ABSTRACT
During a survey conducted in Qassim province, Saudi Arabia, in the year in 2015, 120 samples of carrot (Daucus carota subsp. sativus), onion (Allium cepa), faba bean (Vicia faba), green mustard (Brassica juncea) and alfalfa (Medicago sativa) plants displaying symptoms reminiscent of phytoplasma diseases were collected and tested for phytoplasma infection. Phytoplasma-specific PCR products were only amplified from symptomatic plants by nested-PCR. Disease incidence ranged from 3.14% in alfalfa crop fields 1 year after cultivation to 77.48% in 3-year-old fields. In the five carrot fields sampled in this study, the incidence changed from 3.2% to 100% after 7 months of cultivation. Phylogenetic analysis revealed that all Qassim phytoplasma isolates belong to the 16SrII group. Most of them shared 100% identity with papaya yellow crinkle (16SrII-D Y10097). The results from phylogenetic and virtual restriction fragment length polymorphism analyses of the 16S rRNA gene sequence confirmed that the phytoplasma of Qassim isolates under study is a member of 16SrII-D subgroup. To the best of my knowledge, the onion and green mustard are considered new hosts for the 16SrII group; therefore, this is the first report on the association of phytoplasma with diseases of faba bean, onion, carrot, mustard and alfalfa in Qassim province, Saudi Arabia.
Introduction
Plant-pathogenic phytoplasmas are unique prokaryotic microbes that lack cell walls. Plant-pathogenic phytoplasmas, first described as mycoplasma-like organisms, were discovered by a group of Japanese scientists in 1967 (Doi et al. Citation1967). Taxonomically, they belong to the class Mollicutes, and have been recently classified within the provisional genus “Candidatus phytoplasma” based on 16S rDNA sequence analysis (IRPCM Phytoplasma/Spiroplasma Working Team–Phytoplasma Taxonomy Group Citation2004). They severely affect more than 700 economically important plant species including vegetables, cereals, fruits and ornamentals, as well as forage and forest plants that are found worldwide. Infected plants exhibit symptoms of stunting, shoot proliferation, phyllody, virescence, witches’ broom and fasciation that may be due to the imbalance of plant growth regulators (Lee et al. Citation2000; Bertaccini Citation2007; Hoshi et al. Citation2009; Omar et al. Citation2014). Most diagnoses for the presence of phytoplasma rely mainly on the application of conventional and nested-PCR assays (Lee et al. Citation1994; Bertaccini Citation2007). At present, molecular analysis of conserved genes, in particular 16S rRNA genes, is used for the detection, identification and classification of phytoplasmas (Cai et al. Citation2008; Lee et al. Citation2012; Bertaccini et al. Citation2014). In Al-Qassim region, alfalfa, carrots and onion are very important crops where the cultivated area is 20,069, 3828 and 300 ha with an average annual production of 435,103, 88,023 and 6759 metric tons, respectively (Agricultural Statistical Year Book Citation2012).
Except some reports of phytoplasma members of the 16SrI group affecting lime, some weeds and date palm (Alhudaib et al. Citation2007), no extensive investigations have been conducted on phytoplasma diseases affecting agricultural crops in Saudi Arabia (Alhudaib et al. Citation2009). In 2014, the 16SrII group phytoplasma was identified in tomato in Al-Hassa and Jizan regions, and in alfalfa plants and the associated leafhopper insect vector (Empoasca decipiens) and faba bean in Al-Riyadh region (Al-Saleh et al. Citation2014; Al-Saleh & Amer Citation2014; Alhudaib and Rezk Citation2014). Recently, “Ca. Phytoplasma cynodontis” (16SrXIV) has been reported to associate with Cynodon dactylon, Dodonaea angustifolia, Arundo donax and Acacia salicina in Qassim region (Omar Citation2016). To date, there are no other reports of phytoplasma in Qassim region. The objectives of this study were to detect and characterize phytoplasmas associated with diseases of important agricultural crops in Qassim province based on the 16SrDNA gene sequence.
Materials and methods
Field surveys
A survey of phytoplasma in vegetable and alfalfa crops was conducted during 2015 in three areas (Faculty of Agriculture and Veterinary Medicine farm, Mulayda and Albosor) in Qassim province, Saudi Arabia. The disease incidence was estimated by visual symptoms inspection. Four carrot fields were examined twice; the first examination was after four months of cultivation and the second was after seven months of cultivation. Five one-year-old and five three-year-old alfalfa fields were surveyed. One thousand plants were inspected in each field of carrot and alfalfa using a two-way diagonal pattern as the sampling pattern (Delp et al., Citation1986). However, all of the experimental plantations of onions, faba bean and green mustard at the farm of College of Agriculture and Veterinary Medicine were examined after 4 months from cultivation. A total of 40, 45, 10, 20 and 5 symptomatic plant samples of carrot, alfalfa, onion, faba bean and green mustard, respectively, were collected and transferred to the Department of Plant Production and Protection, College of Agriculture and Veterinary Medicine, Qassim University. Asymptomatic plants grown in the same locations were also collected and used as negative controls. The samples were kept at 4°C until DNA extraction and PCR analysis.
