in IAA and IBA treated plant (Frühling et al. Citation1997; Heidstra et al. Citation1997). The leghemoglobin play crucial role in N2 fixation in Rhizobium-legume interaction (Heidstra et al. Citation1997). But, its role in AM symbiosis is unclear. Some of the nitrate transporter genes get down regulated in AM root (Ludwig-Müller & Güther Citation2007). The endochitnase gene family members are down regulated in AM colonized as well as an IBA treated plant of M. truncatula (Ludwig-Müller & Güther Citation2007). Hanlon and Coenen (2011) carried out experiment in diageotropic (dgt), an auxin-resistant mutant and polycotyledon (pct) mutant with hyperactive polar auxin transport. They found that, G. intraradices stimulate pre-symbiotic root branching in pct but not in dgt root (Hanlon & Coenen Citation2011). In auxin deficient bushy mutant of pea, bsh root has reduced mycorrhizal colonization compared to wild-type plant (Foo, Yoneyama, et al. Citation2013). This mutant also has reduced level of SL content in root due to down-regulation of PsCCD8 gene (Foo et al. Citation2005; Johnson et al. Citation2006). The application of exogenous auxin reverted the effect of bsh mutant and reported to have higher mycorrhizal colonization (Symons et al. Citation2002; Foo Citation2013).
Beside auxin, other phytohormones like gibberellic acid, cytokinin, jasmonic acid, and salicylic acid also plays significant role in AM colonization (Barker & Tagu Citation2000; Foo, Ross, et al. Citation2013; Miransari et al. Citation2014; Yu et al. Citation2014). It is reported that GA (gibberellic acid) involved in root nodulation process (S. Hayashi et al. Citation2014). Floss et al. (Citation2013) demonstrated the role of DELLA protein in gibberellic acid mediated arbuscule formation during mycorrhization process (Floss et al. Citation2013). The gene expression analysis in GA-related genes are up-regulated in M. truncatula as reported earlier in tomato too (Garrido et al. Citation2010; Ortu et al. Citation2012). The jasmonic acid has long been implicated in plant systemic response to pathogen attack, that led to question regarding its involvement in the regulation of AM development (Regvar et al. Citation1996; Hause et al. Citation2002). But it is found that the application of JA or its derivatives can have ranges of effect in the AM colonization process. The effect may be positive or may be inhibitory depending on the species, nutritional conditions, timing and dosage conditions. Regvar et al. (Citation1996) showed a clear promotion of AM colonization in Allium plant by application of low level of jasmonic acid (Regvar et al. Citation1996). Landgraf et al. (Citation2012) reported increased level of endogenous JA content in plants by repeated wounding, and found that, the resulting plants are able to increase AM colonization (Landgraf et al. Citation2012). This suggests the positive role of JA in AM development process. This result supported with the report of Isayenkov et al. (Citation2005), where they found down regulation of JA level by antisense expression of allene oxide cyclase gene and found delay in AM colonization (Isayenkov et al. Citation2005). Tejeda-Sartorius et al. reported, JA deficient spr2 mutant of tomato undergoes reduced AM colonization. Later the AM colonization was restored by exogenous application of methyl jasmonate (Li et al. Citation2002; Tejeda-sartorius et al. Citation2007). Unlike jasmonic acid, salicylic acid (SA) also act to coordinate the plant defense against biotropic pathogen and hence activated during AM colonization (Stacey et al. Citation2006; Zhang et al. Citation2013; Foo, Ross, et al. Citation2013). Blilou et al. (Citation1999) and Herrera-Medina et al. (Citation2003) showed, SA content in plant affect the rate of AM colonization (Blilou et al. Citation1999; Herrera Medina et al. Citation2003).
Calcium signaling
The calcium dependent protein kinases are important calcium binding sensor proteins that play significant role in plant growth and development as well as other biotic and abiotic responses (Kanchiswamy et al. Citation2013). One of the most studied features during early stages of establishment of AM symbiosis is calcium mediated signaling pathway that initiated in host plant by AM fungal diffusible signal. The exudate of germinated fungal spores triggers transient elevation of cytosolic Ca2+ level in Glycine max (Navazio et al. Citation2007). The chemical nature of symbiotic signals released by AM fungi, responsible for calcium spiking for AM symbiosis is carried out by ‘Myc/Nod’ factor ( and ) (Sieberer et al. Citation2009; Chabaud et al. Citation2011; Maillet et al. Citation2011; Genre et al. Citation2013). The LysM receptor-like kinase NFR1/LYK3 and NFR5/NFP are the strongest candidate genes that function as Nod factor receptor (; Broghammer et al. Citation2012). The M. truncatula DOES NOT MAKE INFECTIONS1 (DMI1) and L. japonicus CASTOR and POLLUX that encodes for nuclear ion channel are prominent genes required for initiation of Nod and Myc factor induced Ca2+ spiking. The M. truncatula DMI13 (CCaMK) promotes Ca2+ binding during calcium oscilations (Miller et al. Citation2013). The L. japonicus CASTOR and POLLUX as well as M. truncatula DMI1 are localizes to the nuclear envelope (Riely et al. Citation2007; Charpentier et al. Citation2008). M. truncatula Ca2+ ATPase MCA8 block Nod factor induced calcium spiking (Capoen et al. Citation2011). This MCA8 localize in inner and outer nuclear membrane, suggesting their coordinated role in nuclear calcium spiking (Capoen et al. Citation2011). A study carried out by Venkateshwaran et al. (Citation2012) shown that DMI1 of M. truncatula is a close ortholog of L. japnicus POLLUX and sufficient enough to carry out symbiosis in the absence of CASTOR and POLLUX (Venkateshwaran et al. Citation2012). The L. japonicus castor, pollux and castor pollux double mutant are rescued by presence of DMI1 of M. truncatula, confirming its indispensable role in symbiosis. In another case, both Lj-CASTOR and Lj-POLLUX were required for rescuing dmi1 mutant of M. truncatula (Venkateshwaran et al. Citation2012). When Lj-CASTOR and Lj-POLLUX expressed individually, they failed to rescue dmi1-4 (Venkateshwaran et al. Citation2012). This is due to lack of appropriate promotor that control the expression of these genes. The DMI1 contain ADAGNHA and Lj-CASTOR and Lj-POLLUX have ADSGNHA amino acid residues in the selectivity filter. In this sequence, Ala of DMI1 is substituted by a Ser amino acid. When Lj-CASTORS266A and Lj-POLLUXS329A is replaced by Ala amino acid in exchange of DMI Ser amino acid, the Lj-castor and Lj-pollux mutant able to rescue the plant (Venkateshwaran et al. Citation2012). This substitution of Ser to Ala amino acid reduced the potassium conductance. Roberts et al. (Citation2013) reported LNP (LECTIN NUCLEOTIDE PHOSPHOHYDROLASE) as a Nod factor binding protein that act upstream of calcium signaling event (Roberts et al. Citation2013). The LNP acts in the early stage of symbitic signaling process.
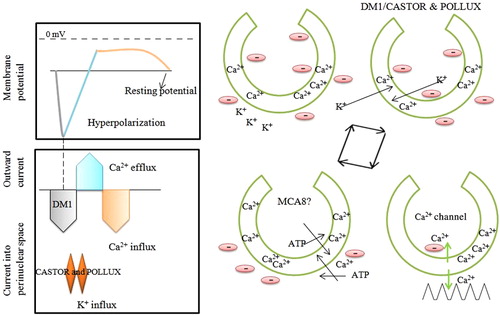
Campos-Sariano et al. (Citation2011) reported expression of seventeen calcium-dependent protein kinase (CPK) genes of rice representative of four distinct phylogenetic groups (Campos-Soriano et al. Citation2011). Among them OsCPK18 and OsCPK4 are transcriptionally activated in response to AM fungus G. intraradices. The OsCPK18 and OsCPK4 gene expression was also up-regulated by fungal produced diffusible molecules. The laser micro-dissection study revealed expression of OsCPK18 in cortical cells, and not in epidermal cells of G. intraradices inoculated rice root, suggesting preferential role of this gene in root cortex. The rapid activation of OsCPK18 expression in response to AM inoculation reflects its roles in perception of AMF signaling. The OsCPK18 gene might be considered as a marker gene of the pre-symbiotic phase. This finding will provide a better understanding of the signaling mechanism process during AM symbiosis and will greatly facilitate their molecular dissection. In L. japonicas, it is reported that seven common symbiotic genes (SYMRK, CASTOR, POLLUX, SYM3, SYM6, SYM15, and SYM24) are required for both fungal and bacterial entry into root epidermal or cortical cells (Stracke et al. Citation2002; Kaneko et al. Citation2005).
