ABSTRACT
Mycorrhizal symbiosis influences plant growth and nutrition and can affect the performance of insect herbivores, but these effects are context-dependent. This study aims to investigate the influence of nitrogen fertilization and mycorrhizal symbiosis on maize and Spodoptera exigua performance and to explore the potential underlying mechanisms. Mycorrhiza promoted maize growth and reduced S. exigua performance, but these effects were dependent on nitrogen availability. We then assessed whether the consequences for S. exigua were mediated by its gut microbiota. Neither nitrogen nor mycorrhization affected S. exigua gut bacterial community. Reduced herbivore performance was instead potentially due to the effects of nitrogen-mycorrhiza interaction on the plant nutritional value.
1. Introduction
In nature, plants coexist and interact with multiple beneficial and antagonistic organisms, including microorganisms and insects (Biere and Tack Citation2013; Heinen et al. Citation2018). Herbivory by phytophagous insects is an example of a two-way interaction with detrimental effects on plant health and notorious consequences for crops (Dhaliwal et al. Citation2010; Deutsch et al. Citation2018). Besides, the mutualistic symbioses between plants and soil-borne beneficial microbes can benefit plant nutrition and resistance, which positively affects yield productivity (Finkel et al. Citation2017; Bargaz et al. Citation2018). Because both phytophagous insect and plant symbionts rely on plant resources, they can simultaneously or sequentially interact with the same host, being potentially affected by each other via the plant (Biere and Bennett Citation2013; Chrétien et al. Citation2018; Gruden et al. Citation2020; de Bobadilla et al. Citation2022).
The mutualistic association between arbuscular mycorrhizal fungi (AMF) and plants, known as arbuscular mycorrhizal (AM) symbiosis, is an ecologically widespread plant-microbe interaction with beneficial effects on plant performance (Jeffries et al. Citation2003). Benefits of this symbiosis include an improved capacity of the host plant to acquire mineral nutrients from the soil, positively affecting its nutritional status (Van Der Heijden et al. Citation2006; Porras-Soriano et al. Citation2009; Balliu et al. Citation2015; Bhantana et al. Citation2021). Moreover, root colonization by AMF can induce resistance in the host plant, protecting both roots and aerial parts against future aggressors (Pozo and Azcón-Aguilar Citation2007; Rivero et al. Citation2021; Manresa-Grao et al. Citation2022). This phenotype, known as Mycorrhiza-Induced Resistance (MIR), is usually characterized by the priming of the plant defenses, a faster and/or more effective activation of the plant defense responses upon challenge (Conrath et al. Citation2006; Pozo and Azcón-Aguilar Citation2007; Jung et al. Citation2012; Pieterse et al. Citation2014). MIR is generally accepted to be effective against aboveground organisms that are sensitive to the plant defenses regulated by jasmonic acid (JA) signaling, including generalist leaf-chewing insects, but it can also negatively affect aphids performance (Vicari et al. Citation2002; Guerrieri et al. Citation2004; Koricheva et al. Citation2009; Pineda et al. Citation2010; Pozo et al. Citation2010; Pineda et al. Citation2013; Song et al. Citation2013). However, abiotic conditions can influence the outcome of plant-AMF interactions, thus impacting plant growth or pest resistance (Borowicz Citation1997; Gange et al. Citation1999; Sanchez-Bel et al. Citation2016; López-Carmona et al. Citation2019; Real-Santillán et al. Citation2019).
Similar to plants, phytophagous insects can establish beneficial associations with microorganisms that confer ecological advantages to exploit their environments (Dubreuil et al. Citation2014; Sugio et al. Citation2015; Giron et al. Citation2017). They may rely on gut bacteria to supplement their nutritional demands by providing novel metabolic pathways or facilitating the digestion of nutrients (Giron et al. Citation2017; Gurung et al. Citation2019; Mason et al. Citation2019). Insect’s gut microbiota can also contribute to counteract plant defenses through detoxification or sequestration of anti-herbivore compounds (Giron et al. Citation2017; Zhang et al. Citation2018; Favery et al. Citation2020; Jing et al. Citation2020). Environmental factors can contribute to shape the gut microbiota of insect herbivores and part of their microbiomes can be actively acquired through their diet or directly from the soil (Hannula et al. Citation2019; Leite-Mondin et al. Citation2021). Despite microorganisms can provide significant benefits to insects, reliance on symbionts for plant resources exploitation can fluctuate across phytophagous insects (Hammer et al. Citation2017; Sudakaran et al. Citation2017; McKenna et al. Citation2019; Frago et al. Citation2020).
Due to the potential impact of beneficial microbes on the performance of both plants and insects, symbionts can be targeted for optimized crop production in sustainable agriculture, either by improving plant resistance and/or by impairing insect performance (Yuan et al. Citation2021). However, since host-microbe interactions are highly context-dependent, studies on particular plant-herbivore systems under different environmental conditions are required for the effective application of symbiont-targeted strategies in pest management.
Lepidoptera includes some of the most destructive and economically relevant agricultural pests (Goldstein Citation2017). Most caterpillars are herbivores, and the composition and abundance of their gut microbial communities are known to vary largely both inter- and intraspecifically (Hammer et al. Citation2019; Mason et al. Citation2020). Interconnection between the gut microbiota and the performance of caterpillars is a current controversial topic (Hammer et al. Citation2017, Citation2019). Their gut morphology and physicochemical properties, together with their rapid digestive transit, have been proposed to impede potential symbionts to colonize digestive tracts (Engel and Moran Citation2013; Hammer et al. Citation2017; Hammer and Moran Citation2019). However, antibiotic suppression of gut bacterial communities in some caterpillar species can affect their nutrition (Kaiser et al. Citation2010) or affect their innate immunity (Krams et al. Citation2017).
Soil nutrients availability affects both plants and insect herbivores, either directly or indirectly (Panda and Khush Citation1995). In this context, biologically available nitrogen (N) is one of the major growth-limiting factors for both plants and insects (LeBauer and Treseder Citation2008; Tripathi et al. Citation2014; Chaudhary et al. Citation2018). N supply is often correlated with several benefits for plants, such as higher biomass and faster developmental cycles (Burns Citation1995; Leghari et al. Citation2016). Moreover, N is necessary for plants to synthetize metabolites with important anti-herbivore functions (Chen Citation2008; Anulika et al. Citation2016). From the herbivore’s perspective, N availability in the soil can modify plant’s nutritional value, digestibility and toxicity, thus reducing its quality as food (Dormann Citation2003). High N fertilization has also been proposed to enrich insect gut microbiota, coinciding with higher larval weight and insecticide tolerance (Hu et al. Citation2022). AMF-modulated improved capacity of the host plant to acquire N from the soil can therefore have cascading effects on insect herbivores (García-Gómez et al. Citation2021).
Nitrogen availability has been previously reported to interfere with MIR against pathogens (Sanchez-Bel et al. Citation2016), but its impact on MIR against pests and whether insect symbionts and plant symbionts can be functionally linked via the plant are still unknown. Our study aims to fill this gap by investigating the influence of N availability and AM symbiosis on the performance of the generalist chewing insect Spodoptera exigua (Hübner) (Lepidoptera: Noctuidae), using two different species of AMF. We hypothesized that N availability and AMF interact in their influence on plant nutrition and defenses, thus reducing the performance of insect herbivores. We explored the potential underlying mechanism through the characterization of the insect microbiota, the plant defense and the plant nutritional value.
2. Materials and methods
2.1. Experimental design
A completely randomized 3 × 2 factorial design was conducted to study the interaction between two factors: AMF and nitrogen availability. We used two different AMF species. Thus, the factor ‘AMF’ had three levels: non-inoculated plants (Ni), plants inoculated with Funneliformis mosseae (Fm) and plants inoculated with Rhizophagus irregularis (Ri). The factor ‘N’ had two levels: 0% (N0) and 100% (N100, regular growing conditions), to test the effect of N deficiency and N-full availability. This resulted in six experimental treatments: (1) NiN100, (2) FmN100, (3) RiN100, (4) NiN0, (5) FmN0, (6) RiN0. Eight replicates per treatment were established.
2.2. Fungal materials, plant materials and growing conditions
Two species of AMF were used to conduct this experiment. F. mosseae BEG12 (International Bank of Glomeromycota, https://www.i-beg.eu/cultures/BEG12.htm) was obtained and inoculated as described by Rivero et al. (Citation2021). Rhizophagus irregularis MUCL 57021 in vitro culture, spores extraction and inoculation were performed as described by Minchev et al. (Citation2021).
Maize seeds (Zea mays hybrid DK-2061) were surface disinfected by immersion in 5% NaHClO during 10 min, followed by multiple sterile water washes. Disinfected seeds were sown in 1.1 L pots filled with 1 L of nutrient-poor sandy soil (BVB, The Netherlands) sterilized by gamma irradiation. AMF inoculation was performed during sowing. Seed germination failed for 1 replicate of treatments FmN0 and RiN0 (n = 7), while germination succeeded for all NiN100, FmN100, RiN100 and NiN0 replicates (n = 8), giving 46 plants in total. Plants were maintained in a greenhouse at 25/16°C with a diurnal photoperiod of 16/8 h and watered twice a week with a 1 mM in phosphorous and N-full strength (20 mM) Long Ashton nutrient solution (Hewitt Citation1966) for 3 weeks. After this period, plants were watered with either N-full strength Long Ashton solution (N100, normal growing conditions) or N-free Long Ashton solution (N0, N deprivation growing conditions) depending on their treatment group.
2.3. Herbivory bioassay
Nine weeks after sowing, Ni, Fm and Ri plants grown under the different N fertilization regimes were challenged with S. exigua caterpillars. Eggs were provided by Dr Salvador Herrero Sendra of the Biotechnological Control of Pest Laboratory of the Universidad de Valencia, Spain. S. exigua larvae were reared on artificial diet (Greene et al. Citation1976) without antibiotics until they reached the L3 instar. The fifth leaf of each plant was harvested as a basal level of reference before herbivory (T0), frozen in liquid nitrogen and stored at −80°C. Then, two L3 larvae were placed in the sixth leaf of each plant after recording their initial weight. Clip-cages of 30 mm Ø were used to limit the movement of caterpillars. Clip-cages were moved across the same leaf every day to ensure food accessibility and herbivory was maintained for 5 days. The survival and weight of the caterpillars were daily recorded. At the end of the bioassay, surviving larvae were collected and preserved at −80°C to characterize their gut bacterial communities.