DNA extraction and PCR assays
Total nucleic acids were extracted from the midribs of all plants except onion. The tissue used to extract DNA from onion plants was leaf. All samples were ground in liquid nitrogen using mortars and pestles and 0.1 g powder of each healthy and symptomatic plant sample was placed into 1.5 ml sterilized Eppendorf tubes (Bio Basic Canada Inc., Canada). DNA was isolated from each sample using an i-genomic plant DNA extraction mini kit (iNtRON Biotechnology, Inc., Cat. No. 17371) according to the manufacturer’s instructions. Finally, DNA was eluted into 100 μl of elution buffer (supplied with the kit) and stored at −20°C. The extracted nucleic acids from different samples were used as templates in PCRs using universal primer pair P1/P7 in the first round (Deng & Hiruki Citation1991; Schneider et al. Citation1995) followed by R16F2n/R16R2 (Lee et al. Citation1993) in nested-PCR assays. fU5/rU3 primers (Seemüller et al. Citation1994) were used with samples which failed in the amplification using pair primers R16F2n/R16R2. PCRs were performed for 34 cycles in an automated thermal cycler (SwiftTM MaxPro Thermal Cycler, ESCO healthcare). Each 40 µl PCR mixture contained 1 µl of 20 ng of nucleic acids, 1 µl of each primer (10 pmol), 8 µl of 5× FIREPol® Master Mix (Solis BioDyne, Estonia) and 29 µl of nuclease-free water (Promega, USA). PCR assays were performed as previously described elsewhere (Deng & Hiruki Citation1991; Schneider et al. Citation1995; Gundersen & Lee Citation1996). Fifteen microliters of all PCR products were analyzed by electrophoresis through a 1.5% agarose gel, stained with ethidium bromide and DNA bands were visualized using a UV transilluminator (G:BOX F3 system, Syngene).
Sequencing PCR products and phylogenetic analysis
The amplicons generated from the 18 samples collected in the Qassim area () that were amplified by the primer pairs R16F2n/R16R2 (alfalfa, faba bean and carrot) and fU5/rU3 (onion and green mustard) were directly sequenced in both directions (Macrogen Inc., Korea). The sequences were assembled and edited using GAP4 (Bonfield et al. Citation1995). Nucleotide sequence similarity and multiple alignments were assessed using ClustalW (Thompson et al. Citation1994). Phylogenetic analyses were developed with partial 16S rDNA sequences from the 18 phytoplasma Qassim isolates (carrot, onion, green mustard, faba bean and alfalfa) and 22 “Candidatus Phytoplasma” strains from GenBank database () using Acholeplasma laidlawii as the outgroup. A phylogenetic tree was constructed using the neighbor joining phylogenetic method with MEGA4 program (Tamura et al. Citation2007) and 1000 bootstrap replications.
Table 1. Host plant, no. of sequenced samples, symptoms, isolate no., seq. accession no. and collection locations of phytoplasmas collected from Qassim province, Saudi Arabia.
In silico restriction enzyme digestions and virtual gel plotting
Seventeen restriction enzymes (AluI, BamHI, BfaI, BstUI (ThaI), DraI, EcoRI, HaeIII, HhaI, HinfI, HpaI, HpaII, KpnI, Sau3AI (MboI), MseI, RsaI, SspI, TaqI) were used to digest in silico the R16F2n/R16R2 region of the 16S rDNA gene sequences detected in isolates Carrot 1 (LN898421), carrot 6 (LN898422), onion 113 (LN898436), onion 114 (LN898437), faba bean 36 (LN898425), green mustard 108 (LN898432) and Alfalfa 96 (LN898428) for phytoplasma classification (Lee et al. Citation1998). Virtual gel plotting was achieved using iPhyclassifier (Zhao et al. Citation2009).