miRNA in mycorrhizal symbiosis
MicroRNAs (miRNAs) are small non-coding RNA gene product of about 22 nt long that are processed by dicer from precursor with a characteristic hairpin secondary structure (Chen et al. Citation2010; Ameres & Zamore Citation2013; Martin et al. Citation2014). The miRNAs are typically inactivate developmentally important mRNA by inhibiting their translation or bringing to their cleavage and degradation (Ossowski et al. Citation2008; Warthmann et al. Citation2008; Djuranovic et al. Citation2012). Devers et al. (Citation2011) suggested the important role of miRNA in AM symbiosis (Devers et al. Citation2011). They studied miRNA mediated mRNA cleavage in root cell reprogramming during AM symbiosis. High throughput (Illumina) sequencing of small RNAs and dendrogram tag of M. truncatula roots led to annotation of 243 novel miRNAs. An increased accumulation of several novel and conserved miRNAs in mycorrhizal roots suggests their significant roles during AM symbiosis. The dendrogram analysis led to identification of 185 root transcript as mature miRNA. Several of identified miRNA targets are known to be involved in root symbiosis. Increased accumulation of specific miRNA and miRNA mediated cleavage of symbiosis relevant genes indicated that miRNAs are important part of regulatory network that lead to the development of symbiosis (Devers et al. Citation2011). They found higher up-regulation in miR5229a/b and miR5229a/b in mycorrhizal roots and are highly abundant in arbuscule containing cells, suggesting their specific role during arbuscule development. Still no target gene has predicted for this miRNA, but prediction suggests a transcript encoding a haem peroxidase could be a target for this miRNA. It might be suggested that induction of miR5229a/b suppresses the haem peroxidase leading to locally increased hydrogen peroxide accumulation in cells with degenerate arbuscules. Lauressergues et al. (Citation2012) reported that miR171h modulates arbuscular mycorrhizal colonization in M. truncatula by targeting NSP2 gene (Lauressergues et al. Citation2012). Induction of miR169d/I also reported in mycorrhizal roots. The miR169 target the transcription factor MtHAP2-1, that reported to play important role in nodule differentiation by restricting MtHAP2-1 expression in the nodule meristematic zone (Combier et al. Citation2008). The miRNA* sequences of miRNA169 family also strongly induced in mycorrhizal roots that can target Mtbcp1 gene. The Mtbcp1 encodes for protein specifically accumulating in periarbuscular membrane and speculated that the miR169* accumulating in mycorrhizal roots are involved in restricting the MtBcp1 expression in arbuscule containing cells (Pumplin & Harrison Citation2009). The miR169* was detected in mycorrhizal colonized phloem, however the function of miRNA169* is yet to elucidate. The miR5204 appears to be phosphate responsive and located around individual arbuscules. They hypothesized that presence miR5204 correlates with phosphate concentration of distinct phosphate containing arbuscule. The miR160f and mycorrhizal induced miR160c predominantly localized in the phloem and targets several transcripts of auxin response factor (ARF) gene family.
The gene MtGst1 encodes for mycorrhizal symbiosis specific glutathione-s-transferase regulated by miR5282 and candidate miRNA new_miRc_275 (Wulf et al. Citation2003a). Earlier it has described that Vitis venifera miR156 and miR535 regulate squamosa promoter binding transcription factor (Mica et al. Citation2009). Some other symbiotic related genes that are regulated by miRNA are MtNsp2, a GRAS transcription factor that essential for root nodule development. The GRAS transcription factor is cleaved by miR5204*. In addition, nsp2-2 mutant show decrease mycorrhizal colonization (Gobbato et al. Citation2012). Significant modulation of MtNsp2 transcript levels was reported in mycorrhizal roots that cleaved by miR171h (Branscheid et al. Citation2011). The second most up-regulated transcript encodes a major facilitator protein PHO2 that is target by miR399 (Kim et al. Citation2011). The strong transcriptional induction of this major facilitator gene and its regulation by miR399 supports the role of miR399 during cellular phosphate homeostasis regulation in mycorrhizal roots (Branscheid et al. Citation2011).
Proteomics of mycorrhizal symbiosis
Proteins are well known effector molecule of plants that responds to different environmental cues. But very little information’s are known about the changes in protein expression in root during AMF symbiosis. In recent year outstanding molecular approaches are used to identify genes and proteins those involved in mycorrhization process. Originally, in 1995 Marc Wilkin coined the term ‘proteome’ which describes the ‘protein complement of genome’. So, it is very important to understand the role of genome complement (proteome) during AMF symbiosis. Although the proteomics study of AMF symbiosis is not significant enough, there are few reports that discussed the proteomic events of AMF symbiosis. Recent report by Abdallah et al. (Citation2014) in M. truncatula root membrane proteome study colonized with Rhizophagus irrgularis revealed differential expression of 1226 candidate proteins (Abdallah et al. Citation2014). Among them abscisic acid 8'- hydroxylase 4, 60S ribosomal protein L6, RuBisco large subunit alpha, pre-mRNA processing ribonucleoprotein, translation factor proteins, flotillin like protein 4 are accumulated much higher than others. The abundance of β- barrel domain containing proteins and palmitate modified proteins are also detected significantly. The palmitate modified proteins are involved in membrane trafficking, protein sorting and other signaling cascades. The model plant M. truncatula inoculated with AMF G. mosseae shows significant modulation in proteome level (Bestel-Corre et al. Citation2002). Major differentially expressed proteins found when M. truncatula colonized with G. mosseae are RNA helicase, phytochrome A1, leghemoglobin, nitorgenase iron protein. The leghemoglobin and nitorgenase iron protein are also differentially modulated when M. truncatula was treated with nitrogen fixing bacteria Sinorhizobium meliloti (Bestel-Corre et al. Citation2002). This shows that leghemoglobin and nitorgenase iron proteins are indispensible for symbiosis. Some other differentially modulated proteins found during this study are glutathione-s-transferase, cytochrome c-oxidase subunit 6b, polygalacturonase inhibitor protein, myosin heavy chain like protein, profucosidase precursor, elongation factor Tu, enolase, malate dehydrogenase, and superoxide dismutase. The cytochrome c-oxidase is an integral membrane protein that play major role in respiratory metabolism. Induction of cytochrome c-oxidase during mycorrhization could directly relate with mitochondrial respiration in arbuscule containing cells. Dumas-Gaudot et al. (Citation2004) performed an experiment in transferred-DNA (RiT-DNA) transformed root of carrot (Daucus carota) and colonized with G. intraradices to identify fungal protein involved in arbuscular mycorrhizal symbiosis (Dumas-Gaudot et al. Citation2004). They reported differential protein expression of NmrA-like protein, oxidoreductase, heat shock protein, ATP synthase β-mitochondrial precursor and MYK15-like protein (Dumas-Gaudot et al. Citation2004). Fester et al. (Citation2002) also reported the role of MYK15-like protein in proteomic study of wheat (Triticum aestivum) root infected with G. intraradices (Fester et al. Citation2002). The MYK15-like protein is an indicator of strong mycorrhizal colonization (Fester et al. Citation2002). The molecular mass of MYK15 is 15 kDa and isoelectric point (pI) is 4.5. So, mycorrhizal proteome study above pI 4.5 will automatically subtract this protein and cannot be detected in 2D-gel electrophoresis analysis. We already mentioned that DMI3 plays significant role in mycorrhiza process (Catoira et al. Citation2000). Study by Amiour et al. (Citation2006) demonstrated that mutation in DMI3 and SUNN modifies the appressorium-responsive root proteome in M. truncatula (Amiour et al. Citation2006). A DMI3 dependent M. truncatula increased accumulation of dehydroascorbate reductase, cyclophilin and actin depolymerization factor like proteins those related to signal transduction pathways. The role of actin depolymerization factor can be highly inferred as this protein is highly responsible for cell rearrangement that require for appressorium and arbuscule formation. No changes in proteome level was observed post five days of infection in M. truncatula mutant TRI22 (hyper-mycorrhizal) and TRV25 (mycorrhizal defective) when inoculated with G. intraradices (Amiour et al. Citation2006). In the wild type M. truncatula, infected with G. intraradices displayed an increase level of chalcone reductase, glutathione dependent dehydroascorbate reductase and actin depolymerization factor proteins. In TRI22 and TRV25 mutant, glutathione transferase is down regulated at appressorium stage. In the proteomic study of Lycopersicum esculentum inoculated with G. mosseae, 16 differentially expressed spots are detected during AM colonization stage (Ferrol & Benabdellah Citation2000). Sequence analysis from the selected bands revealed 69 kDa catalytic subunit of vacuolar type H+ ATPase. Recorbet et al. (Citation2010) colonized M.truncatula root with two Glomus (G. mosseae and G. intraradices) species and found 42 overlapping protein spots (Recorbet et al. Citation2010). Some of them are putative groES chaperonin, putative succinate dehydrogenase, putative malate dehydrogenase, leghemoglobin, glutathione and peroxidase.
Beside these proteomics events in mycorrhizal symbiosis, some research group applied toxicity stress to mycorrhiza inoculated plants. Repetto et al. (Citation2003) applied cadmium stress (100 mg Cd/kg substrate) to G. mosseae inoculated pea root (Pisum sativum) and found differential proteome abundance (Repetto et al. Citation2003). Some differentially observed spots are vacuolar ATPase β subunit, annexin, short chain alcohol dehydrogenase, profucosidase and pea disease resistance protein (Repetto et al. Citation2003). In another study, 2 mg Cd/kg was used in M. truncatula plant inoculated with G. irregularis (Aloui et al. Citation2011). The mycorrhiza responsive shoot proteome highly accumulated photosynthesis related proteins coupled to reduction in gluconeogenesis/glycolysis and antioxidant process. The mycorrhizal plant treated with Cd metal accumulated molecular chaperons in shoot relative to metal free inoculation (Aloui et al. Citation2011). When Pteris vittata, a mycorrhizal fern inoculated with G. mosseae or Gigaspora margarita along with 25 ppm/kg arsenic contamination, it resulted into differential accumulation of glyceraldehyde-3-phosphate dehydrogenase, phosphoglycerate kinase, and enolase (Bona et al. Citation2010). These differentially accumulated proteins are majorly glycolytic proteins and plays important roles in arsenic metabolism. Beside this, a putative arsenic transporter gene PgPOR29 also upregulated by arsenic treatment (Bona et al. Citation2010). The arsenic treatment did not induce any changes in morphological parameters when compared with untreated plants. The plants treated with G. margarita showed significant reduction in P. vitata frond dry weight relative to G. mosseae treated and control plants (Bona et al. Citation2010). The pine tree Populus alba inoculated with G. intraradices shows sharp decrease in ATP synthase isoform and enolase protein (Lingua et al. Citation2012). The grapevine root stock SO4 (selection Oppenheim 4) inoculated with Glomus species elicit proteome response opposite of p-starvation (Cangahuala-Inocente et al. Citation2011). Some highly upregulated protein identified during this study are involved in energy production, signaling, protein synthesis etc. These proteins are 32 subtilisin-like protease, putative signal peptidase, proteasome regulatory subunit S5A, TGF-β receptor-interacting protein 1, putative ATP synthase D chain, ubiquitin carrier protein, and RNA-binding glycine-rich protein. From the overall proteome study of AMF symbiosis, it observed that proteins like leghemoglobin, nitrogenase, enolase, glutathione transgerase, ATPases, and MYK15 are accumulated more than other proteins.