2.4. Plant physiological measurements, harvest and mycorrhiza quantification
Before (T0) and after 5 days of continuous herbivory, a portable chlorophyll CL-01 meter was used on the third fully expanded leaf of each plant to measure the relative chlorophyll content. The measurements were collected on three different points of the lamina (upper, medium and bottom) and their average was used. After 5 days of continuous herbivory, plant shoot and root fresh weight were recorded. The attacked leaf (local leaf, LL) and the immediately upper leaf (systemic leaf, SL) of each plant were harvested as after herbivory samples and preserved in liquid nitrogen and stored at −80°C. Roots were harvested, washed, cleared in KOH (10%) and stained with 5% ink (Lamy, Germany) in 2% acetic acid (García et al. Citation2020). The percentage of root length colonized by AMF was determined according to the gridline intersection method (Giovannetti and Mosse Citation1980).
2.5. Spodoptera exigua DNA extraction
Larvae were surface-disinfected with 75% ethanol during 90 s, rinsed with sterile water and the whole gut, including the gut content, was dissected. Total DNA was then extracted using the DNeasy Blood & Tissue Kit (Qiagen), following the manufacturer’s instructions with a few modifications. In brief, lysis of the samples with Proteinase K was left overnight and final elution volume (buffer AE) was 100 µL. DNA extracts were stored at −20°C until further processing.
DNA extracts were amplified using the pair of primers S-D-Bact-0341-b-S-17 (5′-CCTACGGGNGGCWGCAG-3′) and S-D-Bact-0785-a-A-21 (5′-GACTACHVGGGTATCTAATCC-3′) (Herlemann et al. Citation2011). The amplicon sequencing protocol targets the 16S rRNA gene V3 and V4 regions (465 bp), with the primers designed surrounding conserved regions. Molecular identification (MID) tags were incorporated into the primers in order to generate uniquely MID-tagged PCR products for each amplification. DNA libraries were generated using a limited cycle PCR: initial denaturation at 95°C for 5 min, followed by 25 cycles of annealing (95°C 40 s, 55°C 2 min, 72°C 1 min), extension at 72°C for 7 min. PCR success was checked through gel electrophoresis (2%) and purifiied using MagSi-NGSprep Plus beads (Steinbrenner Laborsysteme GmbH, Wiesenbach, Germany).
After DNA amplification, Illumina sequencing adaptors (Nextera XT index kit v2) were added to the amplicon. The adaptor ligation was conducted by PCR (9 cycles of annealing, following the same sample annealing temperature previously described). Ligation success was confirmed by gel electrophoresis and DNA concentrations were measured using a Qubit fluorometer (Life Technologies, Carlsbad, USA). Samples were then combined into 40 µL pools containing equimolar concentrations of 100 ng. Pooled DNA was purified and selected by size using MagSi-NGSprep Plus beads (Steinbrenner Laborsysteme GmbH, Wiesenbach, Germany) and sequencing was performed at the core facility of Fraunhofer IZI (Leipzig).
2.6. Metabarcoding library filtering and taxonomic classification
Prior to filtering of metabarcoding libraries, the quality of the libraries was checked using FastQC (Andrews Citation2010) on, both, forward and reverse reads (previously merged in one fastq file). Next, we merged complementary forward and reverse reads using PEAR (Zhang et al. Citation2014), with a minimum quality score of 30 (Phred score). Unassembled reads were discarded and successfully merged reads were joined for subsequent analyses in a single fastq file using the sed command. The remaining filtering steps were performed using vsearch v2.8.1 (Rognes et al. Citation2016). The fastx_filter command was used to remove the potential sequencing mistakes by quality rate filter (fastq_maxee = 1), producing and output file in fasta format. Next, dereplication (only unique sequences were kept) was done applying the derep_fulllength command and we removed frequency errors by denoising (UNOISE3 algorithm) using cluster_unoise command. This was followed by the removal of insertions and deletions (filter by length) using the fastx_filter command, allowing amplicon lengths from 250 to 350 bp, and the removing of potential chimeras ‘de novo’ using the command uchime3_denovo. At this point, we obtained a set of biological sequences of bacterial communities that correspond to our communities, which the Amplicon Single Variants (ASV), that are stored in a fasta file. These sequences were mapped to the reads using the search_exact command.
The sequences were classified against Silva database (v138.1) (Quast et al. Citation2012) using the command classify.seqs() from software mothur (Schloss et al. Citation2009; Schloss Citation2020), setting a bootstrap threshold of 80%. Moreover, we performed a classification against NCBI using BLAST algorithm (Johnson et al. Citation2008) and R package taxonomizr (https://github.com/sherrillmix/taxonomizr/; last accessed on October 2021) to obtain the taxonomy table.
The ASV table was filtered to remove the samples with less than 1000 reads using prune_samples() from phyloseq R package. Filtered data was then normalized by CSS (cumulative-sum scaling) to avoid technical bias related to different sequencing depth using the command phyloseq_transform_css(), which is available in metagenomeSeq Bioconductor R package. Potential contaminants were also removed using the subset_taxa() function from phyloseq. Filtered data (ASV table, fasta file and taxonomy table) were used for subsequent biodiversity analyses. The raw dataset generated and analyzed during the current study was submitted to the NCBI Sequence Read Archive (SRA) under the BioProject PRJNA843567 (16S rRNA genes).
2.7. Microbiota biodiversity analyses
A rarefaction curve was constructed to assess the sequencing depth and the species richness of each sample. Individual rarefaction curves were then diluted to obtain the inter-treatment curves. Rarefaction curves were generated using the function rarecurve() from vegan R package. Relative Read Abundance (RRA) metric, the percentage of reads that are present in each treatment, was used to characterize S. exigua bacterial composition across treatments (Deagle et al. Citation2019). This metric was computed using a customized R script and phyloseq R package (Wickham et al. Citation2021).
We calculated the differences in ASV richness (alpha diversity) using two different indices (Chao1 and Shannon), applying the function estimate_richness() from phyloseq R package. To test the significance of alpha diversity measures across treatments, we applied a Kruskal–Wallis using the function kruskal.test(). Finally, we used adonis2() function with Bray–Curtis dissimilarity (PERMANOVA using distance matrices, vegan R package) to test whether microbial composition differed between treatments. PCoA was used to visualize separations among treatments.
2.8. Benzoxazinoids quantification
Before and after herbivory (LL) samples were used to analyze foliar levels of 2,4-dihydroxy-7-methoxy-2H-1,4-benzoxazin-3(4H)-one (DIMBOA) and 6-methoxy-2-benzoxazolinone (MBOA) and 2-hydroxy-4,7-dimethoxy-1,4-benzoxazin-3-one glucoside (HDMBOA-glc), major secondary metabolites involved in maize defense. Three biological replicates per treatment were analyzed, each replicate consisting of a pool of 2 samples from independent plants. DIMBOA, MBOA and HDMBOA-glc were extracted from the samples according to Ahmad et al. (Citation2011). A 50 mg of freeze-dried leaf material were homogenized in 1 mL of extraction buffer (methanol/acetic acid; 49/1; v/v), sonicated for 10 min and centrifuged (12,600 rpm, 10 min). The extraction buffer contained N-Benzoyl-L-tyrosine (100 ppb) as an internal standard for BXs quantification (Sasai et al. Citation2009). Supernatants were recovered, filtered through 0.22 µm cellulose filters (Regenerated Cellulose Filter, 0.20 μm, 13 mm D. pk/100; Teknokroma) and collected for analysis. Ten µl of extracted solution were injected into an Acquity UPLC system coupled to a XEVO TQD Triple quadrupole instrument, UPLC_MS/MS (Waters, Milford, MA, USA). Chromatographic separation was conducted through a UPLC Kinetex EVO C18 column (2.6 μm particle size; 100 A, 50 × 2.1 mm; Phenomenex) and the mobile phase consisted of a mixture of solvent A (pure water) and solvent B (methanol/isopropanol/acetic acid; 3800/200/1; v/v). The solvent gradient was initiated with isocratic conditions at 10% B for 3 min, ramped to 90% B for 2 min and then ramped back to 10% B for 3 min.
Retention times of DIMBOA, MBOA and HDMBOA-glc were determined from synthetic standards (provided by Professor Jurriaan Ton). Standard curves, which showed linearity between peak area and compound concentration, and N-Benzoyl-L-tyrosine were used to determine the content of the three compounds (ng/g dry weight).
2.9. Anti-herbivory related enzymatic activities
Six independent biological replicates of leaf samples after herbivory (LL) were used for leucyl aminopeptidase (LAP) and polyphenol oxidase (PPO) activity assays. LAP activity assay was performed according to Gamir et al. (Citation2021). For protein extraction, 50 mg of grinded fresh leaf material were homogenized by vortex with 900 µL of extraction buffer 50 mM Tris-HCl, 0.5 mM MgCl2, pH 8. Homogenized samples were centrifuged at 10,000 g (4°C) for 20 min. Supernatant was recovered and used as protein extract. For LAP activity, 40 µL of protein extract and 200 µl of enzyme substrate (3 mM Leu-p-nitroanilide in 50 mM Tris-HCl, 0.5 mM MgCl2, pH 8) were incubated at 37°C for 15 min. The absorbance was measured in a 96-well microtiter plate at 410 nm.
PPO activity assay was carried out as described by Shikano et al. (Citation2018). For protein extraction, 50 mg of grinded fresh leaf material were homogenized by vortex with 1.25 mL of ice-cold extraction buffer (5% polyvinylpyrrolidone in 0.1 M potassium phosphate buffer, pH 7) and incubated in ice for 5 min. Samples were centrifuged at 11,000 g (4°C) for 10 min. Supernatant was recovered and used as protein extract. To measure PPO activity 5 µL of protein extract and 200 μL of 3 mM caffeic acid in 0.1 M potassium phosphate buffer were added to a 96 well microtiter plate and after 10 min the absorbance was measured at 450 nm.
Total protein concentration for each sample was determined by the Bradford protein assay (Bradford Citation1976). LAP and PPO activities were expressed as Absorbance/mg of protein.