Results and discussions
Symptoms and disease incidence
Phytoplasma diseases were observed in many vegetables (carrot, onion, green mustard and faba bean) and alfalfa crops in Al-Qassim region with various symptoms, which are summarized in . The incidence reached 100% in delayed harvested carrot fields after 7 months of cultivation, whereas it was initially 3.2% after three months of cultivation (). Therefore, carrot crops suffering severe deformation made carrots unmarketable ((D)). A high incidence of 77.48% was also observed in the three-year-old alfalfa crop fields, whereas it was only 3.14% in a one-year-old field. Although the experimental plant areas of onion, faba bean and green mustard are very close to each other, the incidence of disease varied according to the crop, which was for onion, faba bean and green mustard 1.3%, 47.5% and 4.5%, respectively (). The high incidence of this disease in carrot, alfalfa and faba bean crops may be due to the feeding preferences of insect vectors. Moreover, the age of the field appeared to influence the incidence of the disease in carrot and alfalfa plants. Alfalfa was previously reported to be infected with phytoplasmas in association with witches’ broom, yellows and phyllody symptoms, not only in Al-Riyadh, Saudi Arabia (AL-Saleh et al. Citation2014), but also in many other areas around the world, including Italy (Marcone et al. Citation1997), Canada (Khadhair et al. Citation1997), Oman (Khan et al. Citation2002), Bolivia (Jones et al. Citation2005) and Iran (Esmailzadeh Hosseini et al. Citation2015). Leaf reddening, purpling and yellowing; formation of chlorotic adventitious shoots; proliferation and reduction of size and quality of roots were observed in carrot plants infected with phytoplasma in Serbia (Duduk et al. Citation2007), Israel (Orenstein et al. Citation1999), India (Arocha et al. Citation2009), the UK (Nisbet et al. Citation2014), Lithuania (Valiunas et al. Citation2001), Canada (Wally et al. Citation2004) and China (Li et al. Citation2012). Phytoplasmas were detected in faba bean plants in many countries, such as India, Saudi Arabia, Sudan, Spain, Cuba and Egypt, showing phyllody, witches’ broom, and mild yellow symptoms (Castro and Romero Citation2004; Arocha et al. Citation2007; Alfaro-Fernández et al. Citation2012; Omar & Foissac Citation2012; Singh et al. Citation2013; Al-Saleh & Amer Citation2014; Hamed et al. Citation2014). It was reported in particular countries including Japan, Lithuania, Canada, Pakistan, Mauritius, Iran and Tonga that onion is susceptible to phytoplasma with diverse symptoms including yellows, stunting, twisting and proliferation of flowers (Oshima et al. Citation2001; Khadhair et al. Citation2002; Lee et al. Citation2003; Davis et al. Citation2006; Gungoosingh-Bunwaree et al. Citation2010; Jomantiene et al. Citation2010; Sichani et al. Citation2014; Ahmad et al. Citation2015). Green mustard was surrounded by infected plants of faba bean and onion at the experimental station of College of Agriculture, so it was infected by phytoplasma with an incidence of 4.5%. Noteworthy, this could be considered the first record of green mustard as a phytoplasma host.
Figure 1. Symptoms associated with phytoplasma-infected plant species collected from Qassim region, Saudi Arabia: carrot: (Aa) fasciation, (B) phyllody, (C) healthy plant, (D) hairy roots, (E) proliferation, (F) healthy roots, (G) field view showing yellow and purple leaves, (H) healthy plant; green mustard: (I) healthy plant (left) and diseased plant (right) displaying stunting and curly leaf edges; onion: (J) healthy plant (back) and diseased plant (front) showing yellow and twisting leaves; faba bean: (K) and (L) phyllody, (M) healthy plant; alfalfa: (N) stunting and yellows, (O) healthy plant, (P) witches’ broom and phyllody, (Q) healthy plant.
Table 2. Incidence of phytoplasma in carrot, alfalfa, onion, faba bean and green mustard collected from different areas in Qassim province from open fields during 2015.
DNA amplification
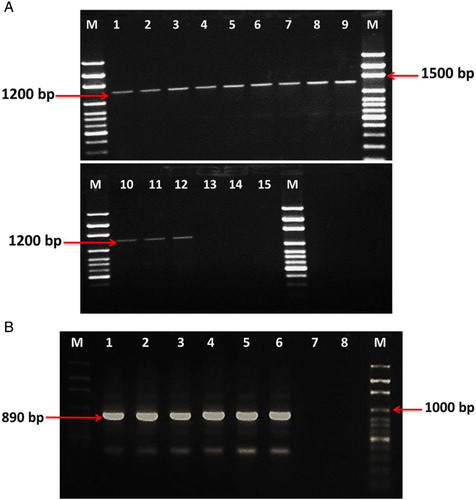
Nested-PCR analyses with universal phytoplasma primers R16F2n/R16R2 provided positive bands of about 1200 bp for carrot, alfalfa and faba bean isolates ((A)), and fU5/rU3 primers succeeded in amplifying 900 bp bands of onion and mustard green isolates ((B)). DNA amplification failure using R16F2n/R16R2 of onion and mustard green isolates may be due to the low concentration of phytoplasma infection which was not detectable by such primers. No band was observed in the healthy plant samples which served as negative control ((A) and (B)).
Figure 2. (A) Agarose gel electrophoresis of nested-PCR products from the16SrRNA gene using primers R16F2n/R16R2. Lanes 1, 2, 3 and 4 are carrot isolates (1, 5, 6 and 8); lanes 5, 6 and 7 are faba bean isolates (35, 36 and 107); lanes 8, 9, 10, 11 and 12 are alfalfa isolates (95, 96, 97, 98 and 99); lanes 13, 14 and 15 are carrot, faba bean and alfalfa symptomless samples, M: 100 bp DNA ladder (Solis BioDyne). (B) Electrophoresis pattern of nested-PCR products from16SrRNA gene using fU5/rU3 primer pairs: lanes 1, 2, 3 and 4 are onion isolates (111, 112, 113 and 114); lanes 5 and 6 are green mustard isolates 108 and 109; lanes 7 and 8 are healthy samples from onion and green mustard, respectively, M: 100 bp DNA ladder (Solis BioDyne).