Conclusion and future perspectives
Symbiotic events are evolutionary conserved phenomena present since the days of evolutionary history. Although the core set of symbiotic genes is conserved in both symbiotic partners, conservation of biochemical function seems to be insufficient to understand their biological function. The reverse genetics study will be very useful to understand these events very efficiently. Characterization of different mutants from cereal crops as well as from other important model organisms by applying genomics, proteomics, and other aspects will be very helpful in understanding the conserved evolutionary aspects of symbiotic event. Beside the AM fungal symbiotic research on legume plants, it is very important to shift the focus to other alternative model organism to gain more insight into the different aspects of symbiotic event. Besides this, genomics and proteomics data are increasing enormously from the plant as well as its fungal partner. This will provide most crucial base to understand different genomics and proteomics aspects of symbiosis.
Additional information
Funding
References
- Abdallah C, Valot B, Guillier C, Mounier A, Balliau T, Zivy M, van Tuinen D, Renaut J, Wipf D, Dumas-Gaudot E, Recorbet G. 2014. The membrane proteome of Medicago truncatula roots displays qualitative and quantitative changes in response to arbuscular mycorrhizal symbiosis. J Proteomics. 108:354–368.10.1016/j.jprot.2014.05.028
- Akiyama K, Matsuzaki K, Hayashi H. 2005. Plant sesquiterpenes induce hyphal branching in arbuscular mycorrhizal fungi. Nature. 435:824–827.
- Akiyama K, Ogasawara S, Ito S, Hayashi H. 2010. Structural requirements of strigolactones for hyphal branching in AM fungi. Plant Cell Physiol. 51:1104–1117.10.1093/pcp/pcq058
- Albrecht C, Geurts R, Lapeyrie F, Bisseling, T. 1998. Endomycorrhizae and rhizobial Nod factors both require SYM8 to induce the expression of the early nodulin genes PsENOD5 and PsENOD12A. The Plant Journal. 15:605–614.10.1046/j.1365-313x.1998.00228.x
- Alder A, Jamil M, Marzorati M, Bruno M, Vermathen M, Bigler P, Ghisla S, Bouwmeester H, Beyer P, Al-Babili S. 2012. The path from β-carotene to carlactone, a strigolactone-like plant hormone. Science. 335:1348–1351.10.1126/science.1218094
- Aloui A, Recorbet G, Robert F, Schoefs B, Bertrand M, Henry C, Gianinazzi-Pearson V, Dumas-Gaudot E, Aschi-Smiti S. 2011. Arbuscular mycorrhizal symbiosis elicits shoot proteome changes that are modified during cadmium stress alleviation in Medicago truncatula. BMC Plant Biol. 11:75.
- Ameres SL, Zamore PD. 2013. Diversifying microRNA sequence and function. Nat Rev Mol Cell Biol. 14:475–488.10.1038/nrm3611
- Amiour N, Recorbet G, Robert F, Gianinazzi S, Dumas-Gaudot E. 2006. Mutations in DMI3 and SUNN modify the appressorium-responsive root proteome in arbuscular mycorrhiza. Plant Cell. 19:988–997.
- Andrade SL, Dickmanns A, Ficner R, Einsle O. 2005. Crystal structure of the archaeal ammonium transporter Amt-1 from Archaeoglobus fulgidus. Proc Natl Acad Sci USA. 102:14994–14999.
- Ané J-M, Kiss GB, Riely BK, Penmetsa RV, Oldroyd GED, Ayax C, Lévy J, Debellé F, Baek J-M, Kalo P, et al. 2004. Medicago truncatula DMI1 required for bacterial and fungal symbioses in legumes. Science. 303:1364–1367.
- Aono T, Maldonado-Mendoza IE, Dewbre GR, Harrison MJ, Saito M. 2004. Expression of alkaline phosphatase genes in arbuscular mycorrhizas. New Phytol. 162:525–534.10.1111/j.1469-8137.2004.01041.x
- Balestrini R, Bonfante P. 2014. Cell wall remodeling in mycorrhizal symbiosis: a way towards biotrophism. Front Plant Sci. 5:237.
- Balestrini R, Gómez-Ariza J, Lanfranco L, Bonfante P. 2007. Laser microdissection reveals that transcripts for five plant and one fungal phosphate transporter genes are contemporaneously present in arbusculated cells. Mol Plant Microbe Interact. 20:1055–1062.10.1094/MPMI-20-9-1055
- Balestrini R, Lanfranco L. 2006. Fungal and plant gene expression in arbuscular mycorrhizal symbiosis. Mycorrhiza. 16:509–524.10.1007/s00572-006-0069-2
- Balestrini R, Jose-Estanyol M, Puigdomenech P, Bonfante P. 1997. Hydroxyproline-rich glycoprotein mRNA accumulation in maize root cells colonized by an arbuscular mycorrhizal fungus as revealed by in situ hybridization. Protoplasma. 198:36–42.
- Balzergue C, Chabaud M, Barker DG, Bécard G, Rochange SF. 2013. High phosphate reduces host ability to develop arbuscular mycorrhizal symbiosis without affecting root calcium spiking responses to the fungus. Front Plant Sci. 4:426.
- Banba M, Gutjahr C, Miyao A, Hirochika H, Paszkowski U, Kouchi H, Imaizumi-Anraku H. 2008. Divergence of evolutionary ways among common sym genes: CASTOR and CCaMK show functional conservation between two symbiosis systems and constitute the root of a common signaling pathway. Plant Cell Physiol. 49:1659–1671.10.1093/pcp/pcn153
- Barker SJ, Tagu D. 2000. The roles of auxins and cytokinins in mycorrhizal symbioses. J Plant Growth Regul. 19:144–154.
- Benedetto A, Magurno F, Bonfante P, Lanfranco L. 2005. Expression profiles of a phosphate transporter gene (GmosPT) from the endomycorrhizal fungus Glomus mosseae. Mycorrhiza. 15:620–627.10.1007/s00572-005-0006-9
- Besserer A, Puech-Pagès V, Kiefer P, Gomez-Roldan V, Jauneau A, Roy S, Portais J-C, Roux C, Bécard G, Séjalon-Delmas N. 2006. Strigolactones stimulate arbuscular mycorrhizal fungi by activating mitochondria. PLoS Biol. 4:e226.
- Bestel-Corre G, Dumas-Gaudot E, Poinsot V, Dieu M, Dierick J-F, Van TD, Remacle J, Gianinazzi-Pearson V, Gianinazzi S. 2002. Proteome analysis and identification of symbiosis-related proteins from Medicago truncatula Gaertn. by two-dimensional electrophoresis and mass spectrometry. Electrophoresis. 23:122–37.10.1002/1522-2683(200201)23:1<122::AID-ELPS122>3.0.CO;2-4
- Bever JD, Kang H, Kaonongbua W, Wang M. 2008. Genomic organization and mechanisms of inheritance in arbuscular mycorrhizal fungi : contrasting the evidence and implications of current theories. Mycorrhiza. 135–148.10.1007/978-3-540-78826-3_7
- Bhattacharya C, Bonfante P, Deagostino A, Kapulnik Y, Larini P, Occhiato EG, Prandi C, Venturello P. 2009. A new class of conjugated strigolactone analogues with fluorescent properties: synthesis and biological activity. Org Biomol Chem. 7:3413–3420.
- Blilou I., Ocampo JA, Garcia-Garrido JM. 1999. Resistance of pea roots to endomycorrhizal fungus or Rhizobium correlates with enhanced levels of endogenous salicylic acid. J Exp Bot. 50:1663–1668.10.1093/jxb/50.340.1663
- Bona E, Cattaneo C, Cesaro P, Marsano F, Lingua G, Cavaletto M, Berta G. 2010. Proteomic analysis of Pteris vittata fronds: two arbuscular mycorrhizal fungi differentially modulate protein expression under arsenic contamination. Proteomics. 10:3811–3834.10.1002/pmic.200900436
- Bonfante P, Genre A. 2008. Plants and arbuscular mycorrhizal fungi: an evolutionary-developmental perspective. Trends Plant Sci. 13:492–498.
- Bonfante P, Requena N. 2011. Dating in the dark: how roots respond to fungal signals to establish arbuscular mycorrhizal symbiosis. Curr Opin Plant Biol. 14:451–457.10.1016/j.pbi.2011.03.014
- Bouwmeester HJ, Roux C, Lopez-Raez JA, Bécard G. 2007. Rhizosphere communication of plants, parasitic plants and AM fungi. Trends Plant Sci. 12:224–230.
- Branscheid A, Devers EA, May P, Krajinski F. 2011. Distribution pattern of small RNA and degradome reads provides information on miRNA gene structure and regulation. Plant Signal Behav. 6:1609–1611.10.4161/psb.6.10.17305
- Branscheid A, Sieh D, Pant BD, May P, Devers EA, Elkrog A, Schauser L, Scheible W-R, Krajinski F. 2010. Expression pattern suggests a role of MiR399 in the regulation of the cellular response to local Pi increase during arbuscular mycorrhizal symbiosis. Mol Plant Microbe Interact. 23:915–926.10.1094/MPMI-23-7-0915
- Brechenmacher L, Weidmann S, van Tuinen D, Chatagnier O, Gianinazzi S, Franken P, Gianinazzi-Pearson V. 2004. Expression profiling of up-regulated plant and fungal genes in early and late stages of Medicago truncatula-Glomus mosseae interactions. Mycorrhiza. 14:253–262.10.1007/s00572-003-0263-4
- Broghammer A, Krusell L, Blaise M, Sauer J, Sullivan JT, Maolanon N, Vinther M, Lorentzen A, Madsen EB, Jensen KJ, et al. 2012. Legume receptors perceive the rhizobial lipochitin oligosaccharide signal molecules by direct binding. Proc Natl Acad Sci USA. 109:13859–13864.10.1073/pnas.1205171109
- Bucher M, Hause B, Krajinski F, Küster H. 2014. Through the doors of perception to function in arbuscular mycorrhizal symbioses. New Phytol. 204:833–840.10.1111/nph.12862
- Büttner M, Sauer N. 2000. Monosaccharide transporters in plants: structure, function and physiology. Biochim Biophys Acta. 1465:263–274.
