ABSTRACT
Considering the more frequent and longer drought events due to climate change, improving plant drought tolerance became a priority. The search for plant growth promoting rhizobacteria (PGPR) able to improve plant drought tolerance has been long addressed, but with inconsistent results. Here, we summarize the PGPR mechanisms that improve plant drought tolerance, identify the pitfalls in current PGPR isolation and selection routines, and discuss the key points to define new strategies to get optimal PGPR for plant drought tolerance. Drought and host genotype impact rhizo-communities, and host-mediated selection strategies may be used to obtain a drought-adapted rhizomicrobiome that can be a source for PGPR isolation. Alternatively, an integrated omics-level analysis can improve our knowledge on the mechanisms of rhizomicrobiome construction, and a targeted approach can be designed, which will be focused on key PGP traits. New strategies to build PGPR consortia for improvement of plant drought tolerance are also suggested.
1. Why the urgent need for ‘new' fertilization strategies?
Currently, drought affects 64% of the global land area, and is estimated to increase to about 80% by 2050, presenting a serious threat to food security worldwide (Kasim et al. Citation2013; Meena et al. Citation2017). Drought reduces nutrient availability to plants because nutrient diffusion and mass flow of water-soluble nutrients decreases under soil water deficit (Vurukonda et al. Citation2016). This results in a multidimensional stress with negative effects on plant metabolism and growth, especially during grain filling of cereal crops and the reproductive phase of field grown crops (Shrivastava and Kumar Citation2015). Under drought conditions, several changes on plant morphology and physiology occur; morphological traits such as root length, leaf morphology and number, fruit number and size, number of seeds, are affected by drought stress (Jaleel et al. Citation2009). Drought accentuates the biosynthesis of ethylene, a phytohormone, whose accumulation has negative impacts on plant growth (Vurukonda et al. Citation2016). Drought weakens the photosynthesis process (Chaves et al. Citation2009) and leads to the accumulation of free radicals and reactive oxygen species (ROS) such as superoxide radicals, hydrogen peroxide and hydroxyl radicals, therefore inducing oxidative stress. A high ROS level causes lipid peroxidation, membrane deterioration, and degradation of proteins and nucleic acids, leading to cell death (Tiwari et al. Citation2016).
Before the negative impacts of increased drought on agriculture became so widespread around the world, the ability to synthesize ammonium from air (i.e. the Haber–Bosch process) since the beginning of the twentieth century increased the availability and use of synthetic fertilizers, which increased crop yields and enabled a dramatic increase in the world population (Rouwenhorst et al. Citation2021). However, the massive use of synthetic fertilizers, also called chemical or inorganic fertilizers, containing nitrogen and other nutrients, has severe negative environmental impacts, as nutrients such as nitrogen and phosphorus are lost from the agrosystems due to run-off and leaching, impacting streams and rivers and causing eutrophication and destruction of aquatic life (Dubeux and Sollenberger Citation2020). These fertilizers stimulate soil microbes, which feast on organic matter, reducing soil organic matter and soil’s ability to store organic nitrogen (Mulvaney et al. Citation2009), and increasing nitrogen leaching and N2O (a potent greenhouse gas) emissions. The loss of soil organic matter negatively affects soil’s carbon sequestration and soil fertility, as soil organic matter: (i) acts as a nutrient storage, gradually providing essential elements; (ii) buffers plants against sudden environmental changes; (iii) preserves moisture during drought periods; (iv) keeps soil physical conditions compatible for seedling growth; and (v) supports a greater biodiversity (Mahmoudi et al. Citation2021). Thus, the loss of soil organic matter turns soils less resilient to climate changes, more vulnerable to erosion and more dependent on irrigation; as water becomes scarcer due to climate change and increased water demand, maintaining the current widespread use of chemical fertilizers is turning agriculture more and more unsustainable.
Therefore, the twentieth century agricultural innovations no longer meet the twenty-first century agricultural challenges and the trend is to reduce chemical fertilizer application rates (and therefore its negative impacts) without hampering food security. Within the ongoing climate crisis, ensuring food security, sustainable crop production and environmental protection, demands for an integrated workplan, which includes ‘new’ options. One of these ‘new’ options is Precision Agriculture (PA), which is emerging as a sustainable solution for the management of farm input resources (e.g. fertilizers, water, seeds, pesticides). It aims at applying the right input, in the right amount, at the right place and right time using advanced sensing, modelling and control technologies. Several studies have shown that variable rate applications of farming input resources according to crop needs result in increased crop yield and profitability at minimized input cost and environmental footprint for chemical fertilization, manure application and seeding (e.g. Searcy Citation1997; Guerrero et al. Citation2021; Zhang et al. Citation2021; Munnaf et al. Citation2021). As water is becoming an increasingly critical factor for farming in many areas of the world, the use of monitoring plant water status as part of PA implementation is increasing to support farmers in making the decision on variable rate irrigation and irrigation scheduling.
Other very important, non-exclusive options to decrease the input of chemical fertilizers include the use of alternative, ‘new’ fertilization strategies such as organic amendments (e.g. organic fractions of agricultural and municipal solid wastes, compost) (Kochakinezhad et al. Citation2012; Ribas-Agustí et al. Citation2017; Ulm et al. Citation2019), and crop rotations, which take advantage of niche complementarity, enabling the optimization of nutrient use (Dias et al. Citation2015); recent meta-analyses reported increases in soil microbial diversity and richness from 3% to 27% when cover cropping or crop rotations were carried out relative to monoculture or soils with less crop diversity (Kim et al. Citation2020). Finally, the use of environmentally friendly plant growth promoting rhizobacteria (PGRR) and mycorrhizal fungi as biofertilizers is another strategy to minimize the application of chemical fertilizers, as various examples are reported showing similar crop yields when they partially replace the later (section 3). Defined as microbial inoculant preparations formed by live or latent cells from specific microbes applied to seed, plant surfaces, or soil to significantly increase growth and yield of crop production (Mitter et al. Citation2021), biofertilizers are now inserted in the broader definition of biostimulants provided in the Regulation (EU) n°2019/1009; this defines a biostimulant as ‘a product stimulating plant nutrition processes independently of the product’s nutrient content with the sole aim of improving one or more of the following characteristics of the plant or the plant rhizosphere: (a) nutrient use efficiency (b) tolerance to abiotic stress (c) quality traits, (d) availability of confined nutrients in soil or rhizosphere.’ In the next sections 3. and 4.1, we summarize PGPR mechanisms leading to these outputs, namely those leading to plant drought tolerance, of utmost significance in a context of increased desertification due to climate change.
2. Many unanswered questions
PGPR-based biofertilizers global market is certainly gaining momentum worldwide and it is expected to reach USD 4.5 billion by 2026 (Global Biofertilizers Market report 2021, ID: 5334050) due to its advantages in promoting soil health and in reducing the pollution caused using agro-chemicals (synthetic fertilizers, pesticides), and due to the recent significant rises in synthetic fertilizers’ prices (https://asmith.ucdavis.edu/news/story-rising-fertilizer-prices). Despite their great potential, the use of biofertilizers by farmers is still limited due to: (i) inconsistent results over different soils, crops and environmental conditions; (ii) practical aspects related to mass production, e.g. definition and control of manufacturing parameters such as the physiological state of the microorganisms when harvested, and the dehydration process (Herrmann and Lesueur Citation2013); (iii) short shelf-life, which compromises microbial viability; and (iv) appropriate recommendations and ease of use for farmers (Mitter et al. Citation2021; Basílio et al. Citation2022).
Here, we will focus on the aspects related with inconsistent results over different soil-crop systems and in response to a specific environmental stress (drought). Since we consider that PGPR isolation and selection procedures are crucial initial processes to develop optimal PGPR-based biofertilizers for field application, we will discuss key questions on how to choose the appropriate methods to find optimal PGPR. For that, we used the available literature to evaluate the parameters considered in sample collection and PGPR isolation and selection, highlighting current pitfalls in these procedures (section 4.1); methods for isolating and screening of PGPR are mostly random, i.e. do not target a specific stress factor or plant-soil system, which certainly hinders finding optimal PGPR. To propose new methods, we must consider the present knowledge (and acquire further) on plant-bacteria interactions (section 4.2), namely on the plant recruitment of soil bacteria, done primarily by root exudates, and on the necessary factors for efficient rhizosphere bacterial colonization, which will depend on the bacteria and host traits and the environmental conditions. What could be the primordial factor for recruitment under drought? These are vast research fields, presently at their infancy, which require a deeper comparative study of the rhizomicrobiome (here defined as the root-associated microbiome, comprising the rhizosphere and root endosphere microbial communities) composition and functionality. The rhizomicrobiome assembly is controlled by both host genotype and drought (section 5.1), and the rhizomicrobiome manipulation through host-mediated selection of microbial communities adapted to drought can be considered for PGPR selection (section 5.2). However, several issues can be pointed out regarding such manipulation, namely if the selected PGPR are effective under the spatial and temporal variation of soil chemistry and climate. Furthermore, PGPR persistence in the rhizosphere and their impact on the native communities at short and long-term (section 5.2) must be considered. Our reflection led to the proposal of an integrated omics approach to design a PGPR screening based on target indicators and to the construction of PGPR consortia with unfamiliar rhizo- and soil bacteria with relevant properties (section 6).
Despite the above questions, many PGPR already commercialized as biofertilizers can improve plant drought tolerance, as exemplified below.