2.10. Carbon and nitrogen plant content
Foliar C/N ratio was used as proxy of plant nutritional quality to insect herbivores. Total carbon (C) and total nitrogen (N) before and after herbivory (SL) were determined using an Elemental Analyzer (LECO TruSpec CN) according to standard procedures at the Ionomic Laboratory of the Technical Services of the Estación Experimental del Zaidín (EEZ-CSIC) in Granada, Spain. Four independent biological replicates were analyzed per treatment, each one consisting of a pool of 2 freeze-dried leaf samples from independent plants.
2.11. Amino acid profile
Foliar accumulation of Protein-Bound Amino Acids (PBAAs) and Free Amino Acids (FAAs) after herbivory (LL) were determined by gases chromatography – mass detection (GC-MS). Powdered freeze-dried H+ samples were divided into two subsets of 5 mg, one for FAAs analysis and one for PBAAs analysis. FAAs were extracted with 1 mL acetonitrile 25% in HCl 0.01 M (1:3, v:v). Using other subsets, proteins were hydrolyzed into their protein-bound amino acids with 500 μL of 4 M methanesulfonic acid at 150°C for 2 h. Either FAAs or PBAAs extracts were transferred into new vials and pH was confirmed to be between 1.5 and 5.0. Extraction and derivatization were then conducted by using EZ:faast GC-ms kit for free amino acids or protein hydrosilates (Phenomenex Torrance, California, USA) according to the manufacturer’s specifications. Norvaline at 200 nmol·mL−1, used as an internal standard (IS), and standards for each physiological amino acids at 200 nmol·mL−1 were provided by the kits. Extracts were then analyzed by an Agilent 7890B gas chromatograph coupled to an Agilent 7000C mass spectrometer Triple Quad (Agilent Technologies, Santa Clara, CA, USA). We used an autosampler (Gerstel, Mühleim an der Ruhr, Germany) to inject 2 µL of extracted solution into the column (Zebron ZH-5HT inferno, 30 m × 0.25 mm × 0.25 µm, Phenomenex, Torrance, CA, USA) in splitless mode; the injector was heated at 250°C. The following temperature program was used: a temperature ramp from 110°C to 320°C at 15°C/min with a final hold time of 7 min. Helium was used as a carrier gas, with a constant flow of 1.2 mL/min. The transfer line was set at 320°C. We used Electron Ionization with electron energy of −70 eV and an ionization source at 230°C. The scan range was 45–450 m/z with 3.7 scans/s.
Compounds were then identified based on the mass spectra provided by the EZ:faast kit and NIST (National Institute of Standards and Technology) libraries. The IS was used to quantify each identified amino acid. The peak area of each amino acid was multiplied by the concentration of the IS and divided by the peak area of the IS.
2.12. Statistical analyses
All statistical analyses were conducted using the ‘R’ software version 4.1.0 (R Development Core Team Citation2021). Figures were obtained using the package ggplot2 (Wickham Citation2011). Treatment effects on larval survival were assessed by comparing the survival curves using the Kaplan-Meier estimator (Kaplan and Meier Citation1958). Survival distribution comparison between treatments was performed using the non-parametric Log-rank test (Mantel–Cox). Survival analyses were performed using survival and survminer R packages. Multifactorial ANOVAs and post hoc tests were conducted to analyze the impact of the studied factors on the larval weight gain, plant growth, plant defenses, total and individual HAAs and total FAAs. Model validations were performed using Shapiro–Wilk and Levene’s tests. If required, data was transformed to meet the model assumptions. Spearman correlation between relative chlorophyll content and foliar N content was calculated using the cor.test() function of stats R package.
3. Results
3.1. Impact of N and AMF on plant growth and relative chlorophyll content
Visible differences in the aspect of the plants according N supply (N100, regular growing conditions or N0, N-deficiency) and AMF inoculation (Ni, non-inoculated plants; Fm, F. mosseae-inoculated plants or Ri, R. irregularis-inoculated plants) were observed at the beginning of the herbivory bioassay (T0). Plants supplied with N100 were bigger, greener and developed more leaves than N0 plants and mycorrhizal N100 plants were bigger than NiN100 plants ((A), Supporting ).
Figure 1. Impact of N and AMF on plant growth and relative chlorophyll content. (A) Picture of plants from each treatment, (B) shoot fresh weight and (C) relative chlorophyll content before herbivory. Non-inoculated control plants (Ni), plants inoculated with Funneliformis mosseae (Fm), or plants inoculated with Rhizophagus irregularis (Ri) and supplied with regular nitrogen (N100) or deprived of nitrogen (N0). Dots represent raw data, boxes represent the interquartile range, inner lines in bold represent the median, black ‘+’ represents the mean, whiskers represent maxima and minima within 1.5 times the interquartile range and empty dots represent outliers. Treatments not sharing a letter are statistically different based on two-way ANOVA followed by Tukey-Kramer’s (HSD) post hoc for unbalanced sample size (p < 0.05, n = 7–8).
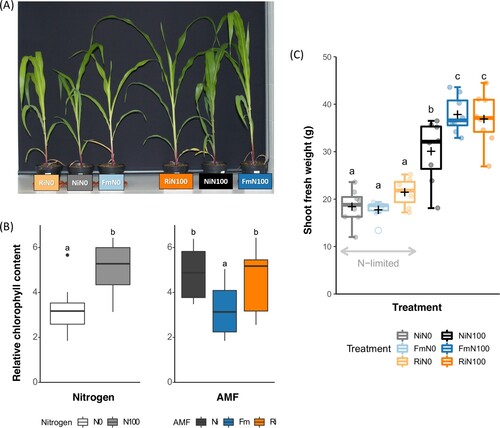
Chlorophyll meter measurements at T0 confirmed the visual differences observed in the leaf greenness of the plants. No significant effects of N-AMF interaction were found according to multifactorial ANOVA (F = 2.79, p = 0.07), while main effects of N (F = 160.65, p < 0.05) and AMF (F = 37.62, p < 0.05) on the relative chlorophyll content were observed. On average, the relative chlorophyll content of N100 plants was higher than in N0 plants ((B)). Ni plants showed a higher chlorophyll content than Fm plants, while no differences between Ri and Ni plants were found ((B)).
At harvest, the shoot fresh weight (SFW) of plants was compared across treatments. N-AMF interaction on plant growth was significant according to multifactorial ANOVA (F = 144.13, p < 0.05). N deprivation led to a reduction of the SFW of the plants and AMF inoculation promoted plant growth under regular growing conditions (N100) ((C)).
Mycorrhizal symbiosis was well-established in all AMF inoculated plants and both N levels, with values ranging 46% and 53% of root length colonized by the fungi (53% and 51% for Fm, and 46 and 50% for Ri at N0 and N100 respectively). Lack of mycorrhizal colonization was confirmed in non-inoculated plants.
3.2. Impact of N and AMF colonization on S. exigua performance
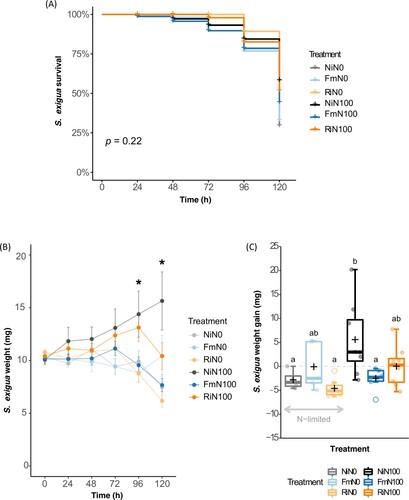
Larval survival and weight were compared across treatments to study the effects of the AMF inoculation and N levels on S. exigua performance. No differences in the survival curves were detected across treatments ((A)). Treatments gradually affected S. exigua weight and significant effects were observed after 96 h of herbivory. A continuous increase in weight was only observed for larvae fed on NiN100 plants ((B)). Indeed, at the end of the bioassay (120 h), both factors and their interaction significantly affected the larval weight gain (F = 4.28, p < 0.05). Caterpillars fed on NiN0 plants showed a reduced weight gain compared to larvae fed on NiN100 plants and AMF inoculation did not influence S. exigua weight gain under N deficiency ((C)). FmN100 significantly reduced the weight gain of S. exigua compared to Ni100, while this reduction was not significant for RiN100 ((C)).
Figure 2. Impact of N and AMF on S. exigua performance. (A) Larval survival (B) absolute larval weights across time (C) S. exigua weight gain after 5 days feeding on maize plants. Non-inoculated control plants (Ni), plants inoculated with Funneliformis mosseae (Fm), or plants inoculated with Rhizophagus irregularis (Ri) and supplied with regular nitrogen (N100) or deprived of nitrogen (N0). Dots represent raw data, boxes represent the interquartile range, inner lines in bold represent the median, black ‘+’ represents the mean, whiskers represent maxima and minima within 1.5 times the interquartile range and empty dots represent outliers. Black ‘*’ represents statistical significance. Treatments not sharing a letter are statistically different based on two-way ANOVA followed by Tukey-Kramer’s (HSD) post hoc for unbalanced sample size (p < 0.05, n = 4–8).
3.3. Impact of N and AMF colonization on S. exigua microbiota
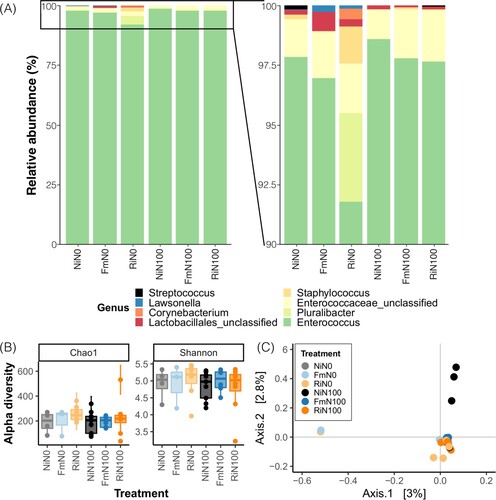
To determine whether changes in larval performance are associated with differences in S. exigua microbiota, the gut bacterial composition of the caterpillars was compared across treatments. Regarding 16 S rRNA gene library quality, we obtained 3,374,278 raw reads, out of which, 1,683,244 were retained (Supporting Table 1). After quality filtering steps and contaminant removal, the library retained 7,681 ASVs, which corresponded to gut bacterial community of S. exigua. Out of the 49 S. exigua samples, 39 were kept (>1000 reads, Supporting Table 2). Regarding the completeness of the sampling, rarefaction curves reached a plateau (Supporting ).