- Campos-Soriano L, Gómez-Ariza J, Bonfante P, San Segundo B. 2011. A rice calcium-dependent protein kinase is expressed in cortical root cells during the presymbiotic phase of the arbuscular mycorrhizal symbiosis. BMC Plant Biol. 11:90.
- Cangahuala-Inocente GC, Da Silva MF, Johnson J-M, Manga A, van Tuinen D, Henry C, Lovato PE, Dumas-Gaudot E. 2011. Arbuscular mycorrhizal symbiosis elicits proteome responses opposite of P-starvation in SO4 grapevine rootstock upon root colonisation with two Glomus species. Mycorrhiza. 21:473–493.10.1007/s00572-010-0352-0
- Capoen W, Sun J, Wysham D, Otegui MS, Venkateshwaran M, Hirsch S, Miwa H, Downie JA, Morris RJ, Ané J-M, Oldroyd GED. 2011. Nuclear membranes control symbiotic calcium signaling of legumes. Proc Natl Acad Sci USA. 108:14348–14353.10.1073/pnas.1107912108
- Cathy A, Guerts R, Bisseling T. 1999. Legume nodulation and mycorrhizae formation ; two extremes in host specificity meet. EMBO J. 18:281–288.10.1093/emboj/18.2.281
- Catoira R, Galera C, de Billy F, Penmetsa RV, Journet EP, Maillet F, Rosenberg C, Cook D, Gough C, Dénarié J. 2000. Four genes of Medicago truncatula controlling components of a nod factor transduction pathway. Plant Cell. 12:1647–1666.10.1105/tpc.12.9.1647
- Chabaud M, Genre A, Sieberer BJ, Faccio A, Fournier J, Novero M, Barker DG, Bonfante P. 2011. Arbuscular mycorrhizal hyphopodia and germinated spore exudates trigger Ca2+ spiking in the legume and nonlegume root epidermis. New Phytol. 189:347–355.10.1111/j.1469-8137.2010.03464.x
- Charpentier M, Bredemeier R, Wanner G, Takeda N, Schleiff E, Parniske M. 2008. Lotus japonicus CASTOR and POLLUX are ion channels essential for perinuclear calcium spiking in legume root endosymbiosis. Plant Cell. 20:3467–3479.10.1105/tpc.108.063255
- Chen H-M, Chen L-T, Patel K, Li Y-H, Baulcombe DC, Wu S-H. 2010. 22-Nucleotide RNAs trigger secondary siRNA biogenesis in plants. Proc Natl Acad Sci USA. 107:15269–15274.
- Colard A, Angelard C, Sanders IR. 2011. Genetic exchange in an arbuscular mycorrhizal fungus results in increased rice growth and altered mycorrhiza-specific gene transcription. Appl Environ Microbiol. 77:6510–6515.
- Combier JP, de Billy F, Gamas P, Niebel A, Rivas S. 2008. Trans-regulation of the expression of the transcription factor MtHAP2-1 by a uORF controls root nodule development. Genes Dev. 22:1549–1559.10.1101/gad.461808
- Coppo M, Damiani F, Drocco M, Grassi E, Guether M, Troina A. 2011. Modelling ammonium transporters in arbuscular mycorrhiza symbiosis. Trans Comput Syst Biol XIII. 6575:85–109.10.1007/978-3-642-19748-2_5
- Czarnecki O, Yang J, Weston DJ, Tuskan GA, Chen J-G. 2013. A dual role of strigolactones in phosphate acquisition and utilization in plants. Int J Mol Sci. 14:7681–7701.10.3390/ijms14047681
- Dahiya P, Kardailsky IV, Brewin NJ. 1997. Immunolocalization of PsNLEC-1, a lectin-like glycoprotein expressed in developing pea nodules. Plant Physiol. 115:1431–1442.
- Delaux P, Guillaume B. 2013. Rapid report NSP1 is a component of the Myc signaling pathway. New Phytol. 11:59–65.
- Delaux P-M, Séjalon-Delmas N, Bécard G, Ané J-M. 2013. Evolution of the plant–microbe symbiotic ‘toolkit’. Trends Plant Sci. 18:298–304.10.1016/j.tplants.2013.01.008
- Devers EA, Branscheid A, May P, Krajinski F. 2011. Stars and symbiosis: microRNA- and microRNA*-mediated transcript cleavage involved in arbuscular mycorrhizal symbiosis. Plant Physiol. 156:1990–2010.10.1104/pp.111.172627
- Devers EA, Teply J, Reinert A, Gaude N, Krajinski F. 2013. An endogenous artificial microRNA system for unraveling the function of root endosymbioses related genes in Medicago truncatula. BMC Plant Biol. 13:82.
- Djuranovic S, Nahvi A, Green R. 2012. miRNA-mediated gene silencing by translational repression followed by mRNA deadenylation and decay. Science. 336:237–240.10.1126/science.1215691
- Domagalska MA, Leyser O. 2011. Signal integration in the control of shoot branching. Nat Rev Mol Cell Biol. 12:211–221.10.1038/nrm3088
- Dumas-Gaudot E, Valot B, Bestel-Corre G, Recorbet G, St-Arnaud M, Fontaine B, Dieu M, Raes M, Saravanan RS, Gianinazzi S. 2004. Proteomics as a way to identify extra-radicular fungal proteins from Glomus intraradices – RiT-DNA carrot root mycorrhizas. FEMS Microbiol Ecol. 48:401–411.10.1016/j.femsec.2004.02.015
- Dun EA, Brewer PB, Beveridge CA. 2009. Strigolactones: discovery of the elusive shoot branching hormone. Trends Plant Sci. 14:364–372.
- Ellerbeck M, Schüßler A, Brucker D, Dafinger C, Loos F, Brachmann A. 2013. Characterization of three ammonium transporters of the glomeromycotan fungus Geosiphon pyriformis. Eukaryot Cell. 12:1554–1562.10.1128/EC.00139-13
- Ferrol N, Benabdellah K. 2000. Alterations in the plasma membrane polypeptide pattern of tomato roots (Lycopersicon esculentum) during the development of arbuscular mycorrhiza. J Exp Bot. 51:747–754.10.1093/jexbot/51.345.747
- Fester T, Kiess M, Strack D. 2002. A mycorrhiza-responsive protein in wheat roots. Mycorrhiza. 12:219–222.
- Floss DS, Levy JG, Lévesque-Tremblay V, Pumplin N, Harrison MJ. 2013. DELLA proteins regulate arbuscule formation in arbuscular mycorrhizal symbiosis. Proc Natl Acad Sci USA. 110:E5025–E5034.10.1073/pnas.1308973110
- Foo E. 2013. Auxin influences strigolactones in pea mycorrhizal symbiosis. J Plant Physiol. 170:523–528.
- Foo E, Bullier E, Goussot M, Foucher F, Rameau C. 2005. The branching gene RAMOSUS1 mediates interactions among two novel signals and auxin in Pea. Plant Cell. 17:464–474.10.1105/tpc.104.026716
- Foo E, Ross JJ, Jones WT, Reid JB. 2013. Plant hormones in arbuscular mycorrhizal symbioses: an emerging role for gibberellins. Ann Bot. 111:769–779.
- Foo E, Yoneyama K, Hugill CJ, Quittenden LJ, Reid JB. 2013. Strigolactones and the regulation of pea symbioses in response to nitrate and phosphate deficiency. Mol Plant. 6:76–87.10.1093/mp/sss115
- Franzini VI, Azcón R, Méndes FL, Aroca R. 2013. Different interaction among Glomus and Rhizobium species on Phaseolus vulgaris and Zea mays plant growth, physiology and symbiotic development under moderate drought stress conditions. Plant Growth Regul. 70:265–273.10.1007/s10725-013-9798-3
- Frühling M, Roussel H, Gianinazzi P, Pühler A, Perlick A. 1997. The Vicia faba leghemoglobin gene VfLb29 is induced in root nodules and in roots colonized by the arbuscular mycorrhizal fungus Glomus fasciculatum. Mol Plant Microbe Interact. 10:124–131.10.1094/MPMI.1997.10.1.124
- Gallou A, Declerck S, Cranenbrouck S. 2012. Transcriptional regulation of defence genes and involvement of the WRKY transcription factor in arbuscular mycorrhizal potato root colonization. Funct Integr Genomics. 12:183–198.10.1007/s10142-011-0241-4
- Garrido JMG, Morcillo RJL, Rodríguez JAM, Bote JAO. 2010. Variations in the mycorrhization characteristics in roots of wild-type and ABA-deficient tomato are accompanied by specific transcriptomic alterations. Mol Plant Microbe Interact. 23:651–64.
- Genre A, Bonfante P. 2010. The making of symbiotic cells in arbuscular mycorrhizal roots. Arbuscular mycorrhiza: physiology and function. In: Koltai H, Kapulnik Y, editors. Springer; p. 57–71.