Sequences and phylogenetic analysis
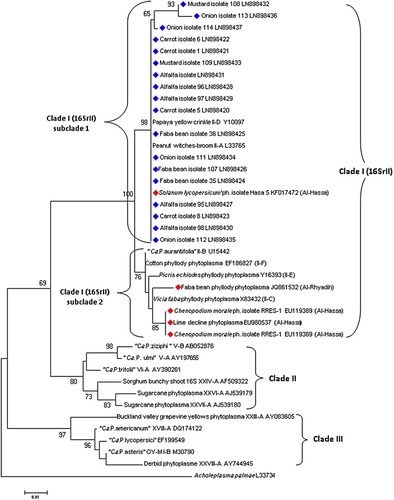
The sequences of the 18 vegetable and alfalfa phytoplasma isolates were compared to each other and to 10 16SrII phytoplasma strains, 5 of them belonging to 16SrII subgroups (A–E), and 11 phytoplasma members of different groups and subgroups reported in GenBank. The phylogenetic tree showed that all compared phytoplasmas are divided into three clades: clade I, clade II and clade III (). Clade I which was separated from the other two groups of phytoplasma with bootstrap 100% () includes vegetable and alfalfa phytoplasma isolates from Qassim and members of 16SrII subgroups A–E, including those reported in Riyadh province from tomato, lime and faba bean plants. Therefore, 16SrII was separated into two sub-clades; one of them (sub-clade 2) includes 16SrII subgroups B, C, E and F; the phytoplasma isolates reported from Al-Hassa province (isolate RRES-1 EU119389, isolate RRES-3 EU119391 and lime isolate EU980537) and faba bean phyllody phytoplasma (JQ861532 Al-Rhyadih) with bootstrap 97%. The other sub-clade (sub-clade 1) includes all Qassim phytoplasma isolates, papaya yellow crinkle (16SrII-D Y10097) and peanut witches’ broom (16SrII-A L33765). Three phytoplasma isolates from Qassim (mustard 108, onion 113 and 144 isolates) shared identities of 98.8%, 98.2% and 99.7% with phytoplasma subgroups, papaya yellow crinkle (16SrII-D Y10097) and peanut witches’ broom (16SrII-A L33765), respectively. However, they have identities of 97.6%, 97% and 98.8% with “Ca. Phytoplasma aurantifolia”. The rest of the Qassim phytoplasma isolates have identity of 100% with papaya yellow crinkle (16SrII-D Y10097) and Solanum lycopersicum phytoplasma (Hasa 5 KF017472, Al-Hassa). The Qassim phytoplasma isolates have identities ranging from 95.7% to 98.5% with other phytoplasma isolates identified in Al-Rhyadih and Al-Hassa provinces. Carrot has been reported to be infected by a phytoplasma of the 16SrI group in many countries (Orenstein et al. Citation1999; Duduk et al. Citation2007; Nisbet et al. Citation2014), 16SrIII and 16SrV groups in Israel and 16SrXII-A group in Serbia (Weintraub et al. Citation2007; Duduk et al. Citation2008); however, its infection by 16SrII was only reported in India (Arocha et al., Citation2009). Although 16SrII has been reported in Al-Rhyadih and Al-Hassa provinces in aflalfa and faba bean plants, respectively (Al-Saleh et al. Citation2014; Al-Saleh & Amer Citation2014), the results of this research are considered to be the first report of 16SrII phytoplasma infection in carrot in Saudi Arabia. Phytoplasma group 16SrII was also previously reported to infect faba bean in Sudan (Alfaro-Fernández et al. Citation2012) and alfalfa with little leaf and witches’ broom in Iran and Oman, respectively (Khan et al. Citation2002; Hosseini et al. Citation2013). Phytoplasma group 16SrI was associated with alfalfa plants in Bolivia, Lithuania and the USA (Peters et al. Citation1999; Jomantiene et al. Citation2000; Jones et al. Citation2005). Although there are many reports about onion infection by phytoplasma group 16SrI around the world (Khadhair et al. Citation2002; Lee et al. Citation2003; Jomantiene et al. Citation2010), the association of 16SrII phytoplasma with onion is considered as the first report not only in Saudi Arabia, but also in the world. While information about the association of phytoplasma with green mustard is very limited, it is hereby demonstrated to be a host for 16SrII phytoplasma. These infections may result from the proximity of highly infected faba bean fields from which the disease had spread. It can be concluded that this is the first report of the association of phytoplasma group 16SrII with the disease of faba bean, onion, carrot, mustard and alfalfa in Qassim province. Therefore, onion and green mustard are considered as new hosts for the 16SrII phytoplasma group. Because of the diversity of phytoplasma host plants in the Qassim region, epidemiological studies will concentrate on using other genes for finer differentiations among the phytoplasma isolates in the Qassim area.