3. First steps towards developing drought-specific biofertilizers
Many different bacterial genera have been incorporated in commercialized PGPR-based biofertilizers or biocontrol formulations (Jiao et al. Citation2021). PGPR may synthesize phytohormones (e.g. auxins which stimulate root proliferation) and/or increase nutrient availability to the plant through atmospheric nitrogen (N) fixation, phosphate solubilization and synthesis of siderophores for iron and other metals sequestration from the soil (Backer et al. Citation2018; Schalk et al. Citation2011). Thus, PGPR improve nutrient acquisition efficiency, whereas only a fraction of the applied nutrients in the form of chemical fertilizers (50% for N and 30% for P) are taken up by plants. PGPR may also enhance nutrient use efficiency [described as yield (biomass) per unit input (fertilizer, nutrient content)] (e.g. Pacheco et al. Citation2021; Basílio et al. Citation2022). Could the effects of PGPR become larger than those reported until now, reducing even further our dependence on chemical fertilizers and reducing irrigation water use? This is an important question particularly regarding drought stress.
There are several studies on the PGPR benefits to the host plant under drought conditions (). Many of these PGPR are Bacillus strains, such as B. licheniformis used in the study by Akhtar et al. (Citation2020). B. licheniformis (and other Bacillus) spores have long stability, do not germinate in tap water, are easy to apply, and are not affected by conventional pesticides. The spores remain dormant until the presence of root exudates trigger their germination and switch to vegetative metabolically active cells (Akhtar et al. Citation2020). These attributes allow Bacillus to be formulated together with most of the chemical additives normally employed in agriculture, as seed coating agents or in liquid media for in-furrow applications. Being a facultative anaerobic organism capable of anaerobic respiration and fermentative growth (Clements et al. Citation2002), B. licheniformis it suited to life in the rhizosphere, where oxygen level fluctuates, from high (drought) to very low (flooding conditions). B. licheniformis, and probably other Gram-positive versatile spore-forming bacteria, could thus be used for enhancing water use efficiency (WUE) under normal and drought conditions (see ), so these microorganisms can be important farming tools under the ongoing climate changes. Also noteworthy, many PGPR that improve plant drought tolerance have been selected based on the presence of 1-aminocyclopropane-1-carboxylate (ACC) deaminase activity (e.g. Niu et al. Citation2018; Duan et al. Citation2021). The ACC, the ethylene precursor, and a plant signalling molecule (Li et al. Citation2022), can be exuded by plant roots, sequestered, and hydrolyzed into ammonia and α-ketobutyrate by ACC deaminase-producing PGPR for nitrogen and energy supply. By removing ACC, these PGPR can lower ethylene levels, thus reducing the negative effects of ethylene accumulation, improving plant stress tolerance, and promoting plant growth despite the stressful conditions (Glick Citation2014). As an example, Gowtham et al. (Citation2020) showed that a PGPR containing ACC deaminase (Bacillus subtilis Rhizo SF 48) enhanced plant growth and relative water content in tomato plants even under severe drought conditions and proposed this strain as a useful bio-inoculant for sustainable tomato production in arid and semi-arid regions.
Table 1. Examples of the effects of PGPR and PGPR-based biofertilizers on different crops under non-stress and abiotic stress. The quantification of the effect of inoculation on a given parameter was based on the comparison between inoculation and non-inoculation.
4. Are we selecting optimal PGPR?
4.1. Considering PGP traits – the current strategy for PGPR selection
The distinct PGPR-based biofertilizers performance between greenhouse and field-tested conditions, and their short shelf life when compared to inorganic fertilizers, are main aspects in PGPR research and development (Adedeji et al. Citation2020). However, ahead steps, i.e. the current PGPR isolation and selection procedures should also be evaluated. Thus, the question presented earlier in section 3, regarding a further reduction of our dependence on chemical fertilizers, can be reformulated: Can an optimal PGPR be found, leading to the production of biofertilizers that could be applied with reduced dependence on mineral fertilizers or irrigation? This might be possible if considering microbial ecology when selecting PGPR isolates.
Although there is consensus that in the search for PGPR many biotic and abiotic variables (e.g. soil types, soil physico-chemical characteristics, plant species, seasons, and status of the host plant) and their interaction should be considered to ensure the successful isolation of putative beneficial rhizobacteria, a search among the literature shows that it is rarely the case. For example, the rhizosphere of wild populations of plants is proposed as an optimal source to isolate PGPR, as a plant in its habitat exerts a high selective pressure in the rhizozone, where it selects for beneficial bacteria (Barriuso et al. Citation2005). However, the isolated PGPR might not be responsive to the rhizo-environment of a targeted crop. The broader habitat for sample collection is also of importance, but many times disregarded. If plant drought tolerance is intended, the beneficial bacteria must themselves be tolerant to drought conditions. As so, the isolation and screening for putative PGPR from soils from chronically dry regions, described in several publications (section 5.2), or even from extreme conditions (section 6) may recover a high number of PGPR, able of withstanding severe drought conditions.
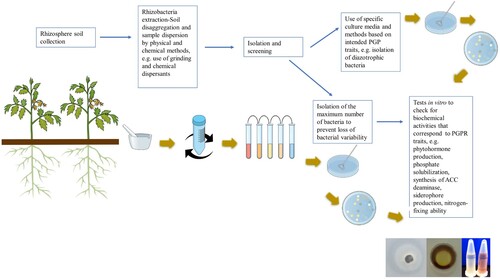
Rhizobacteria extraction () starts with sampling of the rhizosphere soil fraction (considered to be the soil volume at 1–3 mm from the root and the soil adhering to the root) by shaking vigorously the root, with the soil still adherent being collected as the rhizosphere. Then, the rhizospheric soil sample is suspended in water, in saline solution or in phosphate buffer. Bacterial extraction is performed following soil disaggregation and sample dispersion to reduce the association between the soil and the bacterial cells, which occurs, for instance, through adherence by extracellular polymers to the soil particles (Barriuso et al. Citation2008). Chemical extraction methods may be combined with physical methods, such as homogenizing or grinding, and ultrasonics. Each of those methods has advantages and disadvantages and may result in a selective extraction where less resistant bacteria could be damaged (Barriuso et al. Citation2008). The extraction step is thus of great importance.
Bacterial isolation from soil-extracted microbial suspensions follows classical microbiological procedures, and screening of putative PGPR is performed using two different strategies: 1) specific culture media and growth conditions are used for the isolation of intended specific plant beneficial bacteria, or 2) after isolation of the maximum number of bacteria to avoid the loss of variability, different in vitro tests are carried out to inspect for biochemical activities that correspond to known PGP traits (Barriuso et al. Citation2008) (). Those tests, being usual procedures in PGPR screening, are meant to select beneficial bacteria to act in different contexts (for instance, distinct abiotic stresses) and the most common are (i) production of plant growth regulators (e.g. auxins, and cytokinins); (ii) synthesis of ACC deaminase (iii) phosphate solubilization (iv) siderophore production (v) nitrogen-fixation and (vi) production of enzymes that can degrade pathogenic fungi cell walls (i.e. chitinase or β-1,3-glucanase) (Barriuso et al. Citation2008).
Several problems are inherent to this PGPR isolation, for example:
the use of nonselective media, generally of nutrient-rich media, to avoid the loss of bacterial variability, will in fact bias the selection to easily cultivable and fast-growing abundant soil bacteria.
It does not account for the isolation of low-abundant bacteria, which are difficult to culture but can be metabolically very active; these bacteria can synthesize high amounts of proteins, use different substrates (Jousset et al. Citation2017) and play fundamental processes in soils. As an example, dilution-to-extinction experiments for different soils revealed that low-abundance plant-associated bacteria are involved in the production of antagonistic volatile compounds that protect the host plant against pathogens (Hol et al. Citation2015). Soil microstructure (Totsche et al. Citation2018) might be a major factor affecting the detection of low-abundant microorganisms, as many microbes thrive under tight association to specific properties of soil structure, exploiting highly specific microenvironments on or within soil particles (Or et al. Citation2007; Datta et al. Citation2017). Therefore, albeit more laborious, extraction procedures that separate the soil particles according to their size will result in an extended list of putative beneficial isolates. Indeed, Hemkemeyer et al. (Citation2018) revealed bacterial preferences for specific soil particle size fractions by amplicon sequence analysis of microbial communities within each fraction. For instance, Actinobacteria and Nitrosospira members preferred fine silt. The authors used gentle ultrasonication, wet-sieving and centrifugation to isolate particle size fractions (clay <2 µm; fine silt 2–20 µm; coarse silt 20–63 µm; sand 63–2000µm) with attached cells. A similar protocol could be adapted into a hierarchical nested strategy for microbial extraction, starting with soil fractionation, bacterial isolation with choice of specific or several different media, and subsequent testing of the isolates from each fraction for chosen traits (see point iii). An intermediate step, before isolation, could be choosing the fraction for the isolation, after tracking a target microbial activity, for instance the presence of diazotrophic activity through measurement of nitrogenase activity in situ (Pérez et al. Citation2022). Such hierarchical nested strategy for microbial extraction from soils would be more suitable for the isolation of low-abundant bacteria.
It does not account for inducible microbial traits. Regarding the standard biochemical tests, some of the traits shown in vitro are inducible, i.e. they are only expressed in certain conditions, so that a PGP trait could be expressed in the laboratory but not in the rhizosphere and vice versa. For example, phosphate solubilization and siderophore production may not be expressed in phosphorus-rich and iron-rich soils, respectively (Barriuso et al. Citation2008). Within this context, the choice of culture conditions for the identification of PGP biochemical traits must be carefully considered, ideally meeting the conditions of the target environment for PGPR application. For example, very low phosphate solubilization was detected for Rhizobium sp. strains in a broth containing yeast extract and mannitol. However, replacement of yeast extract with NH4SO4 as a nitrogen source led to a huge increase in the solubilization of phosphate by the strains, which was also affected by the carbon source present in the medium with NH4SO4 (Amaya-Gómez et al. Citation2020).