Practically the totality of filtered ASVs (99.5%) was assigned to Firmicutes phylum, representing 100% of the relative abundance of phyla observed in larvae fed on plants grew under regular N conditions and NiN0 plants, 99% in larvae fed on FmN0 and 96% in larvae fed on RiN0. Further analyses at the genus level revealed a low diversity and great similarity in S. exigua gut bacterial composition across treatments (). Enterococcus spp. represented more than 90% of the relative abundance of observed genera in S. exigua gut microbiota, independently of the treatment ((A)). Alpha diversity did not show differences between treatments (Shannon: R2 = 1.47, p = 0.92; Chao1: R2 = 3.30, p = 0.65; (B)). Principal coordinate analysis (PCoA) was used to visualize S. exigua gut community composition and structure by clustering individual samples ((C)). PERMANOVA confirmed that N and AMF inoculation did not influence the bacterial community composition of S. exigua (R2 = 0.12, F = 0.99, p = 0.30). Thus, results suggest that neither N nutrition nor mycorrhizal symbiosis modify S. exigua microbiota.
3.4. Impact of N and AMF colonization on plant defenses
Foliar accumulation of DIMBOA, MBOA and HDMBOA-glc before and after herbivory was compared across treatments to determine the impact of N and AM symbiosis on the accumulation of these defensive compounds.
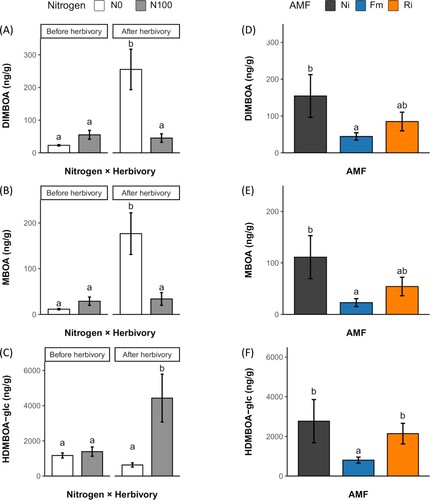
N availability influenced the plant defense responses to S. exigua chewing. DIMBOA and MBOA accumulation was induced in response to herbivory only under N deprivation, while HDMBOA-glc was induced only under N availability ((A–C)). No significant interactions between AMF and the rest of the analyzed factors were found (Supporting Table 3). Foliar accumulation of DIMBOA, MBOA and HDMBOA-glc was similar for Ni and Ri, while Fm tend to limit the accumulation of these compounds ((D–F)).
Figure 4. Effects of nitrogen availability, AMF and herbivory by S. exigua on DIMBOA, MBOA and HDMBOA-glc foliar accumulation. (A) Interactive effect of nitrogen availability and herbivory on the foliar accumulation of DIMBOA, (B) MBOA and (C) HDMBOA-glc in plants supplied with regular nitrogen (N100) or deprived of nitrogen (N0). (D) Main single effect of AMF inoculation on the foliar accumulation of DIMBOA and (E) MBOA and (F) HDMBOA-glc in non-inoculated plants (Ni), plants inoculated with Funneliformis mosseae (Fm) or plants inoculated with Rhizophagus irregularis (Ri). Bars represent the mean ± standard error. Treatments not sharing a letter are statistically different based on multifactorial ANOVA followed by Tukey’s (HSD) post hoc (p < 0.05, n = 3).
To further explore whether the negative impact of F. mosseae on the insect performance under regular N fertilization is associated with changes in plant defenses, we also analyzed the activity of anti-herbivory defense-related enzymes LAP and PPO. No significant differences were found in LAP or PPO (Supporting ).
3.5. Impact of N and AMF colonization on plant nutritional status
Three different parameters, plant C/N ratio, PBAAs and FAAs were used as a proxy of plant nutritional quality to insect herbivores.
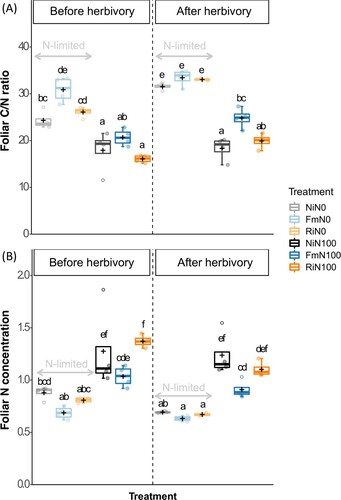
3.5.1. Plant C/N ratio
Significant differences between plant C/N ratios before and after S. exigua herbivory were observed across treatments. N levels influenced the plant C/N ratio. The ratio was significantly higher for N0 plants ((A)). AMF effects on plant C/N ratio depended on the interaction of AMF levels with the different levels of N and herbivory (F = 4.09, p < 0.05). After herbivory, FmN100 had significantly higher C to N after herbivory than NiN100 plants, despite not presenting significant basal differences ((A)). This increased C/N ratio was due to a reduction of the N content of FmN100 maize compared to NiN100 ((B)). Measurements obtained with the chlorophyll CL-01 meter have been previously proposed as a proxy of the plant nitrogen content (Kalaji et al. Citation2016). Our results showed a positive correlation between total plant nitrogen and the relative chlorophyll content (R = 0.88, Supporting ).
Figure 5. Foliar C/N and N concentration before and after herbivory by S. exigua. (A) Carbon to nitrogen and (B) N concentration before or after herbivory by S. exigua, in non-inoculated control plants (Ni), plants inoculated with Funneliformis mosseae (Fm), or plants inoculated with Rhizophagus irregularis (Ri) and supplied with regular nitrogen (N100) or deprived of nitrogen (N0). Boxes represent the interquartile range, inner lines in bold represent the median, black ‘+’ represents the mean, whiskers represent maxima and minima within 1.5 times the interquartile range and empty dots represent outliers. Treatments not sharing a letter are statistically different based on multifactorial ANOVA followed by Tukey’s (HSD) post hoc (p < 0.05, n = 4).
3.5.2. Protein-bound amino acids and total protein
PBAAs content was lower in plants grown under N deficiency ((A)). AMF effects on PBAAs depended on the interaction between AMF and N levels (F = 10.48, p < 0.05). No significant differences in total PBAAs between mycorrhizal and non-mycorrhizal plants were detected independently on N levels, although levels in FmN100 were lower than RiN100 ((A)). In N100 plants, the leaf protein content of Fm plants was significantly lower than the total protein content of Ni and Ri plants ((B)). Effects of N, AMF and AMF × N interaction on individual PBAAs and their patterns of accumulation in leaves across treatments are shown in Supporting Table 4. Glutamic acid + phenylalanine was the most abundant PBAAs in all treatments, representing 39.50% of the total PBAAs detected. Alanine was lower in FmN100 than in NiN100 (Supporting Table 4).
Figure 6. Total protein-bound amino acids, free amino acids and proteins in attacked leaves. Effects of N × AMF on the accumulation of (A) total protein-bound amino acids and (B) free amino acids and (C) total protein content. Non-inoculated control plants (Ni), plants inoculated with Funneliformis mosseae (Fm), or plants inoculated with Rhizophagus irregularis (Ri) and supplied with regular nitrogen (N100) or deprived of nitrogen (N0). Dots represent raw data, boxes represent the interquartile range, inner lines in bold represent the median, black ‘+’ represents the mean and whiskers represent maxima and minima within 1.5 times the interquartile range. Treatments not sharing a letter are statistically different based on two-way ANOVA followed by Tukey-Kramer’s (HSD) post hoc for unbalanced sample size (p < 0.05, n = 7–8).
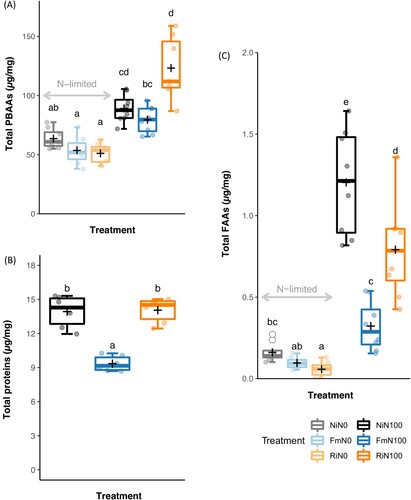
3.5.2. Free amino acids
Levels of FAAs significantly differed across treatments. Plants supplied with N displayed higher amounts of FAAs than N-deprived plants and NiN100 plants showed the highest values ((C)). The effect of AMF on FAAs accumulation depended on N levels (F = 14.73, p < 0.05), but mycorrhizal plants showed less FAAs than Ni plants ((C)). FmN100 had less FAAs than NiN100 and RiN100 plants, showing similar quantities to those found for N-deficient non-mycorrhizal plants ((C)). Aspartic acid + asparagine and alanine were the most represented FAAs and non-proteinogenic amino acids were not detected in any sample (Supporting Table 5).
4. Discussion
This study aimed to investigate the influence of nitrogen fertilization and mycorrhizal symbiosis on plant growth and resistance to pests and to explore the potential underlying mechanisms. Our results show that beneficial effects on plant growth and protection against pests conferred by AMF depend on the availability of N in the soil.
In accordance with the importance of N availability on plant’s growth and development, complete deprivation of N led to a reduction in maize growth. Both AMF species promoted maize growth but only under full N availability. AM symbiosis improves the capacity of the host plant to acquire mineral nutrients from the soil but, under total N deprivation, plants and AMF are likely to compete for the N resources. Growth promotion is of interest when studying mycorrhiza-induced resistance because trade-offs between growth and defense usually occur in plants (Koricheva et al. Citation2009). In our study, growth promotion by F. mosseae coincided with a reduction of S. exigua weight gain, suggesting that AMF-induced maize growth did not compromise plant protection by F. mosseae against S. exigua. A similar trend was observed for Rhizophagus irregularis. In this context, F. mosseae can double-benefit sustainable agriculture, promoting both plant growth and defense.
Several factors have been proposed as potential drivers of larval performance, including insect gut microbiota, plant defenses and plant nutritional value (War et al. Citation2012; Bala et al. Citation2018; Frago et al. Citation2020). Here we explore their contribution to the phenotype observed as an attempt to decipher mechanisms underlying the impact of N supply and AM symbiosis on the performance of S. exigua.