Figure 3. Phylogenetic tree based on 16Sr DNA gene sequences constructed by the neighbor-joining method from 18 phytoplasma isolates from Qassim province (marked in blue color) and 21 phytoplasma members belonging to different subgroups available in the GenBank database. Acholeplasma palmae (L33734) was used as the outgroup to root the tree. Bootstrap analyses were done in 1000 replicates. Phylogenetic analyses were performed using the MEGA4 software.
Virtual RFLPs
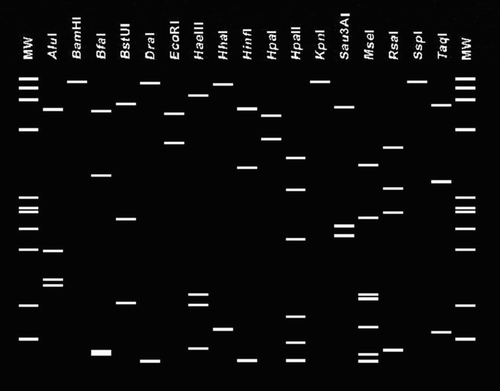
Computer-simulated, virtual RFLP analyses were carried out on R16F2n/R16R2 sequences from seven Qassim phytoplasma isolates: carrot 1 (LN898421), carrot 6 (LN898422), onion 113 (LN898436), onion 114 (LN898437), faba bean 36 (LN898425), green mustard 108 (LN898432) and alfalfa 96 (LN898428), using iPhyClassifier (Zhao et al. Citation2009). The virtual RFLP patterns () derived from the query 16S rDNA F2nR2 fragment of all seven Qassim phytoplasma isolates are identical (similarity coefficient 1.00) to the reference pattern of 16Sr group II, subgroup D (GenBank accession: Y10097). Phytoplasmas were identical to those of “Candidatus Phytoplasma australasia”, representative of the 16SrXII-D subgroup (White et al. Citation1998).
Figure 4. Virtual restriction fragment length polymorphism patterns of 1.2 kb derived from in silico digestions of R16F2n/R16R2 fragments of Qassim isolates: carrot 1 (LN898421), carrot 6 (LN898422), onion 113 (LN898436), onion 114 (LN898437), faba bean 36 (LN898425), green mustard 108 (LN898432) and alfalfa 96 (LN898428).
Acknowledgements
I would like to thank Mr Khaled Al-Jamhan (Department of Plant Production and Protection, College of Agriculture and Veterinary Medicine, Qassim University) for his help during the survey and sample collection.
Disclosure statement
No potential conflict of interest was reported by the author.
References
- Agricultural Statistical Year Book. 2012. Ministry of agriculture, Vol. 25. Kingdom of Saudi Arabia [cited 2015 Nov 7]; Available from: http://www.moa.gov.sa/files/stat25/1/1.htm.
- Ahmad SJN, Ahmad JN, Irfan M, Ahmad M, Aslam M. 2015. Report on phytoplasma new host plants in Pakistan. Phytopathogenic Mollicutes. 5:S71–S72. doi: 10.5958/2249-4677.2015.00030.4
- Alfaro-Fernández A, Ali MA, Mobarak Abdelraheem F, Abd Elhameed Saeed E, Isabel Font San Ambrosio M. 2012. Molecular identification of 16SrII-D subgroup phytoplasmas associated with chickpea and faba bean in Sudan. Eur J Pant Pathol. 133:791–795. doi: 10.1007/s10658-012-9975-7
- Alhudaib K, Arocha Y, Wilson M, Jones P. 2007. “Al-Wijam”, a new phytoplasma disease of date palm in Saudi Arabia. Bull Insectol. 60:285–286.
- Alhudaib K, Arocha Y, Wilson M, Jones P. 2009. Molecular identification, potential vectors and alternative hosts of the phytoplasma associated with a lime decline disease in Saudi Arabia. Crop Prot. 28:13–18. doi: 10.1016/j.cropro.2008.08.007
- Alhudaib k, Rezk A. 2014. Molecular characterization of phytoplasma associated disease in tomato (Lycopersicun esculentum) in Saudi Arabia. Int J Virol. 10:180–191. doi: 10.3923/ijv.2014.180.191
- Al-Saleh MA, Amer MA. 2014. Molecular characterization of the 16Sr II group of phytoplasma associated with faba bean (Vicia Faba L.) in Saudi Arabia. J Anim Plant Sci. 24:221–228.
- Al-Saleh MA, Amer MA, AL-Shahwan IM, Abdalla OA, Damiri BV. 2014. Detection and molecular characterization of alfalfa witches’-broom phytoplasma and its leafhopper vector in Riyadh Region of Saudi Arabia. Int J Agr Biol. 16:300–306.