Thus, although it can be argued that several post-inoculation factors (e.g. soil chemistry, competitive rhizospheric interactions with indigenous microbes, climate fluctuations, and variation across plant genotypes) (Sessitsch et al. Citation2019) can reduce PGPR efficiency in the field compared to the greenhouse, obtaining optimal PGPR may be largely dependent on a correct planning of the isolation and screening procedures that anticipate those post-inoculation factors. Indeed, a major explanation for the inconsistent results at the field-scale is variation in the capacity of bacterial colonization and survival within the rhizosphere, i.e. in the rhizosphere competence, which will depend on the bacterial traits, the host plant and the environmental abiotic and biotic conditions. Therefore, all these factors should be considered in PGPR isolation and selection.
Several molecular approaches (e.g. gene disruption and gene activation) showed that bacterial functions such as motility, attachment, growth, stress resistance and production of secondary metabolites are linked to rhizosphere competence (in Barret et al. Citation2011). But the major traits and corresponding genes relevant in rhizosphere competence under drought are largely unknown, and so unknown which PGP biochemical traits may be crucial for selecting PGPR that improve plant drought tolerance in each plant-soil system.
Reports show that the presence of commonly tested PGP traits improve plant drought tolerance (e.g. Vurukonda et al. Citation2016 and ref. therein). These traits include the synthesis of phytohormones, the nutrient acquisition from soil minerals and organic complexes (e.g. by solubilization of inorganic phosphate and mineralization of organic phosphate), and the inhibition of plant ethylene synthesis (). For instance, auxin/IAA (indole-3-acetic acid)-producing bacteria increase plant root growth and/or enhance the formation of lateral roots and roots hairs, thereby increasing water and nutrient uptake, and thus improve plant drought tolerance (in Vurukonda et al. Citation2016). Bacterial biosynthesis of the phytohormone abscisic acid (ABA), which is involved in signal transduction pathways that regulate stress defense mechanisms, including the regulation of water loss by controlling stomatal closure (Yamaguchi-Shinozaki and Shinozaki Citation1994), contributes to the alleviation of drought stress in maize (Cohen et al. Citation2009). Since drought results in a multidimensional stress, it is easily understood that the presence of those, commonly tested, multiple traits in single PGPR or in a consortium of several PGPR translate into multiple modes of action capable of benefiting crops. Although the selection of multiple traits is important, some modes of action might be more relevant than others for a particular crop/soil/stress setting.
Figure 2. Some PGP characteristics important for the induction of plant drought stress tolerance mechanisms. PGPR attributes and effects on the plant (green boxes) are summarized. ACC:1-aminocyclopropane-1-carboxylate; APX: ascorbate peroxidase, CAT: catalase, EPS: exopolysaccharides, IAA: indole-3-acetic acid, SOD: superoxide dismutase, VOCs: volatile organic compounds.
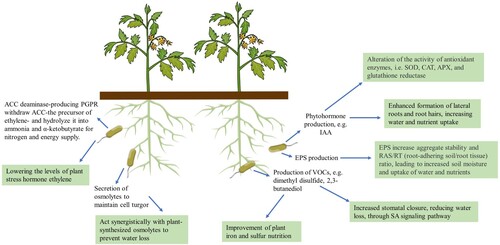
Other PGP traits can be tested by less standardized procedures, or indirectly by biochemical analysis of the plant host. For instance, PGPR may alter the activity of plants’ antioxidant enzymes SOD, CAT, APX and glutathione reductase (GR) (Chiappero et al. Citation2019; Akhtar et al. Citation2020), which scavenge ROS and therefore alleviate the oxidative damage (Kaushal and Wani Citation2016) (). The plant-bacteria interaction mechanism(s) responsible for such output is still unclear and might involve bacterial-produced phytohormones (Clarke et al. Citation2002; Khan et al. Citation2020). Some PGPR strains release volatile organic compounds (VOCs) that mediate increases in plant biomass, disease resistance, and abiotic stress tolerance (Liu and Zhang Citation2015). Bacterial VOC 2,3-butanediol promotes plant growth and induces disease resistance (Ryu et al. Citation2003, Citation2004). Arabidopsis plants exposed to 2,3-butanediol showed increased drought tolerance as a result of stomatal closure and consequently reduced water loss (Cho et al. Citation2008). The application of 2,3-butanediol to plant lines defective in various hormone signalling pathways indicated that the induced drought tolerance is regulated by phytohormones salicylic acid (SA), jasmonic acid (JA), and ethylene, with SA playing a primary role, as free SA levels significantly increased in plants treated with 2,3-butanediol (Cho et al. Citation2008). PGPR VOCS can improve iron and sulfur nutrition in plants (Liu and Zhang Citation2015). Plants largely acquire S through root uptake of SO42– from soil (Santana et al. Citation2021), but they can also assimilate S from S-containing compounds in the air, including VOCs emitted by soil microbes (Meldau et al. Citation2013). Emission of dimethyl disulfide (DMDS), an S-containing VOC commonly produced by many soil bacteria and fungi (Kanchiswamy et al. Citation2015), from Bacillus sp. strain B55 rescued Nicotiana attenuata growth retardation caused by S deprivation (Meldau et al. Citation2013). Noteworthy, sulfur plays a significant role in drought stress responses (Abuelsoud et al. Citation2016), which will be discussed below (section 6).
Although produced in response to several biotic and abiotic stress factors, exopolysaccharides (EPS) biosynthesis (Morcillo and Manzanera Citation2021) and osmolyte accumulation (Signorelli et al. Citation2021), have been described most extensively for their role under water depletion. EPS are hydrated compounds with 97% water in a polymer matrix that confers protection against desiccation (Bhaskar and Bhosle Citation2005). EPS increase aggregate stability and RAS/RT (root-adhering soil/root tissue) ratio, leading to increased uptake of water and nutrients from the rhizosphere (Kaushal and Wani Citation2016). Therefore, EPS-producing PGPR strains contribute to the maintenance of soil moisture content and improve plant drought tolerance (Naseem and Bano Citation2014) (). For example, the inoculation of sunflower plants with an EPS-producing Rhizobium sp. strain increased the RAS/RT ratio, as well as RAS macroporosity, which helped to relieve the effect of drought on sunflower growth (Alami et al. Citation2000). Interestingly, certain bacterial VOCs, such as acetic acid, can induce the formation of biofilms, which have EPS as major constituents (Chen et al. Citation2015). It is then possible that certain PGPR VOCs may indirectly increase plant drought tolerance by mediating EPS synthesis (Liu and Zhang Citation2015). VOCs released by a Bacillus strain stimulated Arabidopsis synthesis of glycine betaine and its precursor chloline under osmotic stress (Zhang et al. Citation2010), both compounds are osmolytes involved in maintaining cell turgor under dehydrating conditions (Rhodes and Hanson Citation1993); the conferred osmo-protection could explain the enhanced tolerance to dehydration in plants treated with the PGPR VOCs or directly inoculated with the strain (Zhang et al. Citation2010). The accumulation of osmolytes, such as betaines, polyols and sugars (e.g. trehalose), polyamines and amino acids (e.g. proline) (Yancey Citation2001), is indeed the most frequent adaptation response observed in plants and bacteria under drought conditions (Sakamoto and Murata Citation2002; Vendruscolo et al. Citation2007; Rodriguez-Salazar et al. Citation2009). PGPR can release osmolytes accumulated in response to drought, which act synergistically with plant-synthesized osmolytes to stimulate plant growth (Paul and Nair Citation2008; Kaushal and Wani Citation2016). Several studies describe the modulation of plant osmolyte production by PGPR, which have been previously characterized or not in terms of osmolyte accumulation (Ghosh et al. Citation2017; Khan and Bano Citation2019). For example, the rise of leaf proline levels in maize under drought, increased even further with the inoculation of Pseudomonas fluorescens (Ansary et al. Citation2012).
Despite the previous examples, and as aforementioned, it is still largely unknow which are the major traits of the PGPR recruited by a given plant that intrinsically relate to the plant’s drought tolerance in each soil environment. da Costa et al. (Citation2014) suggested that under nutrient-rich conditions, plants will favour the recruitment of phytohormone producers, while in nutrient-poor conditions nutrient solubilizers will be favoured. Drought alters soil chemistry and diffusion rates and influence soil microbial communities (section 5.1), hence affecting soil mineralization and plant nutrient uptake (He and Dijkstra Citation2014). Thus, it is possible that nutrient solubilizers will be preferentially recruited under drought to ‘labour’ in micro niches in the rhizosphere. But if the patterns described by da Costa (Citation2014) might be used to direct the bioprospection of PGPR in different rhizospheric soils, the selected strains should be compatible with the recruitment mechanisms of the target plant, which can compromise the PGPR persistence in that rhizosphere.