In coherence with previous studies about microbiomes of Lepidoptera, including Manduca sexta (Brinkmann et al. Citation2008), Spodoptera littoralis (Chen et al. Citation2016) or S. exigua (Gao et al. Citation2019), our results show that gut microbiota of late instar larvae of S. exigua fed on maize is low in abundance and in diversity and that Enterococcus spp. represented its major constituent. Several studies have reported the impact of diverse factors (i.e. diet or environment) on the gut microbial communities in caterpillars, including S. exigua (Hannula et al. Citation2019; Martínez-Solís et al. Citation2020; Mason et al. Citation2020). However, it remains unclear whether the alteration of the microbial community of caterpillars can influence their performance (Voirol et al. Citation2018; Mason et al. Citation2020). Our results show that gut bacterial communities of S. exigua remained unchanged across treatments regardless of the larval performance, discarding insect microbiota as an explanatory factor for MIR observed.
To explore the potential mechanisms underlying herbivore performance among treatments, we evaluated changes in several plant defensive traits. The plant anti-herbivory arsenal is complex and involves secondary metabolites and defensive proteins, including peptidases and phenol oxidases (War et al. Citation2012). Benzoxazinoids (BXs) are important N-containing defensive metabolites constitutively produced by Poaceae (Morant et al. Citation2008; Ahmad et al. Citation2011; Zhou et al. Citation2018). Among others, maize BXs with anti-feedant and insecticidal activities include HDMBOA-glc, DIMBOA and the product of its spontaneous degradation, MBOA (Niemeyer Citation2009; Dixon et al. Citation2012). DIMBOA has been reported to reduce the survival, food consumption and weight gain of S. exigua (Rostás Citation2007). HDMBOA, which is released from its corresponding glucoside during herbivore attack, has also been observed to cause deterrent effects to S. littoralis and Spodoptera frugiperda (Glauser et al. Citation2011). Plants severely deprived of nutrients have been previously reported to alter the production of defensive secondary metabolites (Gershenzon Citation1984; Chishaki and Horiguchi Citation1997). In this study, BXs accumulation in damaged leaves is strongly influenced by N availability in the soil. Deprivation of N led to an increase of DIMBOA and MBOA, while N supply enhanced HDMOA-glc supporting that N availability is a determining factor in the defense response of maize plants against herbivores. Patterns of induction of the analyzed anti-herbivore compounds cannot explain the differences observed in S. exigua performance across treatments. These results suggest that, even though DIMBOA, HDMBOA-glc and MBOA may mediate maize responses to S. exigua attack in relation to N availability, other factors that are affected by N-AMF interaction contribute to determine the caterpillars’ performance. Besides BXs, enzymes such as leucyl aminopeptidase and polyphenol oxidase are important players in plant defense against herbivores. These enzymes can impair insect nutrition by reducing the availability of essential nutrients in the diet (Felton et al. Citation1989; Chen Citation2008; Zhu-Salzman et al. Citation2008). Our results show that these enzymatic activities in herbivore-attacked leaves were not significantly different in mycorrhizal and non-mycorrhizal plants. Overall, our results do not support that enhanced levels of plant defenses are responsible for the reduced performance of the larvae observed in Fm plants under full N nutrition.
According to the nitrogen limitation hypothesis (White Citation1993), phytophagous insects are positively influenced by the N content of plant tissues. A positive correlation between plant N and herbivore’s performance has been observed for several insects, including caterpillars (Mattson Citation1980; Lu et al. Citation2004; Cornelissen and Stiling Citation2009; Han et al. Citation2014). In addition to elemental limitation per se, stoichiometric mismatches between scarce and abundant nutrients have been shown to impair herbivore’s growth (Filipiak and Weiner Citation2017). Indeed, it is well established for numerous herbivores that high C/N ratios reduce the dietary quality of plants, negatively affecting their performances (Srinivasa Rao et al. Citation2012; Xie et al. Citation2015; Dáder et al. Citation2016; Filipiak and Weiner Citation2017).
In agreement with this, our results show that deprivation of N negatively affects the nutritional quality of maize plants for S. exigua by decreasing the total N content and increasing the C/N ratio of the leaves. Full N-supply leads to a better food quality (lower C/N ratio) which may explain the better performance of S. exigua, when feeding on these plants. In addition, relative chlorophyll content positively correlated with plant N. Thus, higher relative chlorophyll content and increased plant N were associated with an improved herbivore performance, as previously described by Ho and Pennings (Citation2013). Plants inoculated with F. mosseae differed in their ability to lower the C/N ratio compared to Ni- and Ri-plants, but this ability was N-dependent, matching observed effects in S. exigua weight gain. Under full N supply, Fm-colonized plants had a lower concentration of foliar N than non-inoculated plants. This might be due to nutrient dilution caused by the promoted growth of mycorrhizal plants (Supporting Table 6). This lower foliar concentration of N in Fm plants correlate with the reduced weight observed in S. exigua larvae fed on these plants, pointing to altered C/N of Fm plants as a potential mechanism mediating the decreased larval performance.
Moving from elemental composition to biological functionality, N is an essential constituent of proteins, one of the most important biomolecules for insects (Raubenheimer and Simpson Citation1993; Simpson and Raubenheimer Citation1995; Behmer Citation2009). Optimal concentrations of proteins have been reported to have beneficial effects on diverse aspects of caterpillar’s health, including biomass gain, development and immunity (Broadway and Duffey Citation1986; Cotter et al. Citation2011; Truzi et al. Citation2021). In this experiment, under N availability, plant inoculation with F. mosseae led to a reduction in the foliar concentration of soluble proteins, which was associated with a decreased weight gain of S. exigua caterpillars. Furthermore, absorption of N in insect’s gut is carried through simple molecules, such as free amino acids (Brodbeck and Strong Citation1987). Thus, their direct assimilation can save metabolic costs associated with proteolysis (Brodbeck and Strong Citation1987) and improve herbivore’s performance (Calatayud et al. Citation2002; Ximénez-Embún et al. Citation2017). Moreover, FAAs have been observed to act as phagostimulants for diverse phytophagous insects, including late instar caterpillars of S. exigua (Allsopp Citation1992; Showler Citation2001; Calatayud et al. Citation2002). These observations are supported by results obtained in this study, in which larval weight is negatively affected by lower concentrations of FAAs in N-deprived plants and Fm plants supplied with full N. Overall, MIR against S. exigua seems to be linked to a reduced nutritional quality (increased foliar C/N and reduced concentration of N, soluble proteins and FAAs in leaves) of Fm plants supplied with full N.
Our study confirms that abiotic factors, such as nutrient availability, influence the effect of plant-AMF symbiosis on plant performance and resistance to herbivores. Plant growth promotion and protection were only observed under full N, and the degree of protection differed among the AMF species tested. These findings illustrate that N levels can affect MIR functionality and support the compatibility of AMF with conditions used in agriculture, such as nitrogen fertilization, potentially showing additive effects on crops performance. Nevertheless, further research in MIR context-dependency is needed to better predict AMF effects on crops under specific conditions for optimized crop management in the frame of sustainable agriculture.
Supplemental Material
Download MS Word (4.1 MB)Acknowledgments
This project has received funding from the European Union’s Horizon 2020 research and Innovation programme under grant agreement No 765290. The authors thank Dr Salvador Herrero Sendra for providing S. exigua eggs. The authors thank Advance Identification Methods GmbH for their advice and efficiency in the library sequencing and the Mutualised Platform for Environmental Genomics of the Institut de Recherche sur la Biologie de l'Insecte (IRBI), UMR 7261 CNRS, for the access to its facilities. The authors would also like to thank the Mutualised Platform for Environmental Genomics and the Chemical Ecology Platform of the Institut de Recherche sur la Biologie de l'Insecte for its services and facilities. The authors acknowledge Christophe Lucas for his advice on chemical experiments. The authors thank the Scientific Instrumentation Service (SCIC) of the Universitat Jaume I for their technical support.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Additional information
Funding
Notes on contributors
Beatriz Ramírez-Serrano
Beatriz Ramirez-Serrano is a doctoral student at the University of Tours, within the EU-ITN project MiRA (‘Microbe Induced Resistance to Agricultural Pests’). Currently, she conducts her research at the Insect Biology Research Institute (IRBI). Her research focuses on multi-way plant-microbe-insect interactions and specifically, microbe-induced resistance against pest.
Marina Querejeta
Marina Querejeta is a postdoctoral fellow in Insect Biology Research Institute (IRBI) in Tours, where she has been working since 2019. In the last years, she has focused on evolutionary and biodiversity analysis, mainly using environmental DNA approaches of microbial, insect and bird communities. Currently, she is studying the pollination patterns of wild bees in agroecosystems to protect and conserve their communities.
Zhivko Minchev
Zhivko Minchev is a doctoral student at Koppert Biological Systems within the EU-ITN project MiRA (‘Microbe Induced Resistance to Agricultural Pests’). His research interest is plant-microbe-insect interactions focusing on microbe-induced resistance against pest and pathogens.
Jordi Gamir
Jordi Gamir is an Early Career Scientist at the University Jaume I of Castellón, where he dissects the role of Damage-Associated Molecular Patterns in plant immunity. Currently, he holds an excellence grant from the Valencian regional research program since 2019.
Elfie Perdereau
Elfie Perdereau is a research engineer at the French National Center for Scientific Research (CNRS) since 2019. She manages the chemical ecology platform of the Insect Biology Research Institute -IRBI-, which is dedicated to the analysis of chemical mediators and metabolites of insects and/or the plants with which they are associated.
Maria J. Pozo
Maria J. Pozo is a biologist and scientific researcher at the Scientific Researcher at Spanish National Research Council (EEZ-CSIC, Granada, Spain). She is interested in how beneficial soil fungi – mostly arbuscular mycorrhizal fungi- boost plant immunity by inducing systemic resistance against pathogens and pests. She investigates the molecular mechanisms regulating plant defenses and the context dependency of the symbiosis impact on plant health. The final aim is to optimize mycorrhizal applications for sustainable crop protection.
Géraldine Dubreuil
Géraldine Dubreuil is an assistant professor at the University of Tours since September 2012. Her research, conducted at the Insect Biology Research Institute (IRBI), focuses on plant/insect/microorganism interactions, more precisely, how insect symbionts can influence their ability to manipulate their plant host.
David Giron
David Giron has several years of expertise in different aspects of entomological research, including nutrients acquisition and allocation strategies in insects and their consequences on life history traits. His integrative approach is at the interface between functional ecology and evolutionary ecology. The current focus of his research is to understand the adaptive mechanisms involved in plant-insect-microorganism relationships. In 2005, he joined the French National Center for Scientific Research (CNRS) and the Insect Biology Research Institute (IRBI), where he is now the director (since 2015).