- Arocha Y, Piñol B, Picornell B, Almeida R, Jones P. 2007. Broad bean and sweet pepper: two new hosts associated with ‘Candidatus Phytoplasma asteris’ (16SrI phytoplasma group) in Cuba. Plant Pathol. 56:345. doi: 10.1111/j.1365-3059.2007.01518.x
- Arocha Y, Singh A, Pandey M, Tripathi AN, Chandra B, Shukla SK, Singh Y, Kumar A, Srivastava RK, Zaidi NW, et al. 2009. New plant hosts for group 16SrII, “Candidatus Phytoplasma aurantifolia”, in India. Plant Pathol. 58:391. doi: 10.1111/j.1365-3059.2008.01969.x
- Bertaccini A. 2007. Phytoplasmas: diversity, taxonomy and epidemiology. Front Biosci. 12:673–689. doi: 10.2741/2092
- Bertaccini A, Duduk B, Paltrinieri S, Contaldo N. 2014. Phytoplasmas and phytoplasma diseases: a severe threat to agriculture. Am J Plant Sci. 5:1763–1788. doi: 10.4236/ajps.2014.512191
- Bonfield JK, Smith KF, Staden R. 1995. A new DNA sequence assembly program. Nucleic Acids Res. 23:4992–4999. doi: 10.1093/nar/23.24.4992
- Cai H, Wei W, Davies RE, Chen H, Zhao Y. 2008. Genetic diversity among phytoplasmas infecting Opuntia species: virtual RFLP analysis identifies new subgroups in the peanut witches, broom phytoplasma group. Int J Syst Evol Microbiol. 58:1448–1457. doi: 10.1099/ijs.0.65615-0
- Castro S, Romero J. 2004. Short communication. First detection of a phytoplasma infecting faba bean (Vicia faba L.) in Spain. Span J Agric Res. 2:253–256. doi: 10.5424/sjar/2004022-83
- Davis RI, Jones P, Holman TJ, Halsey K, Amice R, Tupouniua SK, Seth M. 2006. Phytoplasma disease surveys in Tonga, New Calédonia and Vanuatu. Australas Plant Path. 35:335–340. doi: 10.1071/AP06029
- Delp BR, Stowell LJ, Marios JH. 1986. Evaluation of field sampling techniques for estimation of disease incidence. Phytopathology. 76:1299–1305. doi: 10.1094/Phyto-76-1299
- Deng S, Hiruki C. 1991. Genetic relatedness between two non-culturable mycoplasmalike organism revealed by nucleic acid hybridization and polymerase chain reaction. Phytopathology. 81:1475–1479. doi: 10.1094/Phyto-81-1475
- Doi Y, Teranaka M, Yora K, Asuyama H. 1967. Mycoplasma or PLT grouplike microrganisms found in the phloem elements of plants infected with mulberry dwarf, potato witches’ broom, aster yellows or Pawlownia Witches’ Broom. Jap J Phytopathol. 33:259–266. doi: 10.3186/jjphytopath.33.259
- Duduk B, Bulajić A, Duduk N, Calari A, Paltrinier S, Krstić B, Bertaccini n. 2007. Identification of phytoplasmas belonging to aster yellows ribosomal group (16SrI) in vegetables in Serbia. Bull Insectol. 60:341–342.
- Duduk B, Perić P, Mařuć D, Drobnjaković T, Picuau L, Alma A, Bertaccini A. 2008. Phytoplasma in carrots: disease and potential vectors in Serbia. Bull Insectol. 61:327–331.
- Esmailzadeh Hosseini SA, Khodakaramian G, Salehi M, Fani SR, Bolok Yazdi HR, Raoufi D, Jadidi O, Bertaccini A. 2015. An up to date status of alfalfa witches’ broom disease in Iran. Phytopathogenic Mollicutes. 5:9–18. doi: 10.5958/2249-4677.2015.00057.2
- Gundersen DE, Lee I-M. 1996. Ultrasensitive detection of phytoplasmas by nested-PCR assays using two universal primer pairs. Phytopathol Mediterr. 35:144–151.