- Genre A, Chabaud M, Balzergue C, Puech-Pagès V, Novero M, Rey T, Fournier J, Rochange S, Bécard G, Bonfante P, Barker DG. 2013. Short-chain chitin oligomers from arbuscular mycorrhizal fungi trigger nuclear Ca2+ spiking in Medicago truncatula roots and their production is enhanced by strigolactone. New Phytol. 198:190–202.10.1111/nph.12146
- Genre A, Chabaud M, Timmers T, Bonfante P, Barker DG. 2005. Arbuscular mycorrhizal fungi elicit a novel intracellular apparatus in Medicago truncatula root epidermal cells before infection. Plant Cell. 17:3489–3499.10.1105/tpc.105.035410
- Geurts R, Vleeshouwers VGAA. 2012. Mycorrhizal symbiosis: ancient signalling mechanisms co-opted. Curr Biol. 22:R997–R999.10.1016/j.cub.2012.10.021
- Gianinazzi-Pearson V. 1996. Plant cell responses to arbuscular mycorrhizal fungi: getting to the roots of the symbiosis. Plant Cell. 8:1871–1883.10.1105/tpc.8.10.1871
- Gobbato E, Marsh JF, Vernié T, Wang E, Maillet F, Kim J, Miller JB, Sun J, Bano SA, Ratet P, et al. 2012. A GRAS-type transcription factor with a specific function in mycorrhizal signaling. Curr Biol. 22:2236–2241.10.1016/j.cub.2012.09.044
- Gomez SK, Javot H, Deewatthanawong P, Torres-Jerez I, Tang Y, Blancaflor EB, Udvardi MK, Harrison MJ. 2009. Medicago truncatula and Glomus intraradices gene expression in cortical cells harboring arbuscules in the arbuscular mycorrhizal symbiosis. BMC Plant Biol. 9:10.
- Gomez-Roldan V, Fermas S, Brewer PB, Puech-Pages V, Dun EA, Pillot J-P, Letisse F, Matusova R, Danoun S, Portais J-C. 2008. Strigolactone inhibition of shoot branching. Nature. 455:189–194.10.1038/nature07271
- Govindarajulu M, Pfeffer PE, Jin H, Abubaker J, Douds DD, Allen JW, Bucking H, Lammers PJ, Shachar-Hill Y. 2005. Nitrogen transfer in the arbuscular mycorrhizal symbiosis. Nature. 435:819–823.10.1038/nature03610
- Groth M, Takeda N, Perry J, Uchida H, Dräxl S, Brachmann A, Sato S, Tabata S, Kawaguchi M, Wang TL, Parniske M. 2010. NENA, a Lotus japonicus homolog of Sec13, is required for rhizodermal infection by arbuscular mycorrhiza fungi and rhizobia but dispensable for cortical endosymbiotic development. Plant Cell. 22:2509–2526.10.1105/tpc.109.069807
- Grunwald U, Nyamsuren O, Tamasloukht MB, Lapopin L, Becker A, Mann P, Gianinazzi-Pearson V, Krajinski F, Franken P.. 2004. Identification of mycorrhiza-regulated genes with arbuscule development-related expression profile. Plant Mol Biol. 55:553–566.
- Guether M, Neuhäuser B, Balestrini R, Dynowski M, Ludewig U, Bonfante P. 2009. A mycorrhizal-specific ammonium transporter from Lotus japonicus acquires nitrogen released by arbuscular mycorrhizal fungi. Plant Physiol. 150:73–83.10.1104/pp.109.136390
- Güimil S, Chang H-S, Zhu T, Sesma A, Osbourn A, Roux C, Ioannidis V, Oakeley EJ, Docquier M, Descombes P. 2005. Comparative transcriptomics of rice reveals an ancient pattern of response to microbial colonization. Proc Natl Acad Sci USA. 102:8066–8070.
- Gutjahr C, Banba M, Croset V, An K, Miyao A, An G, Hirochika H, Imaizumi-Anraku H, Paszkowski U. 2008. Arbuscular mycorrhiza-specific signaling in rice transcends the common symbiosis signaling pathway. Plant Cell. 20:2989–3005.10.1105/tpc.108.062414
- Gutjahr C, Parniske M. 2013. Cell and developmental biology of arbuscular mycorrhiza symbiosis. Annu Rev Cell Dev Biol. 29:593–617.
- Gutjahr C, Paszkowski U. 2013. Multiple control levels of root system remodeling in arbuscular mycorrhizal symbiosis. Front Plant Sci. 4:204.
- Gutjahr C, Radovanovic D, Geoffroy J, Zhang Q, Siegler H, Chiapello M, Casieri L, An K, An G, Guiderdoni E. 2012. The half-size ABC transporters STR1 and STR2 are indispensable for mycorrhizal arbuscule formation in rice. Plant J. 69:906–920.
- Gutjahr C. 2014. Phytohormone signaling in arbuscular mycorhiza development. Curr Opin Plant Biol. 20:26–34.10.1016/j.pbi.2014.04.003
- Hanlon MT, Coenen C. 2011. Genetic evidence for auxin involvement in arbuscular mycorrhiza initiation. New Phytol. 189:701–709.10.1111/j.1469-8137.2010.03567.x
- Harrier L. 2001. The arbuscular mycorrhizal symbiosis: a molecular review of the fungal dimension. J Exp Bot. 52:469–478.10.1093/jexbot/52.suppl_1.469
- Harrison M. 1996. A sugar transporter from Medicago truncatula: altered expression pattern in roots during vesicular-arbuscular (VA) mycorrhizal associations. Plant J. 9:491–503.10.1046/j.1365-313X.1996.09040491.x
- Harrison MJ, van Buuren ML. 1995. A phosphate transporter from the mycorrhizal fungus Glomus versiforme. Nature. 378:626–629.
- Harrison MJ, Dewbre GR, Liu J, Samuel T, Noble R, Parkway SN. 2002. A phosphate transporter from Medicago truncatula involved in the acquisition of phosphate released by arbuscular mycorrhizal fungi. Plant Cell. 14:2413–2429.
- Hause B, Maier W, Miersch O, Kramell R, Strack D. 2002. Induction of jasmonate biosynthesis in arbuscular mycorrhizal barley roots. Plant Physiol. 130:1213–1220.10.1104/pp.006007
- Hayashi S, Gresshoff PM, Ferguson BJ. 2014. Mechanistic action of gibberellins in legume nodulation. J Integr Plant Biol. 56:971–978.
- Hayashi T, Shimoda Y, Sato S, Tabata S, Imaizumi-Anraku H, Hayashi M. 2014. Rhizobial infection does not require cortical expression of upstream common symbiosis genes responsible for the induction of Ca2+ spiking. Plant J. 77:146–159.10.1111/tpj.12374
- Heidstra R, Nilsen G, Martinez-Abarca F, van Kammen A, Bisseling T. 1997. Nod factor-induced expression of leghemoglobin to study the mechanism of NH4NO3 inhibition on root hair deformation. Mol Plant Microbe Interact. 10:215–220.
- Helber N, Wippel K, Sauer N, Schaarschmidt S, Hause B, Requena N. 2011. A versatile monosaccharide transporter that operates in the arbuscular mycorrhizal fungus Glomus sp is crucial for the symbiotic relationship with plants. Plant Cell. 23:3812–3823.
- Herrbach V, Remblière C, Gough C, Bensmihen S. 2014. Lateral root formation and patterning in Medicago truncatula. J Plant Physiol. 171:301–310.10.1016/j.jplph.2013.09.006
- Herre E, Knowlton N, Mueller U, Rehner S. 1999. The evolution of mutualisms: exploring the paths between conflict and cooperation. Trends Ecol Evol. 14:49–53.
- Herrera Medina M, Gagnon H, Piché Y, Ocampo JA, García Garrido JM, Vierheilig H. 2003. Root colonization by arbuscular mycorrhizal fungi is affected by the salicylic acid content of the plant. Plant Sci. 164:993–998.
- Hodge A, Campbell CD, Fitter AH. 2001. An arbuscular mycorrhizal fungus accelerates decomposition and acquires nitrogen directly from organic material. Nature. 413:297–299.10.1038/35095041
- Horváth B, Yeun LH, Domonkos A, Halász G, Gobbato E, Ayaydin F, Miró K, Hirsch S, Sun J, Tadege M. 2011. Medicago truncatula IPD3 is a member of the common symbiotic signaling pathway required for rhizobial and mycorrhizal symbioses. Mol Plant Microbe Interact. 24:1345–1358.
- Humphreys CP, Franks PJ, Rees M, Bidartondo MI, Leake JR, Beerling DJ. 2010. Mutualistic mycorrhiza-like symbiosis in the most ancient group of land plants. Nat Commun. 1:103.
- Isayenkov S, Mrosk C, Stenzel I, Strack D, Hause B. 2005. Suppression of allene oxide cyclase in hairy roots of Medicago truncatula reduces jasmonate levels and the degree of mycorrhization with Glomus intraradices. Plant Physiol. 139:1401–1410.
- Ivanov S, Fedorova EE, Limpens E, De Mita S, Genre A, Bonfante P, Bisseling T. 2012. Rhizobium-legume symbiosis shares an exocytotic pathway required for arbuscule formation. Proc Natl Acad Sci USA. 109:8316–8321.10.1073/pnas.1200407109
- Javot H, Penmetsa RV, Terzaghi N, Cook DR, Harrison MJ. 2007. A Medicago truncatula phosphate transporter indispensable for the arbuscular mycorrhizal symbiosis. Proc Natl Acad Sci USA. 104:1720–1725.
- Jin H, Liu J, Liu J, Huang X. 2012. Forms of nitrogen uptake, translocation, and transfer via arbuscular mycorrhizal fungi: a review. Sci China. 55:474–482.
- Johnson X, Brcich T, Dun EA, Goussot M, Haurogné K, Beveridge CA, Rameau C. 2006. Branching genes are conserved across species. Genes controlling a novel signal in pea are coregulated by other long-distance signals. Plant Physiol. 142:1014–1026.