4.2. Integrating plant-bacteria interactions in future PGPR selection strategies
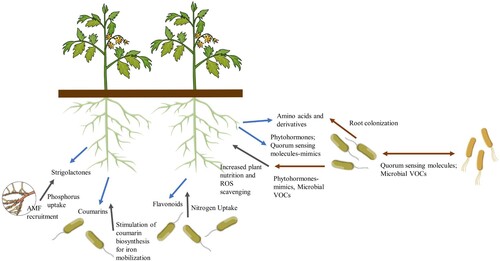
Could there be a primordial plant signal for bacterial recruitment at the onset of drought, causing a specific response of a multiple trait PGPR strain or of a multifunctional PGPR ensemble, which will be translated in efficient rhizospheric colonization? If so, could specific gene(s)/molecule(s) be screened instead of searching among general PGP traits? Plant recruitment of beneficial microbial partners involves a sophisticated chemical interaction, where root exudate is the primary means for ‘microbial capture’ (). Using small molecules, plants and microbes communicate, whereas microbial interspecies and intraspecies signalling molecules are perceived by plants and elicit a response (Venturi and Keel Citation2016); such global interaction results in the construction of the rhizomicrobiome. Coumarins, strigolactones and flavonoids, are secreted by plants under iron, phosphorus, and nitrogen deficiency, respectively, to facilitate the uptake of these nutrients, which results into selective microbial recruitment (Abedini et al. Citation2021). Research has been gradually revealing the existence of other plant signalling molecules, such as benzoxazinoids (BXs), whose production is affected by drought and other abiotic stresses (Abedini et al. Citation2021). The presence of the BX 2,4-dihydroxy-7-methoxy-2H-1,4-benzoxazin-3(4H)-one (DIMBOA) in maize root exudate resulted in the recruitment of the beneficial rhizobacteria Pseudomonas putida KT2440 (Neal et al. Citation2012). By releasing these toxic compounds, e.g. flavonoids and indoles, in the rhizosphere, plant roots exert a selective pressure and trigger a stress response in certain bacteria (Miche et al. Citation2003), affecting rhizobacterial communities. Therefore, potential PGPR must overcome such selective pressure to colonize the roots, i.e. they must therein express rhizosphere competence. Expression profile analysis of several bacterial strains has shown that numerous genes coding for stress response and detoxification proteins were induced in response to exudates (Miche et al. Citation2003; Cheng et al. Citation2009) or in the rhizosphere, with (Barret et al. Citation2009) or without (Barr et al. Citation2008; Barret et al. Citation2011) competing organisms. For example, genes encoding multidrug resistance (MDR) pumps, responsible for the efflux of toxic compounds, are frequently induced in the rhizosphere and present in large number in microorganisms found in association with plants (Konstantinidis and Tiedje Citation2004). Also, reported roles of proteins secreted by T1SS, T3SS and T5SS systems in PGPR rhizocompetence can be found (in Barret et al. Citation2011). For instance, T1SS substrates include proteins that are determinant of biofilm formation (Martinez-Gil et al. Citation2010) and are involved in the bacterial attachment to seeds (Espinosa-Urgel et al. Citation2000) or roots (Martinez-Gil et al. Citation2010), and T3SS of pseudomonads present in the mycorrhizosphere (the soil region around and influenced by mycorrhizal roots) has been proposed to be involved in suppressing the root innate immune response, facilitating the association between PGPR and mycorrhizal fungi/plants (in Barret et al. Citation2011). Both pathogenic and beneficial microbes can express the immunogenic microbe-associated molecular patterns (MAMPs), and thus may trigger an innate immune response in their host plants, known as MAMP-triggered immunity (MTI) (Teixeira et al. Citation2019). Colaianni et al. (Citation2021) proposed that plants monitor the proportion of diverse types of the peptide flagellin (flg22), a MAMP detected by plants, to identify the presence of pathogens; the authors found that plant-associated communities produce different classes of flg22 variants each resulting in a different plant response (e.g. variants that modulate signal-transduction pathways), and are enriched for immune evading flg22 epitopes. This functional diversity is key to plant-microbial interactions and could be inspected as a potential basis for future PGPR screening. Thus, the plant immune system, and defense signalling, is a driver for the selection of microbes that take part of the rhizomicrobiome communities, including the PGPR. For instance, synthesis of the phytohormone SA, whose accumulation is MAMP-induced (Wang et al. Citation2009), can alter the composition of the root and rhizosphere microbial community in A. thaliana (Lebeis et al. Citation2015). Microbe-microbe metabolic interactions through quorum sensing (QS) also play an important role in building the rhizomicrobiome. N-acyl homoserine lactone (AHL), a QS molecule produced by Gram-negative bacteria, acts as a signalling molecule within and between bacterial species, and between bacteria and plants, and has been extensively studied, but other QS molecules were recently identified, e.g. cis2-unsaturated fatty acids, pyrones, etc. (Venturi and Keel Citation2016). Plants can release QS-mimicking compounds, e.g. AHL-mimicking molecules, which can be perceived by bacteria and thus influence rhizomicrobiome assembly (Abedini et al. Citation2021). In addition, plants can produce compounds that modulate the concentration of QS molecules, a mechanism termed ‘Quorum Quenching’(QQ) or ‘Quorum Interfering’ (Dessaux and Faure Citation2018). Aside from QS molecules, microbial-produced VOCs have also been documented for their effect on plants (some examples were already disclosed above) and in the interaction with other microbes (in Abedini et al. Citation2021). In sum, convoluted complex metabolic interactions, involving signalling molecules, occur in the rhizosphere; the recognition of these molecules by the signal-receiving species, many of these unknown, and the cell integration of such signals are still an enigma.
Figure 3. Plant recruitment of soil microbes and their interactions. Signalling (blue and brown arrows from plants and microbes, respectively) and microbial-triggered plant responses (black arrows) are depicted. AMF: arbuscular mycorrhizal fungi; VOCs: volatile organic compounds.
PGPR’s ability to respond to root exudates and to use a large range of nutrients combined with efficient nutrient scavenging systems (section 3), are important for improving plant nutrient acquisition and are certainly decisive factors for successful rhizosphere colonization. The substrate uptake preferences of different bacteria will dictate niche differentiation, competitive exclusion, and cross-feeding phenomena and thus rhizomicrobiome composition (Jacoby and Kopriva Citation2019). In this context, not only specialized metabolites, but also primary metabolites (e.g. amino acids, carbohydrates) in plant root exudates have a role in mediating plant–microbe interactions. Genes involved in bacterial amino acid metabolism, for instance in histidine uptake or catabolism were found to be upregulated by exudates and in the rhizosphere (in Barret et al. Citation2011). Cole et al. (Citation2017) screened a transposon mutant library in Pseudomonas simiae for their ability to colonize Arabidopsis roots; while disruption of genes encoding carbohydrate metabolism resulted in lower root colonization fitness, disruption of genes encoding amino acid synthesis led to enhanced colonization fitness, indicating that amino acid auxotrophs, which will get those primary nutrients from the plant, have a selective advantage in the rhizosphere.
Bacterial chemotaxis response to root exudates is initiated by sensing specific ligands through methyl-accepting chemotaxis proteins (MCP) (for a review see Salah Ud-Din and Roujeinikova Citation2017). Oku et al. (Citation2012) identified and characterized MCPs receptors for sensing amino acids in P. fluorescens Pf0-1 and found these receptors were involved in chemotaxis towards tomato root exudate and root colonization. Despite the complexity of the chemical composition of root exudates, and the fact that the chemotaxis of PGPR to root exudates is an integrative effect of root-secreted chemo-attractants and chemo-repellents, the systematic identification of such compounds and the elucidation of how these are sensed by PGPR MCP chemoreceptors could contribute to the detection of novel PGPR. Feng et al. (Citation2018) identified 39 chemo attractants and 5 chemorepellents among components of cucumber root exudates, including amino acids, organic acids, and sugars, for Bacillus amyloliquefaciens SQR9, a known PGPR that forms biofilms on plant roots (Weng et al. Citation2013; Liu et al. Citation2017). A mutant strain with all eight putative chemoreceptors (predicted by the whole-genome sequence of SQR9) completely deleted, lost the chemotaxis ability to those 44 compounds. It would be interesting to perform similar studies considering root exudate composition, which changes under drought, to find PGPR responsive to drought-related root secreted compounds. For instance, in soybean (Glycine max), an increase in osmolytes concentration (including proline) in root exudates was observed by metabolomic analysis (Canarini et al. Citation2016). However, metabolome analyses are largely missing, namely those to study the root exudome formed under decreasing water availability, an important task to find PGPR recruited at the onset of drought stress, which could prevent extensive plant damage with drought continuation (see section 5.2).
A better understanding of the complex plant-microbial interactions demands for an integrated omics research, both at metabolite and gene-level, to unravel the primordial signals and microbial responsive genes important in the root recruitment of beneficial microbes by a given plant-soil set. Such research will lead to better rationales for the isolation of optimal PGPR. An omics approach can integrate the microbial ecology component, certainly contributing to reduce the inconsistent effects observed when applying PGPR in the field versus the greenhouse. Such approach could also shed light on the following question: is plant recruitment function-based (i.e. selection of functional traits over taxa), with responsive microbes possessing separately or collectively the larger set of traits involved in plant drought tolerance?
5. PGPR as part of the rhizomicrobiome
5.1. The rhizomicrobiome under drought
Drought affects soil and root microbial communities directly (decreasing moisture availability selects for desiccation tolerant taxa) and indirectly (by changing soil chemistry and plant phenotypes). Plant recruitment of bacteria from soil communities will be changed, as plants undergo several physiological responses to drought, which include alterations in root morphology and root exudate profile. Thus, the rhizomicrobiome under drought is determined by how drought formats both the host plant and the surrounding soils, with each of these factors exerting a mutual influence; water-limited soils may be decreased in overall ion content including potassium, phosphorus, and redox-sensitive ions (Bachar et al. Citation2010; Bouskill et al. Citation2016). Drought-induced changes in soil chemistry will modify soil nutrient cycling and shift the soil microbiome, which will in turn influence plant health, as plants depend on microbial biogeochemical transformations to turn soil nutrients bioavailable. On the other hand, drought-induced changes in plant exudome will change the composition and activity of the surrounding soil microbiome, promoting further changes in soil geochemistry. For instance, barley plants under drought had greater proline, potassium, and phytohormone concentrations in root exudates, which have roles in enhancing root growth, osmo-protection, and in stress signalling, if not intervening in bacterial recruitment (Calvo et al. Citation2016). Noteworthy, soil type, climate and anthropogenic activities (e.g. agricultural management practices), are frequently overlooked factors, which will affect microbial communities (Xun et al. Citation2021). Intensive agricultural practices, such as massive chemical inputs, intensification of land use and tillage affect microbial communities and their interactions, and thus crop microbiome structure and function, with a potential shift of the abundance of beneficial microbes that impact plant health. Three major negative effects to crop-associated microbial communities have been observed: i) reduced microbial diversity or overall microbial biomass, ii) altered microbiome functioning, and iii) disruption of beneficial relationships between the host plant and symbiotic microbes (in French et al. Citation2021). In brief, anthropogenic activities will change the soil physico-chemical properties, which are of major importance in shaping microbial communities (Xun et al. Citation2021) and vice versa. Therefore, identical drought conditions applied to chemically distinct soils will result in different responses of the microbial communities therein. As an example, Chodak et al. (Citation2015) found that factors as pH, total nitrogen, organic carbon content and heavy metals influenced the response of different bacterial phyla to drought and rewetting. The overall complexity described here renders the understanding of the effect of drought on the root-associated microbiome extremely difficult to achieve.