- Gungoosingh-Bunwaree A, Contaldo N, Vally V, Madhu SB, Duduk B, Bertaccini A. 2010. Detection of phytoplasmas in watercress and onion plants from Mauritius. Plant Health Prog. doi:10.1094/PHP-2010-0930-01-RS
- Hamed AH, El Attar AK, Om-Hashim M, El-Banna. 2014. First record of a phytoplasma associated with Faba Bean (Vicia faba L.) Witches’-broom in Egypt. Int J Virol. 10:129–135. doi: 10.3923/ijv.2014.129.135
- Hoshi A, Oshima K, Kakizawa S, Ishii Y, Ozeki J, Hashimoto M, Komatsu K, Kagiwada S, Yamaji Y, Namba S. 2009. A unique virulence factor for proliferation and dwarfism in plants identified from a phytopathogenic bacterium. Proc Natl Acad Sci USA. 106:6416–6421. doi: 10.1073/pnas.0813038106
- Hosseini S, Bahar M, Zirak L. 2013. Characterization of phytoplasmas related to peanut witches’-broom and stolbur groups associated with alfalfa diseases in Iran. J Plant Dis Protect. 120:70–76. doi: 10.1007/BF03356456
- IRPCM, Phytoplasma/Spiroplasma Working Team – Phytoplasma Taxonomy Group. 2004. ‘Candidatus Phytoplasma’, a taxon for the wall-less, non-helical prokaryotes that colonize plant phloem and insects. Int J Syst Evol Microbiol. 54:1243–1255. doi: 10.1099/ijs.0.02854-0
- Jomantiene R, Davis RE, Antoniuk L, Staniulis J. 2000. First report of phytoplasmas in soybean, alfalfa, and Lupinus sp. in Lithuania. Plant Dis. 84:198. doi: 10.1094/PDIS.2000.84.2.198C
- Jomantiene R, Davis RE, Lee I-M, Zhao Y, Bottner K, Valiunas D, Petkauskaite R. 2010. Onion is a host for two phytoplasma lineages, subgroups 16SrI-A and 16SrI-L, in Lithuania: a HinfI site revealed a SNP marking divergent branches of evolution. J Plant Pathol. 92:451–460.
- Jones P, Arocha Y, Plata G. 2005. First report of a ‘Candidatus Phytoplasma asteris’ isolate associated with a witches’ broom disease of alfalfa in Bolivia. Plant Pathol. 54:559. doi: 10.1111/j.1365-3059.2005.01231.x
- Khadhair AH, Evans IR, Choban B. 2002. Identification of aster yellows phytoplasma in garlic and green onion by PCR-based methods. Microbiol Res. 157:161–167. doi: 10.1078/0944-5013-00146
- Khadhair AH, Hiruki C, Hwang SF. 1997. Molecular detection of alfalfa witches’ broom phytoplasma in four leafhopper species associated with infected alfalfa plants. Microbiol Res. 152:269–275. doi: 10.1016/S0944-5013(97)80039-0
- Khan AJ, Botti S, Al-Subhi AM, Gundersen-Rindal DE, Bertaccini A. 2002. Molecular identification of a new phytoplasma associated with alfalfa witches’-broom in Oman. Phytopathology. 92:1038–1047. doi: 10.1094/PHYTO.2002.92.10.1038
- Lee I-M, Bottner-Parker KD, Zhao Y, Bertaccini A, Davis RE. 2012. Differentiation and classification of phytoplasmas in the pigeon pea witches’ broom group (16SrIX): an update based on multiple gene sequence analysis. Int J Syst Evol Microbiol. 62:2279–2285. doi: 10.1099/ijs.0.038273-0
- Lee I-M, Davis RE, Gundersen-Rindal DE. 2000. Phytoplasma: phytopathogenic mollicutes. Annu. Rev Phytopathol. 54:221–255.
- Lee I-M, Gundersen DE, Davis RE, Bartoszyk I-M. 1998. Revised classification scheme of phytoplasmas based on RFLP analysis of 16S rRNA and ribosomal protein gene sequences. Int J Syst Bacteriol. 48:1153–1169. doi: 10.1099/00207713-48-4-1153
- Lee I-M, Gundersen DE, Hammond RW, Davis RE. 1994. Use of Mycoplasmalike Organism (MLO) group specific oligonucleotide primers for nested-PCR assays to detect mixed-MLO infections in a single host plant. Phytopathology. 84:559–566. doi: 10.1094/Phyto-84-559
- Lee I-M, Hammond RW, Davis RE, Gundersen DE. 1993. Universal amplification and analysis of pathogen 16S rDNA for classification and identification of mycoplasma like organisms. Phytopathology. 83:834–842. doi: 10.1094/Phyto-83-834
- Lee I-M, Martini M, Bottner KD, Dane RA, Black MC, Troxclair N. 2003. Ecological implications from a molecular analysis of phytoplasmas involved in an aster yellows epidemic in various crops in Texas. Phytopathology. 93:1368–1377. doi: 10.1094/PHYTO.2003.93.11.1368
- Li Z-N, Zhang L, Man J-Y, Wu Y-F. 2012. Detection and identification of elm yellows group phytoplasma (16SrV) associated with alfalfa witches’ broom disease. J Phytopathol. 160:311–313. doi: 10.1111/j.1439-0434.2012.01901.x
- Marcone C, Raggozzino A, Seemüller E. 1997. Detection and identification of phytoplasma infecting vegetable, ornamental and forage crops in southern Italy. J Plant Pathol. 79:211–217.