References
- Ahmad S, Veyrat N, Gordon-Weeks R, Zhang Y, Martin J, Smart L, Glauser G, Erb M, Flors V, Frey M, et al. 2011. Benzoxazinoid metabolites regulate innate immunity against aphids and fungi in maize. Plant Physiol. 157:317–327. doi:10.1104/PP.111.180224.
- Allsopp PG. 1992. Sugars, amino acids, and ascorbic acid as phagostimulants for larvae of Antitrogus parvulus and Lepidiota negatoria (Coleoptera: Scarabaeidae). J Econ Entomol. 85:106–111. doi:10.1093/JEE/85.1.106.
- Andrews S. 2010. FastQC: a quality control tool for high throughput sequence data.
- Anulika NP, Ignatius EO, Raymond ES, Osasere OI, Abiola AH. 2016. The chemistry of natural product: plant secondary metabolites. Int J Technol Enhanc Emerg Eng Res. 4:1–9.
- Bala K, Sood AK, Pathania VS, Thakur S. 2018. Effect of plant nutrition in insect pest management: a review. J Pharmacogn Phytochem. 7:2737–2742.
- Balliu A, Sallaku G, Rewald B. 2015. AMF inoculation enhances growth and improves the nutrient uptake rates of transplanted, salt-stressed tomato seedlings. Sustainability. 7:15967–15981. doi:10.3390/su71215799.
- Bargaz A, Lyamlouli K, Chtouki M, Zeroual Y, Dhiba D. 2018. Soil microbial resources for improving fertilizers efficiency in an integrated plant nutrient management system. Front Microbiol. 9:1–25. doi:10.3389/fmicb.2018.01606.
- Behmer ST. 2009. Insect herbivore nutrient regulation. Annu Rev Entomol. 54:165–187. doi:10.1146/annurev.ento.54.110807.090537.
- Bhantana P, Rana MS, Sun XC, Moussa MG, Saleem MH, Syaifudin M, et al. 2021. Arbuscular mycorrhizal fungi and its major role in plant growth, zinc nutrition, phosphorous regulation and phytoremediation. Symbiosis. 84:19–37. doi:10.1007/s13199-021-00756-6.
- Biere A, Bennett AE. 2013. Three-way interactions between plants, microbes and insects. Funct Ecol. 27:567–573. doi:10.1111/1365-2435.12100.
- Biere A, Tack AJM. 2013. Evolutionary adaptation in three-way interactions between plants, microbes and arthropods. Funct Ecol. 27:646–660. doi:10.1111/1365-2435.12096.
- Borowicz VA. 1997. A fungal root symbiont modifies plant resistance to an insect herbivore. Oecologia. 112:534–542. doi:10.1007/s004420050342.
- Bradford MM. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 72:248–254.
- Brinkmann N, Martens R, Tebbe CC. 2008. Origin and diversity of metabolically active gut bacteria from laboratory-bred larvae of Manduca sexta (Sphingidae, Lepidoptera. Insecta). Appl Environ Microbiol. 74:7189–7196. doi:10.1128/AEM.01464-08.
- Broadway RM, Duffey SS. 1986. The effect of dietary protein on the growth and digestive physiology of larval Heliothis zea and Spodoptera exigua. J Insect Physiol. 32:673–680. doi:10.1016/0022-1910(86)90108-3.
- Brodbeck B, Strong D. 1987. Amino acid nutrition of herbivorous insects and stress to host plants. In: Barbosa P, Schultz J, editors. Insect outbreaks: ecological and evolutionary perspectives. New York: Academic Press; p. 347–364. doi:10.1016/b978-0-12-078148-5.50018-x.
- Burns IG. 1995. Nitrogen supply, growth and development. Ecological aspects of vegetable fertigation in integrated crop production in the field. 428:21–30.
- Calatayud PA, Polanía MA, Guillaud J, Múnera DF, Hamon JC, Belloti AC. 2002. Role of single amino acids in phagostimulation, growth, and development of the cassava mealybug Phenacoccus herreni. Entomol Exp Appl. 104:363–367.
- Chaudhary A, Bala K, Thakur S, Kamboj R, Dumra N. 2018. Plant defenses against herbivorous insects: a review. Int J Chem Stud. 6:681–688.
- Chen B, Teh BS, Sun C, Hu S, Lu X, Boland W, et al. 2016. Biodiversity and activity of the gut microbiota across the life history of the insect herbivore Spodoptera littoralis. Sci Rep. 6:1–14. doi:10.1038/srep29505.
- Chen M. 2008. Inducible direct plant defense against insect herbivores : a review. Insect Sci. 15(2):101–114.
- Chishaki N, Horiguchi T. 1997. Responses of secondary metabolism in plants to nutrient deficiency. Soil Sci Plant Nutr. 43:987–991. doi:10.1080/00380768.1997.11863704.
- Chrétien LTS, David A, Daikou E, Boland W, Gershenzon J, Giron D, et al. 2018. Caterpillars induce jasmonates in flowers and alter plant responses to a second attacker. New Phytol. 217:1279–1291.
- Conrath U, Beckers GJM, Flors V, García-Agustín P, Jakab G, Mauch F, et al. 2006. Priming: getting ready for battle. Mol Plant-Microbe Interact. 19:1062–1071. doi:10.1094/MPMI-19-1062.
- Cornelissen T, Stiling P. 2009. Spatial, bottom-up, and top-down effects on the abundance of a leaf miner. Ecography. 32:459–467. doi:10.1111/j.1600-0587.2008.05590.x.
- Cotter SC, Simpson SJ, Raubenheimer D, Wilson K. 2011. Macronutrient balance mediates trade-offs between immune function and life history traits. Funct Ecol. 25:186–198. doi:10.1111/j.1365-2435.2010.01766.x.
- Dáder B, Fereres A, Moreno A, Tre¸bicki P. 2016. Elevated CO2 impacts bell pepper growth with consequences to myzus persicae life history, feeding behaviour and virus transmission ability. Sci Rep. 6:1–10. doi:10.1038/srep19120.
- Deagle BE, Thomas AC, McInnes JC, Clarke LJ, Vesterinen EJ, Clare EL, et al. 2019. Counting with DNA in metabarcoding studies: how should we convert sequence reads to dietary data? Mol Ecol. 28:391–406.
- de Bobadilla MF, Van Wiechen R, Gort G, Poelman EH. 2022. Plasticity in induced resistance to sequential attack by multiple herbivores in Brassica nigra. Oecologia. 198:11–20. doi:10.1007/s00442-021-05043-1.
- Deutsch CA, Tewksbury JJ, Tigchelaar M, Battisti DS, Merrill SC, Huey RB, et al. 2018. Increase in crop losses to insect pests in a warming climate. Science. 361:916–919. doi:10.1126/science.aat3466.
- Dhaliwal G, Jindal V, Dhawan A. 2010. Insect pest problems and crop losses: changing trends. Indian J Ecol. 37:1–7. doi:10.13140/RG.2.2.25753.47201.
- Dixon DP, Sellars JD, Kenwright AM, Steel PG. 2012. The maize benzoxazinone DIMBOA reacts with glutathione and other thiols to form spirocyclic adducts. Phytochemistry. 77:171–178. doi:10.1016/j.phytochem.2012.01.019.
- Dormann CF. 2003. Consequences of manipulations in carbon and nitrogen supply for concentration of anti-herbivore defence compounds in Salix polaris. Ecoscience. 10:312–318.
- Dubreuil G, Deleury E, Crochard D, Simon JC, Coustau C. 2014. Diversification of MIF immune regulators in aphids: link with agonistic and antagonistic interactions. BMC Genomics. 15:1–12. doi:10.1186/1471-2164-15-762.
- Engel P, Moran NA. 2013. The gut microbiota of insects diversity in structure and function. FEMS Microbiol Rev. 37:699–735. doi:10.1111/1574-6976.12025.
- Favery B, Dubreuil G, Chen MS, Giron D, Abad P. 2020. Gall-inducing parasites: convergent and conserved strategies of plant manipulation by insects and nematodes. Annu Rev Phytopathol. 58:1–22. doi:10.1146/annurev-phyto-010820-012722.
- Felton GW, Donato K, Del Vecchio RJ, Duffey SS. 1989. Activation of plant foliar oxidases by insect feeding reduces nutritive quality of foliage for noctuid herbivores. J Chem Ecol. 15:2667–2694. doi:10.1007/BF01014725.
- Filipiak M, Weiner J. 2017. Nutritional dynamics during the development of xylophagous beetles related to changes in the stoichiometry of 11 elements. Physiol Entomol. 42:73–84. doi:10.1111/phen.12168.
- Finkel OM, Castrillo G, Herrera Paredes S, Salas González I, Dangl JL. 2017. Understanding and exploiting plant beneficial microbes. Curr Opin Plant Biol. 38:155–163. doi:10.1016/J.PBI.2017.04.018.
- Frago E, Zytynska SE, Fatouros NE. 2020. Microbial symbionts of herbivorous species across the insect tree. Adv Insect Physiol. 58:111–159. doi:10.1016/bs.aiip.2020.04.002.
- Gamir J, Minchev Z, Berrio E, García JM, De Lorenzo G, Pozo MJ. 2021. Roots drive oligogalacturonide-induced systemic immunity in tomato. Plant, Cell Environ. 44:275–289. doi:10.1111/pce.13917.
- Gange AC, Bower E, Brown VK. 1999. Positive effects of an arbuscular mycorrhizal fungus on aphid life history traits. Oecologia. 120:123–131. doi:10.1007/s004420050840.
- Gao X, Li W, Luo J, Zhang L, Ji J, Zhu X, et al. 2019. Biodiversity of the microbiota in spodoptera exigua (Lepidoptera: Noctuidae). J Appl Microbiol. 126:1199–1208. doi:10.1111/jam.14190.
- García JM, Pozo MJ, López-Ráez JA. 2020. Histochemical and molecular quantification of arbuscular mycorrhiza symbiosis. In: Rodríguez-Concepció M, Welsch R, editors. Plant and food carotenoids. New York (NY): Humana; p. 293–299. doi:10.1007/978-1-4939-9952-1_22.