Microbiome studies have revealed higher diversity in bulk soil than in rhizosphere soil (Uroz et al. Citation2010; Lundberg et al. Citation2012). A great diversity is considered beneficial for soils, as a high species richness translates into many metabolic activities to facilitate an efficient decomposition of organic matter and nutrient mineralization (Nautiyal and Dion Citation2008). This bacterial diverse bulk soil is the primary repository from which roots recruit their microbiomes. Thus the root microbiome under drought will depend on the response of soil bacterial communities to moisture limitation. Overall, it has been reported that drought has little impact on bacterial phylogenetic diversity of soil communities (Bachar et al. Citation2010; Acosta-Martínez et al. Citation2014; Armstrong et al. Citation2016), but this tendency may depend on drought context, as, for instance, first exposure to drought reduced phylogenetic alpha-diversity in tropical forest soil plots, but no change was observed in pre-exposed plots (Bouskill et al. Citation2013). In contrast, bacterial community composition is significantly impacted by drought, with shifts in relative abundance, rather than fully disappearance of drought-susceptible taxa and appearance of tolerant ones, which helps explain the little effect in alpha-diversity. Often these relative abundance shifts are driven by few groups (Barnard et al. Citation2013). The increase in the ratio of Gram-positive to Gram-negative bacteria under drought is frequently observed, and commonly relative abundance changes include decreases in phyla Proteobacteria, Verrucomicrobia, and Bacteroidetes (Barnard et al. Citation2013; Bouskill et al. Citation2013; Acosta-Martínez et al. Citation2014) and increases in Firmicutes and Actinobacteria (Bouskill et al. Citation2013; Hartmann et al. Citation2017). Differences in metabolic capacities between Gram-positive and -negative bacteria may explain this phenomenon. Dry environments are ‘oligotrophic, i.e. poor in nutrients but abundant in oxygen. Oligotroph microbes, albeit slow-growers, can thrive under poor conditions, being substrate specialists with a narrow substrate utilization profile (Kurm et al. Citation2017), in opposition to substrate generalist copiotrophs that thrive under nutrient- and water-rich environments (Pascault et al. Citation2013; Hartmann et al. Citation2017). A greater abundance of bacterial genes involved in the degradation of complex plant polysaccharides was found in dry soils (Bouskill et al. Citation2016), indicating the proliferation of oligotrophic bacteria. The oligotrophic-copiotrophic dichotomy overlaps with the distinction of Gram-positive and Gram-negative bacteria. For example, Gram-positive bacteria can utilize inorganic nitrogen to support the cost production of extracellular enzymes necessary for the degradation of complex organic compounds present in dry soils, whereas Gram-negative prefer labile carbon compounds and organic nitrogen (Treseder et al. Citation2011). Additionally, other mechanisms that allow bacterial tolerance to drought, as sporulation and synthesis of thick cell walls, are attributes of Gram-positive phyla (Tocheva et al. Citation2016). Differences in the production and accumulation of osmolytes may also contribute to the enrichment of Gram-positive in dry soils. Gram-negative bacteria produce osmolytes as a drought-inducible response, whereas Gram-positive bacteria can produce osmolytes on a constitutive basis (Schimel et al. Citation2007). Either as an output of microbial recruiting from drought-affected soil communities, or/and due to endophytic communities with similar behaviors to those in the bulk soil, changes in the relative abundance in the root-associated microbiome have been shown to be largely similar to those seen in soil; the same taxa trends may be observed between soil and rhizomicrobiome communities when exposed to drought (), although the degree of enrichment and the specific taxa may differ (in Naylor and Coleman-Derr Citation2018).
Figure 4. Mechanisms at the basis of rhizomicrobiome assembly. A. Factors that drive the rhizosphere microbiome assembly. B. Drought has effects on soils, plants, and biotic-abiotic interactions. C. The integration of biotic and abiotic factors drives microbial community assembly at the rhizosphere level under a functional compensatory principle; the pattern of the assembly involves outside-in recruitment of the soil microbiome and inside-out release of the endosphere microbiome. D. Shifts in biotic and abiotic parameters under drought and their interactions influence plants and soils microbiome assembly and functionality, with changes towards the selection of rhizomicrobiomes adapted to drought that promote plant tolerance to this stressor. MAMP: microbe-associated molecular pattern, SA: salicylic acid, VOCs: volatile organic compounds.
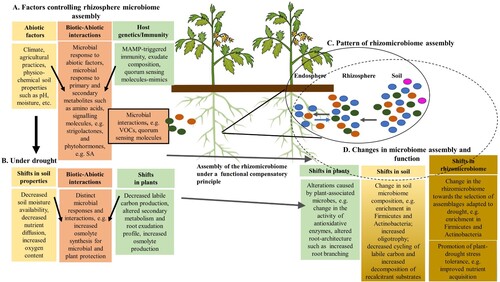
Host genetics should also be considered in the study of root-associated microbiome changes under drought, as it is an important driver of plant microbiome composition (). A variation in microbial community composition, and responsiveness to introduced microbes has been shown for several crop plants, with plant cultivar contributing to 5–20% of overall microbiome variation (French et al. Citation2021). However, such variation has been mostly studied across a small number of genotypes and growing conditions, and large-scale field studies are necessary to understand the heritability (i.e. plant genotype-sensitive microbial colonization) of the root-associated microbiome, having been performed only for maize (Walters et al. Citation2018) and rice (Zhang et al. Citation2019b). Importantly, until now, it is largely unknow if and how heritable variation in the microbiome correlates with beneficial traits and, despite several studies that have identified natural variation for beneficial bacteria root recruitment (Wintermans et al. Citation2016; French et al. Citation2020), few loci related to host-microbiome interactions and to beneficial microbiome traits have been isolated. Advances in this research area will be of major importance for crop breeding for beneficial plant–microbe interactions and cultivar selection. Recently, Deng et al. (Citation2021) performed a genome-wide association study to analyze the host genetic control on the microbiome composition of the sorghum rhizosphere. The authors identified ‘heritable’ taxa, and they were able to correlate the abundance of specific heritable bacterial lineages within the rhizosphere microbiome to specific host genetic loci, namely with allelic variation at a locus located on chromosome 4. The methodology allowed the detection of several lineages with high heritability, where prior evidence of plant-microbe interactions was lacking, including Verrucomicrobiales and Planctomycetales.
Host plants may have unusually high influence on rhizobacterial selection in ecosystems with limited microbial diversity. Recently, a study examining the rhizobacterial communities of date palms grown in heterogeneous soils across a broad geographic range of the Sahara Desert found similar trends in bacterial community composition across sites indicating the plant as major responsible for microbial assembly due to the decreased microbial complexity of the desert soils (Mosqueira et al. Citation2019). Host species had been previously shown to influence the beta-diversity in both watered and dry rhizosphere and root endosphere communities (Bouasria et al. Citation2012; Naylor et al. Citation2017). Naylor et al. (Citation2017) showed a correlation between host phylogenetic distances and microbiome dissimilarity for a broad range of grass plants. However, drought weakened that correlation by inducing conserved shifts in the rhizosphere and root microbiome across different grass hosts. A ‘core root microbiome’ of drought-enriched taxa was indeed found in several studies (Desgarennes et al. Citation2014; Coleman-Derr et al. Citation2016; Naylor et al. Citation2017). The presence in the enriched taxa of outlying lineages to those more commonly drought-associated might result from an active recruitment by the root based on the presence of specific PGP traits, even if those lineages possess a degree of drought sensitivity. In this context, it should be mentioned that the activity of many enzymes is higher in rhizosphere than in bulk soil (Marschner et al. Citation2005; Li et al. Citation2014) and these environments also show differences in their ‘functional gene structure’: the rhizosphere is a hotspot of genes conferring increased functional diversity (Li et al. Citation2014), indicating that more than inspecting for drought-responsive taxa one must study the functional rhizomicrobiome traits under drought. For example, the analysis of maize rhizospheres under drought showed higher protease, CAT, alkaline phosphatase, and invertase activities during most growth stages in a drought-tolerant cultivar compared with a drought-susceptible cultivar, a difference the authors postulated to be caused by differences in root exudates and microbial community composition (Song et al. Citation2012). According to Yan et al. (Citation2017), who studied the structure and functional diversity of the Jacobaea vulgaris rhizosphere and soil bacterial communities, functional traits are a key to the assembly of bacterial rhizosphere communities. However, the selection of such traits will be species-specific, i.e. the rhizosphere will select specific species based on functional traits. Xun et al. (Citation2021) proposed that a functional compensatory principle underlies the rhizosphere microbiome assembly, which is governed by abiotic factors, plant genotype and immunity and biotic interactions. In other words, the rhizosphere microbiome is assembled to compensate for functional requirements for the host plant fitness under given soil conditions, with gathering of the rhizosphere community involving both outside-in recruitment of the soil microbiome and inside-out release of the endosphere microbiome. It is thus valid to consider a positive answer to our last question in section 4.2, that plant-bacterial interactions under drought will promote the establishment of a rhizomicrobiome formed by those microbes that collectively contribute with the larger set of traits needed to cope with drought, and it is therefore of major importance to target community transcriptional and enzymatic activity in rhizosphere research. Additionally, genomic and metagenomic studies should be directed to the functionality of communities through the analysis of the sequences by a process-centric approach using Clusters of Orthologous Groups of proteins (COGs) as functional categories (Barret et al. Citation2011; Tringe et al. Citation2005). With such an approach, COGS overrepresented in one environmental sample are expected to be the result of selection by the local environment and thus related to specific functions of that ecological niche. As an example, changes in functional categories of the microbial gene pool of Populus rhizosphere were found under drought stress; in drought-treated samples, a significant increase in genes associated with signal transduction, membrane transport, and transport and catabolism was observed, whereas the abundance of genes associated with the hydrolysis and/or rearrangement of glycosidic bonds was significantly lower than in control samples. A high-resolution EggNOG (Evolutionary Genealogy of Genes: Non-supervised Orthologous Groups) analysis of the functional categories influenced by drought in the rhizosphere showed that sub-functions of a large number of genes were related to phosphonate ABC transport and sulfate ABC transport (Xie et al. Citation2021).