- Nisbet C, Ross S, Monger WA, Highet F, Jeffries C. 2014. First report of ‘Candidatus Phytoplasma Asteris’ in commercial carrots in the United Kingdom. New Dis Rep. 30:16. doi: 10.5197/j.2044-0588.2014.030.016
- Omar AF. 2016. Association of ‘Candidatus Phytoplasma cynodontis’ with Bermuda grass white leaf disease and its new hosts in Qassim province, Saudi Arabia. J Plant Interact. 11:101–107. doi: 10.1080/17429145.2016.1196401
- Omar AF, Dewir YH, El-Mahrouk ME. 2014. Molecular identification of phytoplasmas in fasciated cacti and succulent species and associated hormonal perturbation. J Plant Interact. 9:632–639. doi: 10.1080/17429145.2014.882421
- Omar AF, Foissac X. 2012. Occurrence and incidence of phytoplasmas of the 16SrII-D subgroup on solanaceous and cucurbit crop in Egypt. Eur J Plant Pathol. 133:353–360. doi: 10.1007/s10658-011-9908-x
- Orenstein S, Franck A, Kuznetzova L, Sela I, Tanne E. 1999. Association of phytoplasmas with a yellows disease of carrot in Israel. J Plant Pathol. 81:193–199.
- Oshima K, Shiomi T, Kuboyama T, Sawayanagi T, Nishigawa H, Kakizawa S, Miyata S, Ugaki M, Namba S. 2001. Isolation and characterization of derivative lines of the onion yellows phytoplasma that do not cause stunting or phloem hyperplasia. Phytopathology. 91:1024–1029. doi: 10.1094/PHYTO.2001.91.11.1024
- Peters RD, Lee ME, Grau CR, Driscoll SJ, Winberg RM, Kurtzweil NC, Lukaesko LA, Lee I-M. 1999. First report of aster yellows phytoplasma in alfalfa. Plant Dis. 83:488. doi: 10.1094/PDIS.1999.83.5.488C
- Schneider B, Seemüller E, Smart CD, Kirkpatrick BC. 1995. Phylogenetic classification of plant pathogenic Mycoplasma like organisms or phytoplasmas. In: S Razin, JG Tully, editor. Molecular and diagnostic procedures in mycoplasmology. San Diego: Academic Press; p. 369–380.
- Seemüller E, Schneider B, Mäurer R, Ahrens U, Daire X, Kison H, Lorenz K-H, Firrao G, Avinent L, Sears BB, Stackebrandt E. 1994. Phylogenetic classification of phytopathogenic mollicutes by sequence analysis of 16S ribosomal DNA. Int J Syst Bacteriol. 44:440–446. doi: 10.1099/00207713-44-3-440
- Sichani FV, Bahar M, Zirak L. 2014. Characterization of phytoplasmas related to aster yellows group infecting annual plants in Iran, based on the studies of 16S rRNA and rp genes. J Plant Prot Res. 54:1–8. doi: 10.2478/jppr-2014-0001
- Singh AK, Bhatt BP, Manibhushan. 2013. Occurrence of phytoplasma phyllody and witches’ broom disease of faba bean in Bihar. J Environ Biol. 34:837–840.
- Tamura K, Dudley J, Nei M, Kumar S. 2007. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol Biol Evol. 24:1596–1599. doi: 10.1093/molbev/msm092
- Thompson JD, Higgins DG, Gibson TJ. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673–4680. doi: 10.1093/nar/22.22.4673
- Valiunas D, Alminaite A, Staniulis J, Jomantiene R. 2001. First report of aster yellows-related subgroup I-A phytoplasma strains in carrot, phlox, sea-lavender, aconitum, and hyacinth in Lithuania. Plant Dis. 85:804. doi: 10.1094/PDIS.2001.85.7.804C
- Wally O, Daayf F, Khadhair AH, Adam L, Elliott B, Shinners-Carnelley T, Iranpour M, Northover P, Keyworth S. 2004. Incidence and molecular detection of yellows-type disease in carrots, associated with leafhoppers in southern Manitoba, Canada. Can J Plant Pathol. 26:498–505. doi: 10.1080/07060660409507170
- Weintraub PG, Zeidan M, Spiegel S, Gera A. 2007. Diversity of the known phytoplasmas in Israel. Bull Insectol. 60:143–144.
- White DT, Blackall LL, Scott PT, Walsh KB. 1998. Phylogenetic positions of phytoplasmas associated with dieback, yellow crinkle and mosaic diseases of papaya, and their proposed inclusion in ‘Candidatus Phytoplasma australiense’ and a new taxon, ‘Candidatus Phytoplasma australasia’. Int J Syst Bacteriol. 48:941–951. doi: 10.1099/00207713-48-3-941
- Zhao Y, Sun Q, Wei W, Davis RE, Wu W, Liu Q. 2009. ‘Candidatus Phytoplasma tamaricis’, a novel taxon discovered in witches’ broom diseased salt cedar (Tamarix chinensis Lour.). Int J Syst Evol Microbiol. 59:2496–2504. doi: 10.1099/ijs.0.010413-0