- Kagiyama M, Hirano Y, Mori T, Kim S-Y, Kyozuka J, Seto Y, Yamaguchi S, Hakoshima T. 2013. Structures of D14 and D14L in the strigolactone and karrikin signaling pathways. Genes cells. 18:147–160.10.1111/gtc.12025
- Kaldorf M, Ludwig-Müller J. 2000. AM fungi might affect the root morphology of maize by increasing indole-3-butyric acid biosynthesis. Physiol Plant. 109:58–67.
- Kanamori N, Madsen LH, Radutoiu S, Frantescu M, Quistgaard EMH, Miwa H, Downie JA, James EK, Felle HH, Haaning LL, et al. 2006. A nucleoporin is required for induction of Ca2+ spiking in legume nodule development and essential for rhizobial and fungal symbiosis. Proc Natl Acad Sci USA. 103:359–364.10.1073/pnas.0508883103
- Kanchiswamy CN, Mohanta TK, Capuzzo A, Occhipinti A, Verrillo F, Maffei ME, Malnoy M. 2013. Differential expression of CPKs and cytosolic Ca2+ variation in resistant and susceptible apple cultivars (Malus × domestica) in response to the pathogen Erwinia amylovora and mechanical wounding. BMC Genomics. 14:760.
- Kaneko T, Tabata S, Sandal N, Stougaard J, Webb KJ, Szczyglowski K, Parniske M. 2005. Seven Lotus japonicus genes required for transcriptional reprogramming of the root during fungal and bacterial symbiosis. Plant Cell. 17:2217–2229.10.1105/tpc.105.032714
- Karandashov V, Bucher M. 2005. Symbiotic phosphate transport in arbuscular mycorrhizas. Trends Plant Sci. 10:22–29.
- Kardailsky V, Sherrier OJ, Brewin NJ. 1994. Identification of a new pea gene, PsNlecl, encoding a lectin-like clycoprotein isolated from the symbiosomes of root nodules. Plant Physiol. 115:49–60.
- Kim W, Ahn HJ, Chiou T-J, Ahn JH. 2011. The role of the miR399-PHO2 module in the regulation of flowering time in response to different ambient temperatures in Arabidopsis thaliana. Mol Cells. 32:83–88.
- Koegel S, Ait Lahmidi N, Arnould C, Chatagnier O, Walder F, Ineichen K, Boller T, Wipf D, Wiemken A, Courty P-E. 2013. The family of ammonium transporters (AMT) in Sorghum bicolor: two AMT members are induced locally, but not systemically in roots colonized by arbuscular mycorrhizal fungi. New Phytol. 198:853–865.10.1111/nph.12199
- Kosuta S, Chabaud M, Gough C, De J, Barker DG. 2003. A diffusible factor from arbuscular mycorrhizal fungi induces symbiosis-specific MtENOD11 expression in roots of Medicago truncatula. Plant Physiol. 131:952–962.
- Krajinski F, Courty P-E, Sieh D, Franken P, Zhang H, Bucher M, Gerlach N, Kryvoruchko I, Zoeller D, Udvardi M, Hause B. 2014. The H+-ATPase HA1 of Medicago truncatula is essential for phosphate transport and plant growth during arbuscular mycorrhizal symbiosis. Plant Cell. 26:1808–1817.
- Kranabetter JM. 2014. Ectomycorrhizal fungi and the nitrogen economy of conifers—implications for genecology and climate change mitigation. Botany. 92:417–423.10.1139/cjb-2013-0198
- Kretzschmar T, Kohlen W, Sasse J, Borghi L, Schlegel M, Bachelier JB, Reinhardt D, Bours R, Bouwmeester HJ, Martinoia E. 2012. A petunia ABC protein controls strigolactone-dependent symbiotic signalling and branching. Nature. 483:341–344.
- Lamoureux G, Javelle A, Baday S, Wang S, Bernèche S. 2010. Transport mechanisms in the ammonium transporter family. Transfus Clin Biol. 17:168–175.
- Landgraf R, Schaarschmidt S, Hause B. 2012. Repeated leaf wounding alters the colonization of Medicago truncatula roots by beneficial and pathogenic microorganisms. Plant Cell Environ. 35:1344–1357.10.1111/j.1365-3040.2012.02495.x
- Lauressergues D, Delaux P-M, Formey D, Lelandais-Brière C, Fort S, Cottaz S, Bécard G, Niebel A, Roux C, Combier J-P. 2012. The microRNA miR171h modulates arbuscular mycorrhizal colonization of Medicago truncatula by targeting NSP2. Plant J. 72:512–522.
- Lefebvre V, North H, Frey A, Sotta B, Seo M, Okamoto M, Nambara E, Marion-Poll A. 2006. Functional analysis of Arabidopsis NCED6 and NCED9 genes indicates that ABA synthesized in the endosperm is involved in the induction of seed dormancy. Plant J. 45:309–319.10.1111/j.1365-313X.2005.02622.x
- Li L, Li C, Lee GI, Howe GA. 2002. Distinct roles for jasmonate synthesis and action in the systemic wound response of tomato. Proc Natl Acad Sci USA. 99:6416–6421.
- Lingua G, Bona E, Todeschini V, Cattaneo C, Marsano F, Berta G, Cavaletto M. 2012. Effects of heavy metals and arbuscular mycorrhiza on the leaf proteome of a selected poplar clone: a time course analysis. PLoS One. 7:e38662.
- Liu W, Kohlen W, Lillo A, Op den Camp R, Ivanov S, Hartog M, Limpens E, Jamil M, Smaczniak C, Kaufmann K. 2011. Strigolactone biosynthesis in Medicago truncatula and rice requires the symbiotic GRAS-type transcription factors NSP1 and NSP2. Plant Cell. 23:3853–65.
- Liu J, Blaylock LA, Endre G, Cho J, Town CD, Vanden Bosch KA, Harrison MJ. 2003. Transcript profiling coupled with spatial expression analyses reveals genes involved in distinct developmental stages of the arbuscular mycorrhizal symbiosis. Plant Cell. 15:2106–2123.
- López-Ráez J, Kohlen W, Charnikhova T, Mulder P, Undas AK, Sergeant MJ, Verstappen F, Bugg TDH, Thompson AJ, Ruyter-Spira C, Bouwmeester H. 2010. Does abscisic acid affect strigolactone biosynthesis? New Phytol. 187:343–354.
- López-ráez JA, Bouwmeester H. 2008. Fine-tuning regulation of strigolactone biosynthesis under phosphate starvation. Plant Signal Behav. 3:963–965.
- Loth-Pereda V, Orsini E, Courty P-E, Lota F, Kohler A, Diss L, Blaudez D, Chalot M, Nehls U, Bucher M, Martin F. 2011. Structure and expression profile of the phosphate Pht1 transporter gene family in mycorrhizal Populus trichocarpa. Plant Physiol. 156:2141–2154.
- Ludwig-Müller J, Güther M. 2007. Auxins as signals in arbuscular mycorrhiza formation. Plant Signal Behav. 2:194–196.
- Ludwig-Müller J, Kaldorf M, Sutter EG, Epstein E. 1997. Indole-3-butyric acid (IBA) is enhanced in young maize (Zea mays L.) roots colonized with the arbuscular mycorrhizal fungus Glomus intraradices. Plant Sci. 125:153–162.10.1016/S0168-9452(97)00064-2
- Maillet F, Poinsot V, Andre O, Puech-Pages V, Haouy A, Gueunier M, Cromer L, Giraudet D, Formey D, Niebel A. 2011. Fungal lipochitooligosaccharide symbiotic signals in arbuscular mycorrhiza. Nature. 469:58–63.
- Martin HC, Wani S, Steptoe AL, Krishnan K, Nones K, Nourbakhsh E, Vlassov A, Grimmond SM, Cloonan N. 2014. Imperfect centered miRNA binding sites are common and can mediate repression of target mRNAs. Genome Biol. 15:R51.
- Matsumura A, Taniguchi S, Yamawaki K, Hattori R, Tarui A. 2013. Nitrogen uptake from amino acids in maize through arbuscular mycorrhizal symbiosis. Am J Plant Sci. 4:2290–2294.
- Matusova R, Rani K, Verstappen FWA, Franssen MCR, Beale MH, Bouwmeester HJ. 2005. The strigolactone germination stimulants of the plant-Parasitic Striga and Orobanche spp. Are Derived from the Carotenoid Pathway. Plant Physiol. 139:920–934.
- Meixner C, Ludwig-Müller J, Miersch O, Gresshoff P, Staehelin C, Vierheilig H. 2005. Lack of mycorrhizal autoregulation and phytohormonal changes in the supernodulating soybean mutant nts1007. Planta. 222:709–715.10.1007/s00425-005-0003-4
- Messinese E, Mun J-H, Yeun LH, Jayaraman D, Rougé P, Barre A, Lougnon G, Schornack S, Bono J-J, Cook DR, Ané J-M. 2007. A novel nuclear protein interacts with the symbiotic DMI3 calcium- and calmodulin-dependent protein kinase of Medicago truncatula. Mol Plant Microbe Interact. 20:912–921.
- Mica E, Piccolo V, Delledonne M, Ferrarini A, Pezzotti M, Casati C, Del Fabbro C, Valle G, Policriti A, Morgante M, et al. 2009. High throughput approaches reveal splicing of primary microRNA transcripts and tissue specific expression of mature microRNAs in Vitis vinifera. BMC Genomics. 10:558.10.1186/1471-2164-10-558
- Miller JB, Pratap A, Miyahara A, Zhou L, Bornemann S, Morris RJ, Oldroyd GED. 2013. Calcium/Calmodulin-dependent protein kinase is negatively and positively regulated by calcium, providing a mechanism for decoding calcium responses during symbiosis signaling. Plant Cell. 25:5053–5066.