- García-Gómez G, Real-Santillán RO, Larsen J, Pérez LL, de la Rosa JIF, Pineda S, et al. 2021. Maize mycorrhizas decrease the susceptibility of the foliar insect herbivore Spodoptera frugiperda to its homologous nucleopolyhedrovirus. Pest Manage Sci. 77:4701–4708. doi:10.1002/ps.6511.
- Gershenzon J. 1984. Changes in the levels of plant secondary metabolites under water and nutrient stress. In: Timmermann BN, Steelink C, Loewus FA, editors. Phytochemical adaptations to stress. Boston (MA): Springer; p. 273–320.
- Giovannetti M, Mosse B. 1980. An evaluation of techniques for measuring vesicular arbuscular mycorrhizal infection in roots. New Phytol. 84:489–500. doi:10.1111/j.1469-8137.1980.tb04556.x.
- Giron D, Dedeine F, Dubreuil G, Huguet E, Mouton L, Outreman Y, Vavre F, Simon JC. 2017. Influence of microbial symbionts on plant–insect interactions. In: Sauvion N, Thiéry D, Calatayud PA, editors. Insect-plant interactions in a crop protection perspective, Vol. 81. London, UK: Elsevier; p. 225–257. doi:10.1016/bs.abr.2016.09.007.
- Glauser G, Marti G, Villard N, Doyen GA, Wolfender JL, Turlings TCJ, et al. 2011. Induction and detoxification of maize 1,4-benzoxazin-3-ones by insect herbivores. Plant J. 68:901–911. doi:10.1111/j.1365-313X.2011.04740.x.
- Goldstein PZ. 2017. Diversity and significance of lepidoptera: a phylogenetic perspective. In: Foottit RG, Adler PH, editors. Insect biodiversity: science and society. Oxford: Wiley-Blackwell; p. 463–495. doi:10.1002/9781118945568.ch13.
- Greene GL, Leppla NC, Dickerson WA. 1976. Velvetbean caterpillar: a rearing procedure and artificial medium. J Econ Entomol. 69:487–488.
- Gruden K, Lidoy J, Petek M, Podpečan V, Flors V, Papadopoulou KK, et al. 2020. Ménage à trois: unraveling the mechanisms regulating plant–microbe–arthropod interactions. Trends Plant Sci. 25:1215–1226. doi:10.1016/j.tplants.2020.07.008.
- Guerrieri E, Lingua G, Digilio MC, Massa N, Berta G. 2004. Do interactions between plant roots and the rhizosphere affect parasitoid behaviour? Ecol Entomol. 29(6):753–756.
- Gurung K, Wertheim B, Falcao Salles J. 2019. The microbiome of pest insects: it is not just bacteria. Entomol Exp Appl. 167:156–170. doi:10.1111/eea.12768.
- Hammer TJ, Janzen DH, Hallwachs W, Jaffe SP, Fierer N. 2017. Caterpillars lack a resident gut microbiome. Proc Natl Acad Sci U S A. 114:9641–9646. doi:10.1073/pnas.1707186114.
- Hammer TJ, Moran NA. 2019. Links between metamorphosis and symbiosis in holometabolous insects. Philos Trans R Soc, B. 374(1783):20190068. doi:10.1098/rstb.2019.0068.
- Hammer TJ, Sanders JG, Fierer N. 2019. Not all animals need a microbiome. FEMS Microbiol Lett. 366:fnz117. doi:10.1093/femsle/fnz117.
- Han P, Lavoir AV, Le Bot J, Amiens-Desneux E, Desneux N. 2014. Nitrogen and water availability to tomato plants triggers bottom-up effects on the leafminer Tuta absoluta. Sci Rep. 4:1–8. doi:10.1038/srep04455.
- Hannula SE, Zhu F, Heinen R, Bezemer TM. 2019. Foliar-feeding insects acquire microbiomes from the soil rather than the host plant. Nat Commun. 10:1–9. doi:10.1038/s41467-019-09284-w.
- Heinen R, Biere A, Harvey JA, Bezemer TM. 2018. Effects of soil organisms on aboveground plant-insect interactions in the field: patterns, mechanisms and the role of methodology. Front Ecol Evol. 6:1–15. doi:10.3389/fevo.2018.00106.
- Herlemann DP, Labrenz M, Jürgens K, Bertilsson S, Waniek JJ, Andersson AF. 2011. Transitions in bacterial communities along the 2000 km salinity gradient of the Baltic Sea. ISME J. 5:1571–1579. doi:10.1038/ismej.2011.41.
- Hewitt EJ. 1966. Sand and water culture methods used in the study of plant nutrition, 2nd ed. Farnham Royal: Commonwealth Agricultural Bureaux. doi:10.1017/S0014479700021852.
- Ho CK, Pennings SC. 2013. Preference and performance in plant-herbivore interactions across latitude – a study in U. S. Atlantic Salt Marshes. PLoS ONE. 8:1–11. doi:10.1371/journal.pone.0059829.
- Hu L, Sun Z, Xu C, Wang J, Mallik AU, Gu C, et al. 2022. High nitrogen in maize enriches gut microbiota conferring insecticide tolerance in lepidopteran pest Spodoptera litura. iScience. 25:103726. doi:10.1016/j.isci.2021.103726.
- Jeffries P, Gianinazzi S, Perotto S, Turnau K, Barea JM. 2003. The contribution of arbuscular mycorrhizal fungi in sustainable maintenance of plant health and soil fertility. Biol Fertil Soils. 37:1–16. doi:10.1007/s00374-002-0546-5.
- Jing TZ, Qi FH, Wang ZY. 2020. Most dominant roles of insect gut bacteria: digestion, detoxification, or essential nutrient provision? Microbiome. 8:1–20. doi:10.1186/s40168-020-00823-y.
- Johnson M, Zaretskaya I, Raytselis Y, Merezhuk Y, McGinnis S, Madden TL. 2008. NCBI BLAST: a better web interface. Nucleic Acids Res. 36(Web Server):W5–W9.
- Jung SC, Martinez-Medina A, Lopez-Raez JA, Pozo MJ. 2012. Mycorrhiza-induced resistance and priming of plant defenses. J Chem Ecol. 38:651–664. doi:10.1007/s10886-012-0134-6.
- Kaiser W, Huguet E, Casas J, Commin C, Giron D. 2010. Plant green-island phenotype induced by leaf-miners is mediated by bacterial symbionts. Proc R Soc B Biol Sci. 277:2311–2319.
- Kalaji HM, Dąbrowski P, Cetner MD, Samborska IA, Łukasik I, Brestic M, et al. 2016. A comparison between different chlorophyll content meters under nutrient deficiency conditions. J Plant Nutr. 40:1024–1034. doi:10.1080/01904167.2016.1263323.
- Kaplan EL, Meier P. 1958. Nonparametric estimation from incomplete observations. J Am Stat Assoc. 53:457–481.
- Koricheva J, Gange AC, Jones T. 2009. Effects of mycorrhizal fungi on insect herbivores: a meta-analysis. Ecology. 90:2088–2097. doi:10.1890/08-1555.1.
- Krams IA, Kecko S, Jõers P, Trakimas G, Elferts D, Krams R, et al. 2017. Microbiome symbionts and diet diversity incur costs on the immune system of insect larvae. J Exp Biol. 220:4204–4212. doi:10.1242/jeb.169227.
- LeBauer DS, Treseder KK. 2008. Nitrogen limitation of net primary productivity in terrestrial ecosystems is globally distributed. Ecology. 89:371–379.
- Leghari SJ, Wahocho NA, Laghari GM, Laghari A, Bhabhan M, HussainTalpur G, et al K. 2016. Role of nitrogen for plant growth and development: a review. Adv Environ Biol. 10:209–219.
- Leite-Mondin M, DiLegge MJ, Manter DK, Weir TL, Silva-Filho MC, Vivanco JM. 2021. The gut microbiota composition of Trichoplusia ni is altered by diet and may influence its polyphagous behavior. Sci Rep. 11:1–16. doi:10.1038/s41598-021-85057-0.
- López-Carmona DA, Alarcón A, Martínez-Romero E, Peña-Cabriales JJ, Larsen J. 2019. Maize plant growth response to whole rhizosphere microbial communities in different mineral N and P fertilization scenarios. Rhizosphere. 9:38–46. doi:10.1016/j.rhisph.2018.11.004.
- Lu ZX, Heong KL, Yu XP, Hu C. 2004. Effects of plant nitrogen on ecological fitness of the brown planthopper, Nilaparvata lugens Stal. in Rice. J Asia Pac Entomol. 7:97–104. doi:10.1016/S1226-8615(08)60204-6.
- Manresa-Grao M, Pastor-Fernández J, Sanchez-Bel P, Jaques JA, Pastor V, Flors V. 2022. Mycorrhizal symbiosis triggers local resistance in citrus plants against spider mites. Front Plant Sci. 13:867778. doi:10.3389/fpls.2022.867778.
- Martínez-Solís M, Collado MC, Herrero S. 2020. Influence of diet, sex, and viral infections on the gut microbiota composition of Spodoptera exigua caterpillars. Front Microbiol. 11:1–11. doi:10.3389/fmicb.2020.00753.
- Mason CJ, Jones AG, Felton GW. 2019. Co-option of microbial associates by insects and their impact on plant–folivore interactions. Plant Cell Environ. 42:1078–1086. doi:10.1111/pce.13430.
- Mason CJ, St. Clair A, Peiffer M, Gomez E, Jones AG, Felton GW, et al. 2020. Diet influences proliferation and stability of gut bacterial populations in herbivorous lepidopteran larvae. PloS one. 15:e0229848.
- Mattson WJ. 1980. Herbivory in relation to plant nitrogen content. Annu Rev Ecol Syst. 11:119–161.
- McKenna DD, Shin S, Ahrens D, Balke M, Beza-Beza C, Clarke DJ, et al. 2019. The evolution and genomic basis of beetle diversity. Proc Natl Acad Sci U S A. 116:24729–24737. doi:10.1073/pnas.1909655116.
- Minchev Z, Kostenko O, Soler R, Pozo MJ. 2021. Microbial consortia for effective biocontrol of root and foliar diseases in tomato. Front Plant Sci. 12:1–12. doi:10.3389/fpls.2021.756368.
- Morant AV, Jørgensen K, Jørgensen C, Paquette SM, Sánchez-Pérez R, Møller BL, et al. 2008. β-Glucosidases as detonators of plant chemical defense. Phytochemistry. 69:1795–1813. doi:10.1016/j.phytochem.2008.03.006.