Genomic sequences can also be used to predict growth rates based on codon-usage bias, and data in wild oat showed that both fast-growing and slow-growing strains were enriched in the rhizosphere (Zhalnina et al. Citation2018), an evidence against the long established assumption that successful rhizosphere colonizers are fast-growing generalists using labile carbon substrates. Instead, the rhizosphere might confer temporal and spatial niches for metabolic specialists, able to take up more recalcitrant substrates (), such partitioning is consistent with the aforementioned enrichment of oligotrophs in bulk and rhizosphere communities under drought. Noteworthy, Nuccio et al. (Citation2016) proposed that ‘roots select less abundant or possibly rare populations in the soil microbial community’, which appear to consist of those bacteria that ‘have made a physiological tradeoff for rhizosphere competence at the expense of their competitiveness in non-rhizosphere soil’. Dawson et al. (Citation2017) showed that a small number of bacteria belonging to the rare biosphere dominated plant species-specific responses in rhizosphere colonization, indicating growth and metabolic activity for low-abundance populations in the rhizosphere. Hence, rhizospheric soils are potential hotspots for the survival of bulk soil low-abundant microbial members (see also section 6), whose selection may be mostly driven by ‘useful rhizospheric traits’, and their metabolic activity therein of importance for plant health. Such selection might explain the presence of unexpected taxa when analyzing vegetated soils. For instance, Firmicutes closely related to Parageobacillus thermoglucosidasius and Geobacillus thermocatenulatus, known thermophiles with an average temperature growth range of ca. 40°C–69°C (Nazina et al. Citation2001), were found associated with the shrub Zygophyllum dumosum (Kaplan et al. Citation2013).
5.2. Rhizomicrobiome-centered approaches for PGPR isolation
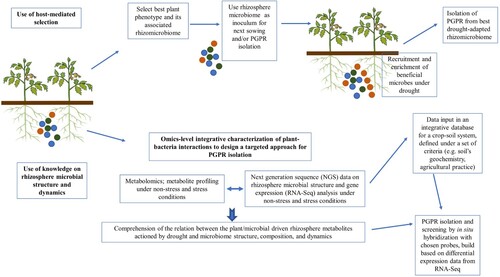
Due to the combined influence of plant host genotype and drought, host-mediated selection strategies to acquire a drought-adapted rhizomicrobiome may be envisaged. For example, the rhizomicrobiome can be selected in a specific host-genotype background and chosen from plants with improved phenotype(s), or changes in the microbiome can parallel selection on the host. The drought-adapted rhizomicrobiome may be used as inoculum for the next sowing (and similar selection) and/or PGPR isolation. This consists in a host-mediated engineering of the microbiome, an example of a ‘steered’ manipulation of the rhizosphere microbiome towards a specific outcome, which has been another global sustainable strategy under focus in recent years, meant to respond to the growing need to increase crop yields while reducing agrochemicals input. An alternative rhizomicrobiome-centered approach for PGPR isolation and selection would be one based on a deeper characterization at an omics-level of plant-rhizobacteria interactions and associated metabolic outputs. The integrated knowledge would be used to design a targeted procedure (). Examples of fundaments and prospects of both approaches follow.
Lau and Lennon (Citation2012) demonstrated that plant fitness under drought was highest when plants were grown in previously dry soils, as the plant will have no choice but to recruit a beneficial drought-tolerant microbiome which has developed through soils exposure to drought. On the other hand, plant recruitment of a drought-specific microbiome can be the result of generations of repeated drought events, that have led to the evolution of stable and beneficial plant-microbe interactions. For example, Brassica rapa plants that had been exposed to generations of drought tended to support higher bacterial abundance and diversity surrounding roots under dry environments (TerHorst et al. Citation2014). As so, screening for putative PGPR from plants rhizosphere (and from bulk soil) subjected to drought is a valid strategy and, in fact, roots and soils from chronically dry regions were found to frequently possess bacteria with PGP traits able to promote drought tolerance (Kaplan et al. Citation2013; Mayak et al. Citation2004; Timmusk et al. Citation2014). The isolation and use of putative PGPR, that coevolved with plant roots in harsh environments has been done for a variety of plant species (Mayak et al. Citation2004; Marasco et al. Citation2012; Timmusk et al. Citation2014; Wang et al. Citation2014; Cherif et al. Citation2015), suggesting that plants change the structure of the rhizomicrobiome towards the selection of drought-adapted assemblages, thereby improving their drought tolerance. Those PGPR may increase drought tolerance in plants different from those plants they were originally isolated from, and in certain cases plant growth is only promoted under drought conditions (Wang et al. Citation2014). These observations suggest that drought’s influence in shaping the rhizomicrobiome may be stronger than that of plant species. Jochum et al. (Citation2019) drew a screening procedure to specifically select PGPR that could rapidly colonize the rhizosphere and benefit multiple grass hosts when inoculated at the onset of drought conditions, so that the selected strains may be added as needed, as compared to current seed coating formulations. The original source for PGPR isolation were the rhizospheres of perennial grasses collected from a semi-arid environment, where those vigorously growing plants were likely to have a microbiome capable of conferring plant drought tolerance. A pre-screening process in a controlled setting focused on the desired host phenotype, which was the delayed onset of drought stress symptoms, instead of analyzing bacterial phenotypes. Rhizosphere samples of plants for which drought symptoms were most delayed under moderate and severe drought conditions were used for the bacterial isolation. Two PGPR strains, identified as Bacillus sp. (12D6) and Enterobacter sp. (16i), producers of IAA and SA in relatively high amounts compared with other profiled phytohormones, caused delays in the onset of drought symptoms when inoculated into the rhizospheres of wheat and maize seedlings. These strains led to alterations in root architecture; both strains increased root branching in wheat, and increased root length, surface area and root tips number in maize, which are phenotypic traits that correlate with better plant productivity under drought (Comas et al. Citation2013).
Using the co-selection for an enhanced crop phenotype with desired traits and associated beneficial microbiome in iterative rounds of plant phenotyping and microbiome transfer for the selection of PGPR and development of consortia thereof, has been already the subject of patents (e.g. Wigley et al. Citation2017). Nevertheless, this microbiome engineering methodology through host-mediated artificial selection, with the microbiome then used as a source for PGPR isolation, may have several drawbacks regarding the finding of optimal PGPR. The procedure should involve selected cultivars of agricultural importance, but the ability of modern crops to interact with PGPR may have become compromised during domestication (Valente et al. Citation2020). Other questions regarding the host-mediated selection of PGPR are: (i) the strength of selection to impose on the target microbial community; (ii) the fact that host-engineering experiments are performed under highly controlled conditions-requiring future work to determine if the selected PGPR are effective under the spatial and temporal variation of soil chemistry, native microbial communities, and climate found in field conditions-; and (iii) the choice of the host trait (French et al. Citation2021).
A major benefit of host-mediated engineering is that it does not rely on a specific mechanism associated with the target microbiome; by contrast, by selecting a host trait associated with a particular microbial community, an increasingly stable plant-associated microbiome, adapted to the host environment, may evolve over time (Mueller and Sachs Citation2015; Morella et al. Citation2020), and might provide an enrichment in native beneficial bacteria. Such bacteria are expected to be efficient in terms of rhizosphere competence and to have a positive impact on rhizosphere ecology. On the other hand, the potential for undesirable effects, namely in an iterative procedure of propagation of a selected host and its adapting microbiome, on host fitness and microbial community dynamics could be significant. Experiments were conducted involving selection of Brassica rapa plants, with repeated seed propagation, in the background of soil microbial communities experiencing the same selection history as the associated B. rapa plants (wet or dry soil conditions) (Lau and Lennon Citation2012; TerHorst et al. Citation2014). Reciprocal transplants were performed, where offspring from plant populations that had experienced wet or dry environments for three generations (‘plant evolutionary history’) where grown with microbial communities that had experienced either wet or dry environments (‘microbe history’) under either wet or dry contemporary environmental conditions. Data showed that microbial history and plant evolutionary history affected soil microbial communities and plant fitness and that interactions between ‘history’ and ecological context affected the direction and magnitude of the observed effects. For example, ‘plants were most fit when their contemporary environmental conditions (wet vs. dry soil) matched the historical environmental conditions of their associated microbial community’ (Lau and Lennon Citation2012). In other words, an eco-evolutionary perspective is required to fully assess plant-microbe interactions. The abovementioned experiments suggest that although field inoculation with PGPR or a community ensemble obtained following microbiome engineering could lead in a relatively short-term period to drought tolerant crops associated with drought-responsive rhizo-communities, that ensemble might not be suited to deal with a different newly imposed stress. If so, we might be forced to include such PGPR within an agricultural management practice to promote soil microbial diversity, such as crop rotation.