- Miransari M, Abrishamchi A, Khoshbakht K, Niknam V. 2014. Plant hormones as signals in arbuscular mycorrhizal symbiosis. Crit Rev Biotechnol. 34:123–133.
- Miransari M. 2010. Contribution of arbuscular mycorrhizal symbiosis to plant growth under different types of soil stress. Plant Biol. 12:563–569.
- Nagy R, Karandashov V, Chague V, Kalinkevich K, Tamasloukht M, Xu G, Jakobsen I, Levy AA, Amrhein N, Bucher M. 2005. The characterization of novel mycorrhiza-specific phosphate transporters from Lycopersicon esculentum and Solanum tuberosum uncovers functional redundancy in symbiotic phosphate transport in solanaceous species. Plant J. 42:236–250.
- Nakamura H, Xue Y-L, Miyakawa T, Hou F, Qin H-M, Fukui K, Shi X, Ito E, Ito S, Park S-H. 2013. Molecular mechanism of strigolactone perception by DWARF14. Nat. Commun. 4:2613.10.1038/ncomms3613
- Navazio L, Moscatiello R, Genre A, Novero M, Baldan B, Bonfante P, Mariani P. 2007. A diffusible signal from arbuscular mycorrhizal fungi elicits a transient cytosolic calcium elevation in host plant cells. Plant Physiol. 144:673–681.
- Oehl F, Sieverding E, Palenzuela J, Ineichen K, Alves da Silva G. 2011. Advances in Glomeromycota taxonomy and classification. IMA Fungus. 2:191–199.
- Ortu G, Balestrini R, Pereira PA, Becker JD, Küster H, Bonfante P. 2012. Plant genes related to gibberellin biosynthesis and signaling are differentially regulated during the early stages of AM fungal interactions. Mol Plant. 5:951–954.
- Ossowski S, Schwab R, Weigel D. 2008. Gene silencing in plants using artificial microRNAs and other small RNAs. Plant J. 53:674–690.10.1111/j.1365-313X.2007.03328.x
- Paszkowski U, Kroken S, Roux C, Briggs SP. 2002. Rice phosphate transporters include an evolutionarily divergent gene specifically activated in arbuscular mycorrhizal symbiosis. Proc Natl Acad Sci USA. 99:13324–13329.
- Pérez-Tienda J, Testillano PS, Balestrini R, Fiorilli V, Azcón-Aguilar C, Ferrol N. 2011. GintAMT2, a new member of the ammonium transporter family in the arbuscular mycorrhizal fungus Glomus intraradices. Fungal Genet Biol. 48:1044–1055.
- Perotto S, Rodda M, Benetti A, Sillo F, Ercole E, Rodda M, Girlanda M, Murat C, Balestrini R. 2014. Gene expression in mycorrhizal orchid protocorms suggests a friendly plant-fungus relationship. Planta. 239:1337–1349.
- Prandi C, Occhiato EG, Tabasso S, Bonfante P, Novero M, Scarpi D, Bova ME, Miletto I. 2011. New potent fluorescent analogues of strigolactones: synthesis and biological activity in parasitic weed germination and fungal branching. European J Org Chem. 2011:3781–3793.10.1002/ejoc.201100616
- Pumplin N, Harrison MJ. 2009. Live-cell imaging reveals periarbuscular membrane domains and organelle location in Medicago truncatula roots during arbuscular mycorrhizal symbiosis. Plant Physiol. 151:809–819.
- Pumplin N, Mondo SJ, Topp S, Starker CG, Gantt JS, Harrison MJ. 2010. Medicago truncatula Vapyrin is a novel protein required for arbuscular mycorrhizal symbiosis. Plant J. 61:482–494.
- Quain MD, Makgopa ME, Márquez-García B, Comadira G, Fernandez-Garcia N, Olmos E, Schnaubelt D, Kunert KJ, Foyer CH. 2014. Ectopic phytocystatin expression leads to enhanced drought stress tolerance in soybean (Glycine max) and Arabidopsis thaliana through effects on strigolactone pathways and can also result in improved seed traits. Plant Biotechnol J.12:903–913.
- Rath AC, Kang I-K, Park C-H, Yoo W-J, Byun J-K. 2006. Foliar application of aminoethoxyvinylglycine (AVG) delays fruit ripening and reduces pre-harvest fruit drop and ethylene production of bagged “Kogetsu” apples. Plant Growth Regul. 50:221–227.
- Ravnskov S, Wu Y, Graham JH. 2003. Arbuscular mycorrhizal fungi differentially affect expression of genes coding for sucrose synthases in maize roots. New Phytol. 157:539–545.10.1046/j.1469-8137.2003.00692.x
- Recorbet G, Valot B, Robert F, Gianinazzi-Pearson V, Dumas-Gaudot E. 2010. Identification of in planta-expressed arbuscular mycorrhizal fungal proteins upon comparison of the root proteomes of Medicago truncatula colonised with two Glomus species. Fungal Genet Biol.47:608–618.
- Regvar M, Gogala N, Zalar P. 1996. Effects of jasmonic acid on mycorrhizal Allium sativum. New Phytol. 134:703–707.
- Remy W, Taylor T, Hass H, Kerp H. 1994. Four hundred-million-year-old vesicular arbuscular mycorrhizae. Proc Natl Acad Sci USA. 91:11841–11843.10.1073/pnas.91.25.11841
- Repetto O, Bestel-corre G, Dumas-gaudot E, Berta G, Gianinazzi-pearson V, Gianinazzi S. 2003. Targeted proteomics to identify cadmium-induced protein modifications in Glomus mosseae-inoculated pea roots. New Phytol. 157:555–567.
- Requena N, Mann P, Franken P. 2000. A homologue of the cell cycle check point TOR2 from Saccharomyces cerevisiae exists in the arbuscular mycorrrhizal fungus Glomus mosseae. Protoplasma. 212:89–98.10.1007/BF01279350
- Requena N, Mann P, Hampp R, Franken P. 2002. Early developmentally regulated genes in the arbuscular mycorrhizal fungus Glomus mosseae : identification of GmGIN1, a novel gene with homology to the C-terminus of metazoan hedgehog proteins. Plant Soil. 244:129–139.
- Rhody D, Stommel M, Roeder C, Mann P, Franken P. 2003. Differential RNA accumulation of two β-tubulin genes in arbuscular mycorrhizal fungi. Mycorrhiza. 13:137–142.
- Rich MK, Schorderet M, Reinhardt D. 2014. The role of the cell wall compartment in mutualistic symbioses of plants. Front Plant Sci. 5:238.
- Riely BK, Lougnon G, Ané J-M, Cook DR. 2007. The symbiotic ion channel homolog DMI1 is localized in the nuclear membrane of Medicago truncatula roots. Plant J. 49:208–216.10.1111/j.1365-313X.2006.02957.x
- Roberts NJ, Morieri G, Kalsi G, Rose A, Stiller J, Edwards A, Xie F, Gresshoff PM, Oldroyd GED, Downie JA, Etzler ME. 2013. Rhizobial and mycorrhizal symbioses in Lotus japonicus require lectin nucleotide phosphohydrolase, which acts upstream of calcium signaling. Plant Physiol. 161:556–567.
- Saito K., Yoshikawa M., Yano K., Miwa H., Uchida H., Asamizu E., Sato S., Tabata S., Imaizumi-Anraku H., Umehara Y., et al. 2007. NUCLEOPORIN85 is required for calcium spiking, fungal and bacterial symbioses, and seed production in Lotus japonicus. Plant Cell. 19:610–624.10.1105/tpc.106.046938
- Sanders I, Croll D. 2010. Arbuscular mycorrhiza: the challenge to understand the genetics of the fungal partner. Annu Rev Genet. 44:271–292.10.1146/annurev-genet-102108-134239
- Scaffidi A, Waters M, Sun YK, Skelton BW, Dixon KW, Ghisalberti EL, Flematti G, Smith S. 2014. Strigolactone hormones and their stereoisomers signal through two related receptor proteins to induce different physiological responses in Arabidopsis. Plant Physiol. 165:1221–1232.
- Schuszler A, Martin H, Cohen D, Fitz M, Wipf D. 2006. Characterization of a carbohydrate transporter from symbiotic glomeromycotan fungi. Nature. 444:933–936.
- Selosse M, Le Tacon F. 1998. The land flora: a phototroph-fungus partnership? Trends Ecol Evol. 13:15–20.
- Seo M, Koshiba T. 2002. Complex regulation of ABA biosynthesis in plants. Trends Plant Sci. 7:41–48.
- Shaul-keinan O, Gadkar V, Ginzberg I, Grunzweig JM, Chet I, Elad Y, Wininger S, Belausov E, Eshed Y, Atzmon N, et al. 2002. Hormone concentrations in tobacco roots change during arbuscular mycorrhizal colonization with Glomus intraradices. New Phytol. 154:501–507.10.1046/j.1469-8137.2002.00388.x
- Siciliano V, Genre A, Balestrini R, Cappellazzo G, DeWit PJGM, Bonfante P. 2007. Transcriptome analysis of arbuscular mycorrhizal roots during development of the prepenetration apparatus. Plant Physiol. 144:1455–1466.
- Sieberer BJ, Chabaud M, Timmers AC, Monin A, Fournier J, Barker DG. 2009. A nuclear-targeted cameleon demonstrates intranuclear Ca2+ spiking in Medicago truncatula root hairs in response to rhizobial nodulation factors. Plant Physiol. 151:1197–1206.
- Singh S, Katzer K, Lambert J, Cerri M, Parniske M. 2014. CYCLOPS, A DNA-binding transcriptional activator, orchestrates symbiotic root nodule development. Cell Host Microbe. 15:139–152.