- Niemeyer HM. 2009. Hydroxamic acids derived from 2-hydroxy-2h-1,4-benzoxazin-3(4 h)-one: key defense chemicals of cereals. J Agric Food Chem. 57:1677–1695. doi:10.1021/jf8034034.
- Panda N, Khush GS. 1995. Host plant resistance to insects. Wallingford: CAB international.
- Pieterse CMJ, Zamioudis C, Berendsen RL, Weller DM, Van Wees SCM, Bakker PAHM. 2014. Induced systemic resistance by beneficial microbes. Annu Rev Phytopathol. 52:347–375. doi:10.1146/annurev-phyto-082712-102340.
- Pineda A, Dicke M, Pieterse CM, Pozo MJ. 2013. Beneficial microbes in a changing environment: are they always helping plants to deal with insects? Funct Ecol. 27(3):574–586. doi:10.1111/1365-2435.12050.
- Pineda A, Zheng SJ, van Loon JJA, Pieterse CMJ, Dicke M. 2010. Helping plants to deal with insects: the role of beneficial soil-borne microbes. Trends Plant Sci. 15:507–514. doi:10.1016/j.tplants.2010.05.007.
- Porras-Soriano A, Soriano-Martín ML, Porras-Piedra A, Azcón R. 2009. Arbuscular mycorrhizal fungi increased growth, nutrient uptake and tolerance to salinity in olive trees under nursery conditions. J Plant Physiol. 166:1350–1359. doi:10.1016/J.JPLPH.2009.02.010.
- Pozo MJ, Azcón-Aguilar C. 2007. Unraveling mycorrhiza-induced resistance. Curr Opin Plant Biol. 10:393–398. doi:10.1016/J.PBI.2007.05.004.
- Pozo MJ, Jung SC, López-Ráez JA, Azcón-Aguilar C. 2010. Impact of arbuscular mycorrhizal symbiosis on plant response to biotic stress: the role of plant defence mechanisms. In: Koltai H, Kapulnik Y, editors. Arbuscular mycorrhizas: physiology and function. Dordrecht: Springer; p. 193–207. doi:10.1007/978-90-481-9489-6_9.
- Quast C, Pruesse E, Yilmaz P, Gerken J, Schweer T, Yarza P, Peplies J, Glöckner FO. 2012. The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res. 41(D1):D590–D596.
- Raubenheimer D, Simpson SJ. 1993. The geometry of compensatory feeding in the locust. Anim Behav. 45:953–964.
- R Development Core Team. 2021. R: a language and environment for statistical. Vienna, Austria: R Foundation for Statistical Computing. https://www.R-project.org/.
- Real-Santillán RO, del-Val E, Cruz-Ortega R, Contreras-Cornejo HÁ, González-Esquivel CE, Larsen J. 2019. Increased maize growth and P uptake promoted by arbuscular mycorrhizal fungi coincide with higher foliar herbivory and larval biomass of the Fall Armyworm Spodoptera frugiperda. Mycorrhiza. 29:615–622. doi:10.1007/s00572-019-00920-3.
- Rivero J, Lidoy J, Llopis-Giménez Á, Herrero S, Flors V, Pozo MJ. 2021. Mycorrhizal symbiosis primes the accumulation of antiherbivore compounds and enhances herbivore mortality in tomato. J Exp Bot. 72:5038–5050. doi:10.1093/jxb/erab171.
- Rognes T, Flouri T, Nichols B, Quince C, Mahé F. 2016. VSEARCH: a versatile open source tool for metagenomics. PeerJ. 4:e2584.
- Rostás M. 2007. The effects of 2,4-dihydroxy-7-methoxy-1,4-benzoxazin-3-one on two species of spodoptera and the growth of setosphaeria turcica in vitro. J Pest Sci (2004). 80:35–41. doi:10.1007/s10340-006-0151-8.
- Sanchez-Bel P, Troncho P, Gamir J, Pozo MJ, Camañes G, Cerezo M, et al. 2016. The nitrogen availability interferes with mycorrhiza-induced resistance against botrytis cinerea in tomato. Front Microbiol. 7:1–16. doi:10.3389/fmicb.2016.01598.
- Sasai H, Ishida M, Murakami K, Tadokoro N, Ishihara A, Nishida R, et al. 2009. Species-specific glucosylation of DIMBOA in larvae of the rice armyworm. Biosci Biotechnol Biochem. 73:1333–1338. doi:10.1271/bbb.80903.
- Schloss PD. 2020. Reintroducing mothur: 10 years later. Appl Environ Microbiol. 86(2):e02343–19.
- Schloss PD, Westcott SL, Ryabin T, Hall JR, Hartmann M, Hollister EB, et al. 2009. Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl Environ Microbiol. 75:7537–7541. doi:10.1128/AEM.01541-09.
- Shikano I, Pan Q, Hoover K, Felton GW. 2018. Herbivore-induced defenses in tomato plants enhance the lethality of the entomopathogenic bacterium, Bacillus thuringiensis var. kurstaki. J Chem Ecol. 44:947–956.
- Showler AT. 2001. Spodoptera exigua oviposition and larval feeding preferences for pigweed, Amaranthus hybridus, over squaring cotton, Gossypium hirsutum, and a comparison of free amino acids in each host plant. J Chem Ecol. 27:2013–2028. doi:10.1023/A:1012238803311.
- Simpson SJ, Raubenheimer D. 1995. The geometric analysis of feeding and nutrition: a user’s guide. J Insect Physiol. 41:545–553.
- Song YY, Ye M, Li CY, Wang RL, Wei XC, Luo SM, et al. 2013. Priming of anti-herbivore defense in tomato by arbuscular mycorrhizal fungus and involvement of the jasmonate pathway. J Chem Ecol. 39:1036–1044. doi:10.1007/s10886-013-0312-1.
- Srinivasa Rao M, Manimanjari D, Vanaja M, Rama Rao CA, Srinivas K, Rao VUM, et al. 2012. Impact of elevated CO2 on tobacco caterpillar, Spodoptera litura on peanut, Arachis hypogea. J Insect Sci. 12:1–10. doi:10.1673/031.012.10301.
- Sudakaran S, Kost C, Kaltenpoth M. 2017. Symbiont acquisition and replacement as a source of ecological innovation. Trends Microbiol. 25:375–390. doi:10.1016/j.tim.2017.02.014.
- Sugio A, Dubreuil G, Giron D, Simon JC. 2015. Plant-insect interactions under bacterial influence: ecological implications and underlying mechanisms. J Exp Bot. 66:467–478. doi:10.1093/jxb/eru435.
- Tripathi DK, Singh VP, Chauhan DK, Prasad SM, Dubey NK. 2014. Role of macronutrients in plant growth and acclimation: recent advances and future prospective. In: Ahmad P, Wani M, Azooz M, Phan Tran LS, editors. Improvement of crops in the era of climatic changes. New York, NY: Springer; p. 197–216.
- Truzi CC, Vieira NF, De Souza JM, De Bortoli SA. 2021. Artificial diets with different protein levels for rearing Spodoptera frugiperda (Lepidoptera: Noctuidae). J Insect Sci. 21:1–7. doi:10.1093/jisesa/ieab041.
- Van Der Heijden MGA, Streitwolf-Engel R, Riedl R, Siegrist S, Neudecker A, Ineichen K, et al. 2006. The mycorrhizal contribution to plant productivity, plant nutrition and soil structure in experimental grassland. New Phytol. 172:739–752. doi:10.1111/j.1469-8137.2006.01862.x.
- Vicari M, Hatcher PE, Ayres PG. 2002. Combined effect of foliar and mycorrhizal endophytes on an insect herbivore. Ecology. 83:2452–2464.
- Voirol LRP, Frago E, Kaltenpoth M, Hilker M, Fatouros NE. 2018. Bacterial symbionts in lepidoptera: their diversity, transmission, and impact on the host. Front Microbiol. 9:1–4. doi:10.3389/fmicb.2018.00556.
- War AR, Paulraj MG, Ahmad T, Buhroo AA, Hussain B, Ignacimuthu S, et al. 2012. Mechanisms of plant defense against insect herbivores. Plant Signal Behav. 7:1306–1320. doi:10.4161/psb.21663.
- White TCR. 1993. The inadequate environment: nitrogen and the abundance of animals. New York: Springer Science and Business Media.
- Wickham H. 2011. Ggplot2. Wiley Interdiscip Rev Comput Stat. 3:180–185.
- Wickham H, François R, Henry L, Müller K. 2021. Dplyr: a grammar of data manipulation. R Package Version. 1(0):5.
- Xie H, Liu K, Sun D, Wang Z, Lu X, He K. 2015. A field experiment with elevated atmospheric CO2-mediated changes to C4 crop-herbivore interactions. Sci Rep. 5:1–9. doi:10.1038/srep13923.
- Ximénez-Embún MG, Castañera P, Ortego F. 2017. Drought stress in tomato increases the performance of adapted and non-adapted strains of Tetranychus urticae. J Insect Physiol. 96:73–81. doi:10.1016/j.jinsphys.2016.10.015.
- Yuan C, Xing L, Wang M, Hu Z, Zou Z. 2021. Microbiota modulates gut immunity and promotes baculovirus infection in Helicoverpa armigera. Insect Sci. 28:1766–1779. doi:10.1111/1744-7917.12894.
- Zhang H, Dubreuil G, Faivre N, Dobrev P, Kaiser W, Huguet E, et al. 2018. Modulation of plant cytokinin levels in the Wolbachia-free leaf-mining species Phyllonorycter mespilella. Entomol Exp Appl. 166:428–438. doi:10.1111/eea.12681.
- Zhang J, Kobert K, Flouri T, Stamatakis A. 2014. PEAR: a fast and accurate illumina paired-End reAd mergeR. Bioinformatics. 30:614–620.
- Zhou S, Richter A, Jander G. 2018. Beyond defense: multiple functions of benzoxazinoids in maize metabolism. Plant Cell Physiol. 59:1528–1533. doi:10.1093/pcp/pcy064.
- Zhu-Salzman K, Luthe DS, Felton GW. 2008. Arthropod-inducible proteins: broad spectrum defenses against multiple herbivores. Plant Physiol. 146:852–858. doi:10.1104/pp.107.112177.