Altogether, it becomes clear that a strategy for isolating optimal PGPR for plant drought tolerance is missing, a strategy that does not require a successive, long-term, microbiome engineering. Any long used or recent management agricultural practice, including biofertilizer addition, will cause shifts in a crop rhizosphere microbiome (Mahmud et al. Citation2021) (section 5.1). French et al. (Citation2021) named host-mediated engineering of microbiomes, crop breeding (including gene-editing strategies) and the use of synthetic communities (section 6) as second-generation microbiome manipulations, while PGPR inoculation was designated as first-generation microbiome manipulation. Therefore, although efforts to optimize microbial inoculants (e.g. section 6), may improve their efficiency, failure to account for complex host-microbiome and microbial community interactions in PGPR selection (section 4.2 and below), and after PGPR application has limited their success. Further research is needed to understand how environmental factors (e.g. native microbial communities, soil type, nutrient levels, and management history) influence the establishment of microbial inoculants and benefits the host under complex field conditions, as exemplified from comparative meta-analysis of collected data (Veresoglou and Menexes Citation2010); in particular, understanding the interactions between native microbial communities and introduced microbes will result in important insights into better, ecology-guided, management practices integrating PGPR inoculation. However, such interactions have been disregarded, because the majority of the (few) studies has shown only transient establishment or low abundance of microbial inoculants during plant growth (Schreiter et al. Citation2014; Eltlbany et al. Citation2019), which led to the assumption that microbial inoculations would have negligible effects on the resident soil microbial communities. Despite a decreased abundance of inoculants following the inoculation, sometimes below the detection limit, microbial community composition can still be impacted (Cordier and Alabouvette Citation2009; Mallon et al. Citation2018). Whereas in some cases, when inoculant’s survival became low, the impact on soil communities was transient (Baudoin et al. Citation2009; Yin et al. Citation2013), in others, community composition changes persisted (Mallon et al. Citation2018; Renoud et al. Citation2022). Thus, quick disappearance of a bacterial inoculum in the soil does not necessarily mean a lack of a legacy on the resident community therein. Also, the magnitude of the impact, either transient or long-lasting, might not relate to the fate of the inoculant populations. In a recent review by Berg et al. Citation2021 several types of microbiome modulation have been described for microbial inoculants, including transient microbiome shifts, stabilization or increase of plant microbiome diversity and evenness- attributes of a healthy microbiome, and targeted shifts towards potential beneficial members of native microbiota. For instance, PGPR inoculation was shown to result in a significant change in pepper rhizosphere microbial community structure and composition, with higher relative abundance of genera commonly associated with crop yield enhancement (Zhang et al. Citation2019a). The mechanisms that underly the alteration in the resident microbial abundance, structure, and activities following introduction of a microbial inoculant were discussed by Mawarda et al. (Citation2020), who highlighted resource competition, antagonistic and synergistic interactions, and changes in root exudation rate and exudate composition. Interestingly, no study so far described an overall negative impact due to inoculant application, whereas many studies described an enrichment of well-known plant beneficial bacteria (in Berg et al. Citation2021). On the other hand, the lack of studies, together with the inconsistency of inoculant application strategies, time of sampling and technical approaches for community analysis, which differ in sensitivity (in Mawarda et al. Citation2020), are factors that impede to fully assess if the microbiome retains function and structure regardless the amount of inoculum (resistance); self-organizes after disturbance, returning to its original state (resilience); or adapts by reaching an alternative stable state. Importantly, many of the community analyses were performed by DNA based methods, where relic DNA obscures microbial diversity analysis, and the impact of microbial inoculation has been assessed mostly from a compositional perspective. Therefore, adding a functional perspective is fundamental to determine if the microbiome retains its functioning despite inoculant-induced changes in its composition, a key perspective considering a functional compensatory principle in rhizomicrobiome assembly (section 5.1). The lack of studies is notorious regarding the joint effect of abiotic stress and inoculant as we found only one report by Armada et al. (Citation2018): the authors showed that one year after the inoculation of autochthonous shrubs of semiarid Mediterranean zones with a native Bacillus thuringiensis strain, nearly no effect on rhizosphere microbial activity and on the rhizosphere microbiome was observed under drought, indicating a resilient community despite measurable contributions to plant growth and nutrient uptake. Like in most studies, the persistence of the inoculum throughout the experiment was not assessed, clouding the weight of direct and indirect contributions of the inoculant on the measured changes. Persisting inoculants may have longer impact not only on plants, but also on native communities compared to short-lived inoculants. Proposed strategies for overcoming environmental-persistence issues include repeated applications, PGPR strain selection based primarily in planta screening instead of in vitro testing, and the use of regionally microbial strains better adapted to local environmental stress conditions (in French et al. Citation2021 and afore discussed in this section). Still, short-term versus long-term effects of reshaping microbiomes, through PGPR establishment (and agricultural practices in general) are largely unknown and underexplored; a short-lived PGPR that will cause high but transient effects on microbial communities, which may recover from disturbance, may be an advantageous choice over an iterative microbiome manipulation, when the latter may lead to non-labile communities, unresponsive to new environmental conditions.
New procedures for PGPR isolation based on rhizosphere knowledge and encompassing the application of moderate or severe drought must be envisaged. The selection of PGPR from plant beneficial microbes recruited at the onset of drought will be of particular interest, since these might be applied as needed, and prevent extensive plant damage with drought continuity, by delaying plant drought symptoms. If abiotic, e.g. drought, and biotic factors intervene in rhizomicrobiome assembly under a functional compensatory principle (section 5.1), the evaluation of rhizomicrobiome diversity and composition must be an integrative process based on rhizosphere ‘functional gene structure’ and activity, to exploit the microbial functions needed for a sustainable agriculture in dry areas. For instance, the characterization of rhizosphere metabolite composition by metabolomics techniques for sample profiling under non-stress and stress conditions and its correlation with next generation sequence (NGS) data on rhizosphere microbial structure and metabolic traits (i.e. amplicon sequencing, RNA-based metatranscriptomics (Turner et al. Citation2013), and metagenomics) and gene expression (i.e. metatranscriptomics) is fundamental to the comprehension of the relation between the plant/microbial driven rhizosphere metabolites actioned by drought and microbiome structure, composition, and dynamics. By inferring the metabolic pathways and corresponding taxa associated to the plant root recruitment under drought, PGPR selection will target key indicators, as these will correspond to the repertoire of those essential genes expressed for effective PGPR response to plant signals, colonization and survival within the rhizosphere and a successful plant-microbial interaction. Although a herculean task, it may be fragmented targeting specific molecular subsets, for instance a correlation study can be done relating amino acid profiling with gene expression analysis of amino acid transport and metabolism. Combining microbiome omics analysis with high-throughput screening of rhizo-isolates and plant bioassays will enable the identification of relevant microbial traits important to plant health (Gu et al. Citation2020). Application of high-throughput methods to directly measure the rhizomicrobial consumption of identified root metabolites (such as amino acids) will provide additional information on rhizospheric interactions to be used for PGPR screening. For instance, using exometabolomics (a sub-field of metabolomics regarding the footprinting of extracellular metabolites) to profile a defined growth medium before and after inoculation with a rhizobacterial ensemble or rhizo-isolates can reveal which metabolites are consumed as growth substrates and which are released for microbial cross-feeding (Jacoby and Kopriva Citation2019).
The results of such studies could become part of an integrative database organized for a crop/reference cultivar(s)-soil system, being this system defined under a set of criteria such as soil geochemistry and agriculture management history. The interacting effects of common agricultural practices (e.g. crop rotations, organic amendments, and environmental factors) on the rhizomicrobiome structure and function are understudied, thus the definition of criteria for each plant-rhizomicrobial crosstalk is important and will contribute to identify when an agricultural practice will be opposite to beneficial plant–microbe relationships. A research consortium bringing together different research centers with distinct but complementary skills could be created to standardize the different analyses to be performed and to build the database.
The above acquired data might be used to draw new PGPR screening procedures. For instance, metabolite profiling of the rhizosphere of stressed plants may reveal distinct carbon compounds at the onset of drought, which might be important for microbial recruitment, thus, isolation media containing those compounds as carbon sources can be conceived. Knowledge of the composition of the active microbial fraction may allow inference of the major taxa recruited under stress, an information that may be used for media elaboration to select putative PGPR. In situ hybridization might be used to detect colonies expressing genes identified by RNA-Seq, found to be up-regulated under drought stress. A protocol can be defined that will streamline media conception and probe construction, with the latter based on differential expression data from mRNA sequencing. RNA-Seq data might be useful as part of a data set of drought stress related genes, which could be used to design a molecular toolbox based on probe conception for detection of homologue genes and similar putative PGPR in several plant-rhizomicrobiome systems; Kaplan et al. (Citation2020) showed that soil microbial communities are compositionally more similar between close plant relatives, suggesting that PGPR detected with a such molecular toolbox could be used for phylogenetic similar plants under similar settings. The putative PGPR may be tested for drought stress relief in consortia with other microbes, for instance STB (soil thermophilic bacteria) (section 6), not classified as PGPR, but important in biogeochemical cycling, and thus in soil fertilization.