- Slewinski TL. 2011. Diverse functional roles of monosaccharide transporters and their homologs in vascular plants: a physiological perspective. Mol Plant. 4:641–662.10.1093/mp/ssr051
- Smith S, Smith F. 2011. Roles of arbuscular mycorrhizas in plant nutrition and growth: new paradigms from cellular to ecosystem scales. Annu Rev Plant Biol. 62:227–250.
- Smith SE, Smith FA. 2012. Fresh perspectives on the roles of arbuscular mycorrhizal fungi in plant nutrition and growth. Mycologia. 104:1–13.10.3852/11-229
- Stacey G, Bickley C, Alvin M, Kim S, Olivares J, Soto M. 2006. Effects of endogenous salicylic acid on nodulation in the model legumes Lotus japonicus. Plant Physiol. 141:1473–1481.
- Stracke S, Kistner C, Yoshida S, Mulder L, Sato S, Kaneko T, Tabata S, Sandal N, Stougaard J, Szczyglowski K, Parniske M. 2002. A plant receptor-like kinase required for both bacterial and fungal symbiosis. Nature. 417:959–962.
- Symons GM, Ross JJ, Murfet IC. 2002. The bushy pea mutant is IAA-deficient. Physiol Plant. 116:389–397.10.1034/j.1399-3054.2002.1160315.x
- Takeda N, Sato S, Asamizu E, Tabata S, Parniske M. 2009. Apoplastic plant subtilases support arbuscular mycorrhiza development in Lotus japonicus. Plant J. 58:766–777.
- Tamura Y, Kobae Y, Mizuno T, Hata S. 2014. Identification and expression analysis of arbuscular mycorrhiza-inducible phosphate transporter genes of soybean. Biosci Biotechnol Biochem. 76:309–313.10.1271/bbb.110684
- Tan BC, Schwartz SH, Zeevaart JA, McCarty DR. 1997. Genetic control of abscisic acid biosynthesis in maize. Proc Natl Acad Sci USA. 94:12235–12240.
- Tejeda-Sartorius M, Martínez-gallardo NA, Olalde-portugal V, Délano-frier JP, Fitopatología RM De, Olalde- V, Investigación C De, Estudios D, Irapuato U, Carr N. 2007. Jasmonic acid accelerates the expression of a pathogen-specific lipoxygenase (POTLX-3) and delays foliar late blight development in potato (Solanum tuberosum L.). Rev Mex Fitopatol. 25:18–25.
- Tisserant B, Gianinazzi-Pearson V, Gianinazzi S, Gollotte A. 1993. In planta histochemical staining of fungal alkaline phosphatase activity for analysis of efficient arbuscular mycorrhizal infections. Mycol Res. 97:245–250.
- Tisserant E, Kohler A, Dozolme-Seddas P, Balestrini R, Benabdellah K, Colard A, Croll D, Da Silva C, Gomez SK, Koul R, et al. 2012. The transcriptome of the arbuscular mycorrhizal fungus Glomus intraradices (DAOM 197198) reveals functional tradeoffs in an obligate symbiont. New Phytol. 193:755–769.10.1111/j.1469-8137.2011.03948.x
- Tisserant E, Malbreil M, Kuo A, Kohler A, Symeonidi A, Balestrini R, Charron P, Duensing N, Frei dit Frey N, Gianinazzi-Pearson V, et al. 2013. Genome of an arbuscular mycorrhizal fungus provides insight into the oldest plant symbiosis. Proc Natl Acad Sci. USA. 110:20117–20122.10.1073/pnas.1313452110
- Torelli A, Trotta A, Acerbi L, Arcidiacono G, Berta G, Branca C. 2000. IAA and ZR content in leek (Allium porrum L.), as influenced by P nutrition and arbuscular mycorrhizae, in relation to plant development Plant Soil. 226:29–35.10.1023/A:1026430019738
- van Buuren ML, Maldonado-Mendosa IE, Trieu AT, Blaylock LA, Harrison MJ. 1999. Novel genes induced during an arbuscular mycorrhizal symbiosis formed between Medicago truncatula and Glomus versiforme. Mol Plant Microbe Interact. 12:171–181.
- Veiga RS, Faccio A, Genre A, Pieterse CMJ, Bonfante P, van der Heijden MGA. 2013. Arbuscular mycorrhizal fungi reduce growth and infect roots of the non-host plant Arabidopsis thaliana. Plant Cell Environ. 36:1926–1937.
- Venkateshwaran M, Cosme A, Han L, Banba M, Satyshur KA, Schleiff E, Parniske M, Imaizumi-Anraku H, Ané J-M. 2012. The recent evolution of a symbiotic ion channel in the legume family altered ion conductance and improved functionality in calcium signaling. Plant Cell. 24:2528–2545.
- Waldie T, McCulloch H, Leyser O. 2014. Strigolactones and the control of plant development: lessons from shoot branching. Plant J. 79:607–622.10.1111/tpj.12488
- Wang E, Schornack S, Marsh JF, Gobbato E, Schwessinger B, Eastmond P, Schultze M, Kamoun S, Oldroyd GED. 2012. A common signaling process that promotes mycorrhizal and oomycete colonization of plants. Curr Biol. 22: 2242–2246.
- Warthmann N, Chen H, Ossowski S, Weigel D, Hervé P. 2008. Highly specific gene silencing by artificial miRNAs in rice. PLoS One. 3:e1829.
- Waters MT, Brewer PB, Bussell JD, Smith SM, Beveridge CA. 2012. The Arabidopsis ortholog of rice DWARF27 acts upstream of MAX1 in the control of plant development by strigolactones. Plant Physiol.159:1073–1085.
- Willmann M, Gerlach N, Buer B, Polatajko A, Nagy R, Koebke E, Jansa J, Flisch R, Bucher M. 2013. Mycorrhizal phosphate uptake pathway in maize: vital for growth and cob development on nutrient poor agricultural and greenhouse soils. Front Plant Sci. 4:533.
- Wulf A, Manthey K, Doll J, Perlick AM, Linke B, Bekel T, Meyer F, Franken P, Küster H, Krajinski F. 2003a. Transcriptional changes in response to arbuscular mycorrhiza development in the model plant Medicago truncatula. Mol Plant Microbe Interact. 16:306–314.10.1094/MPMI.2003.16.4.306
- Wulf A, Manthey K, Doll J, Perlick AM, Linke B, Bekel T, Meyer F, Franken P, Küster H, Krajinski F. 2003b. Transcriptional changes in response to arbuscular mycorrhiza development in the model plant Medicago truncatula. Mol Plant Microbe Interact. 16:306–314.
- Xie X, Kusumoto D, Takeuchi Y, Yoneyama K, Yamada Y, Yoneyama K. 2007. 2’-epi-orobanchol and solanacol, two unique strigolactones, germination stimulants for root parasitic weeds, produced by tobacco. J Agric Food Chem. 55:8067–8072.
- Xie X, Yoneyama K, Kisugi T, Uchida K, Ito S, Akiyama K, Hayashi H, Yokota T, Nomura T, Yoneyama K. 2013. Confirming stereochemical structures of strigolactones produced by rice and tobacco. Mol Plant. 6:153–163.
- Yamada Y, Furusawa S, Nagasaka S, Shimomura K, Yamaguchi S, Umehara M. 2014. Strigolactone signaling regulates rice leaf senescence in response to a phosphate deficiency. Planta. 240:399–408.
- Yano K, Yoshida S, Müller J, Singh S, Banba M, Vickers K, Markmann K, White C, Schuller B, Sato S, et al. 2008. CYCLOPS, a mediator of symbiotic intracellular accommodation. Proc Natl Acad Sci USA. 105:20540–20545.10.1073/pnas.0806858105
- Yoneyama K, Awad AA, Xie X, Yoneyama K, Takeuchi Y. 2010. Strigolactones as germination stimulants for root parasitic plants. Plant Cell Physiol. 51:1095–1103.10.1093/pcp/pcq055
- Yoneyama K, Xie X, Yoneyama K, Takeuchi Y. 2009. Strigolactones: structures and biological activities. Pest Manag Sci. 65:467–470.
- Yoshida S, Kameoka H, Tempo M, Akiyama K, Umehara M, Yamaguchi S, Hayashi H, Kyozuka J, Shirasu K. 2012. The D3 F-box protein is a key component in host strigolactone responses essential for arbuscular mycorrhizal symbiosis. New Phytol. 196:1208–1216.
- Yu N, Luo D, Zhang X, Liu J, Wang W, Jin Y, Dong W, Liu J, Liu H, Yang W, et al. 2014. A DELLA protein complex controls the arbuscular mycorrhizal symbiosis in plants. Cell Res. 24:130–133.10.1038/cr.2013.167
- Zampieri E, Murat C, Cagnasso M, Bonfante P, Mello A. 2010. Soil analysis reveals the presence of an extended mycelial network in a Tuber magnatum truffle-ground. FEMS Microb Ecol. 71:43–49.
- Zhang Q, Blaylock LA, Harrison MJ. 2010. Two Medicago truncatula half-ABC transporters are essential for arbuscule development in arbuscular mycorrhizal symbiosis. Plant Cell. 22:1483–1497.
- Zhang R-Q, Zhu H-H, Zhao H-Q, Yao Q. 2013. Arbuscular mycorrhizal fungal inoculation increases phenolic synthesis in clover roots via hydrogen peroxide, salicylic acid and nitric oxide signaling pathways. J Plant Physiol. 170:74–79.
- Zheng L, Kostrewa D, Bernèche S, Winkler FK, Li X-D. 2004. The mechanism of ammonia transport based on the crystal structure of AmtB of Escherichia coli. Proc Natl Acad Sci USA. 101:17090–17095.
- Zhou F, Lin Q, Zhu L, Ren Y, Zhou K, Shabek N, Wu F, Mao H, Dong W, Gan L, et al. 2013. D14–SCFD3-dependent degradation of D53 regulates strigolactone signalling. Nature. 504:406–410.10.1038/nature12878