6. Planning consortia with unfamiliar rhizo- and soil bacteria for improving plant drought tolerance
An approach to optimize the impact of PGPR involves the development of formulations with multiple microbial strains with different functions, hypothesized to act additively to improve plant performance. Adding multiple culturable strains to build synthetic communities as a simplified proxy of a microbiome in structure and function has been done to study community assembly dynamics and functional interactions, revealing keystone strains in the assembly of beneficial communities (Niu et al. Citation2017; Carlström et al. Citation2019). Predicting higher-order community dynamics has proven difficult (in French et al. Citation2021), but advances in high-throughput screening technologies, as the availability of a recent droplets-based platform for massively parallel construction and screening of synthetic microbial communities, will enable a rapid identification and improved design of beneficial consortia (Kehe et al. Citation2019). The development of reproducible fabricated microbial ecosystems, termed EcoFABs (Zengler et al. Citation2019), with standardized system components and workflows will enable a precise manipulation of communities and analysis of microbial localization, activity and interactions across temporal and spatial scales and future elaboration of ecological theories and predictive models. The use of these emerging technologies will ultimately result in the design of microbial products (e.g. consortia to optimize beneficial plant-microbial partnerships) avoiding the mixed results (in French et al. Citation2021) from consortia that are built simply based on the hypothesis of additive effects of diverse, within compatible PGPR on plant health. Noteworthy, increasing diversity in microbial communities can reduce community functioning and plant health as a result of increased antagonistic interactions (Becker et al. Citation2012), i.e. lower diversity is not always indicative of less healthy microbial communities (Shade Citation2017). This fact reinforces our preference in the search and application of PGPR strains and minimal consortia over the use of rhizomicrobiome manipulations, the latter might be associated with a higher number of undesirable antagonistic interactions, above all in low resource-environments, such as in dry soils.
Despite the multiple beneficial characteristics of described PGPR and consortia, it is largely unknown if these can improve plant fitness under high levels of abiotic stresses such as high salt, temperature, and drought conditions found in local agricultural fields. With the expansion of drylands (semi-arid, arid, and hyper-arid areas), often also experiencing high saline conditions, alternative, unfamiliar bacteria have been searched, hoping they can perform as PGPR under challenging soil conditions. As an example, Mishra et al. (Citation2017) searched for bacteria in volcano soils, expecting for a high probability that soil bacterial isolates from such extreme conditions may present both multiple PGP and relevant abiotic stress tolerance traits. They isolated bacteria capable of withstanding high temperature, salinity and drought, presenting multiple PGP traits such as EPS and auxin production, and phosphate solubilization. The in vivo seed treatment of maize plants with individual selected isolates resulted in biomass enhancement, which was most evident for a bacterial isolate identified as an Ochrobactrum sp.
However, there is no need to look for thermophilic bacteria in extreme environments. Soil thermophilic bacteria of the Firmicutes Phylum (genera Geobacillus, Parageobacillus, Ureibacillus, Brevibacillus, Bacillus), herein named STB, were shown to be ubiquitous (Marchant et al. Citation2002a, Citation2002b) in upper soils worldwide, where they were found in high numbers (Marchant et al. Citation2002a; Portillo et al. Citation2012). The maintenance of these thermophiles in cool temperate soils could be the result of a balance between near-zero growth and negligible death rates (Marchant et al. Citation2008). At medium and low latitudes, where high temperature events on surface soil layers are frequent, STB have therein temporal opportunities for activity and growth (Portillo et al. Citation2012; Santana and Gonzalez Citation2015) and may play an important role in S-cycle, as well as in N and C cycles, being able to release significant quantities of both sulfate (and ammonium) from the consumption of proteinaceous compounds, as a product of a dissimilative organic S-oxidizing process (Portillo et al. Citation2012; Santana et al. Citation2013).
STB sulfate-release activity has been related to seedling growth enhancement (Santana et al. Citation2013). Moreover, STB have been isolated from rhizospheric soils (Pawar and Borkar Citation2018). Although not characterized in terms of phylogeny, thermophilic bacteria were also isolated from the bulk soil and the rhizosphere of the grass Aristida adscensionis L. in a caatinga of the semi-arid Brazilian Northeast (Gorlach-Lira and Coutinho Citation2007). The rhizospheric effect of A. adscensionis on the number of bacterial isolates was more pronounced in case of thermophiles than mesophiles. Among the thermophilic bacteria isolates, five were thermophiles sensu stricto with Tmax > 55°C and Tmin > 30°C while other strains were thermotolerant. More recently, it was shown that wheat root exudate increased the survival of an STB (Ureibacillus sp. 18UE/10) at low temperature and that this strain affected wheat physiology under rhizospheric conditions (Santana et al. Citation2020). Because STB are motile, one can hypothesize their migration between a deeper rhizospheric, subsurface layer and upper soil. At upper soil layers, STB will proliferate and mineralize organic S; the inorganic S formed at the upper soil layers may be easily transported down to deeper soil layers (Edwards Citation1998; Conant et al. Citation2011), representing an S source for plants and microbes. STB could thus profit from the high organic content existing at upper soil layers (top 5 cm) (López-Bellido et al. Citation2010) and the rhizosphere. Notably, although STB represent a minority among soil microbial communities, their extracellular enzymes, which were shown to have optimal activity at high temperature and low water activity (Gomez et al. Citation2020a; Citation2021), persist longer in soils than enzymes of mesophiles, suggesting soils could be a reservoir of these enzymes allowing a fast response by STB to changing conditions (Gomez et al. Citation2020b). Since soil’s upper layers frequently get dry and with high temperatures, and as the activity of microbial extracellular enzymes is a key step for soil organic matter mineralization, STB are of increasingly importance in biogeochemical soil cycles under the ongoing climate change.
The potential importance of STB is particularly relevant when considering the increasing demand for sulfate during plant metabolic adaptation reactions during drought stress, which reflects specific roles of sulfur-containing compounds, for example, the role of cysteine as an S donor for the sulfuration of molybdenum cofactor (Moco) acting in ABA biosynthesis, or the role of sulfated compounds of the secondary metabolism as osmoprotectants (Abuelsoud et al. Citation2016). The ubiquity of STB and proliferation under high temperature events, their maintenance at low temperatures and their potential role in plant S nutrition (Santana et al. Citation2021) make them candidates to include in consortia with PGPR for improving plant drought tolerance. Nonetheless, implementation of such consortia should consider the evaluation of post-inoculation effects on several soil physicochemical parameters, namely on soil organic content, and on soil microbial diversity and abundance. A raw prediction of the effect of STB on soil biodiversity may be presented based on their small proportion among soil microbial communities and on the absence or seasonal STB active proliferation in temperate soils; although soil microbial diversity and relative abundance might change following STB inoculation, these factors might contribute to soil biodiversity resilience at mid- or long-term. Overall, besides considering the typical rhizobacteria populations in the search of PGPR, bacteria from other habitats, or low-abundant soil members can be important PGPR or adjuvants in soil fertility under local high stress events, and thus contribute for establishing a network of integrated functionality.
7. Conclusions-summarizing key points and research gaps
The search for PGPR to improve plant drought tolerance is a key strategy in the context of increasing desertification and ongoing climate changes. Current procedures for PGPR isolation are limited in the search of optimal PGPR, since important parameters as different soil type and (micro)structure, plant species and seasonality, are frequently neglected in sample collection. Also, those procedures bias the selection towards easily cultivable abundant soil bacteria, which may not be the best to improve plant drought tolerance. Moreover, PGPR screening has been mainly based in the identification of general biochemical PGP traits in isolates, which are then tested for PGP activity in the greenhouse. Overall, this ‘physiological shotgun procedure’ misses more fitted PGPR candidates with enhanced ‘drought-solving’ ability, those that are efficiently recruited by the targeted plant-rhizosphere system. Potential PGPR must efficiently colonize the rhizosphere under drought, but the major traits relevant for rhizosphere competence and for plant drought tolerance under a given crop-soil system are largely unknown. For identifying such primordial trait(s), we need a deeper knowledge of the convoluted interactions in the rhizosphere involving plants and microbes, which implies the comparative study of the rhizomicrobiome. Drought and host genotype contribute to modulate the plant’s rhizomicrobiome, whose assembly is ruled by a functional compensatory principle. Such principle might explain the presence of unexpected taxa in the rhizosphere, namely of bulk-soil low-abundant microbial members, whose selection may be mostly driven by ‘useful rhizospheric traits’, important for plant health. Plant-microbiota-abiotic interactions under drought will thus be directed to the establishment of a rhizomicrobiome formed by microbial assemblages adapted to drought. A drought-specific rhizomicrobiome can be achieved after generations of repeated drought events leading to the evolution of stable and beneficial plant-microbe interactions. Hence, a host-mediated selection of a microbiome adapted to drought, to be used as a source for PGPR able to improve plant drought tolerance, is a valid strategy. However, the strategy could have drawbacks, namely if in an iterative procedure of propagation of selected host and associated microbiome, on host fitness and microbial community (including PGPR within) response after the imposition of a different newly stress. Alternatively, we propose an omics-level (metabolomics and metatranscriptomics) comparative analysis to improve our knowledge on the mechanisms of rhizomicrobiome construction under drought, so to design a strategy for selection of PGPR, whose inoculation may lead to a shift to a beneficial rhizosphere microbiome, as revealed by few studies. An integrated meta-evaluation of the rhizomicrobiome composition and of the associated rhizo-metabolites is fundamental to infer the taxa and metabolic pathways associated to the root recruitment under drought and to design a PGPR screening based on target indicators. Selected putative PGPR can then be tested and used either alone or in consortia to take advantage of the additional benefits conferred by multiple PGP traits, and also from the indirect growth promotion given by unfamiliar rhizo-microbes, not classified as PGPR, but important for soil health. For example, STB may be part of such consortia, as they have been shown to have a role in organic S-mineralization and sulfate production, which are important for plant drought tolerance.
Authors’contributions
MMS, APR and AM: Original draft preparation; Review and editing: MMS, TD, and CC. All authors have read and agreed to the published version of the manuscript.
Acknowledgments
This study was financially supported by Portuguese funds through Fundação para a Ciência e a Tecnologia (project UIDB/00329/2020), researcher contracts to MMS and TD, and PhD grant SFRH/BD/136188/2018.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Additional information
Funding
Notes on contributors





Ana Paula Rosa
Ana Paula Rosa has a Degree in Biology and a MSc in Conservation Biology from the Faculty of Sciences, University of Lisbon, and is currently doing a PhD in Microbiology with the Plant-Soil Ecology research group of Centre for Ecology, Evolution and Environmental Changes & Global Change and Sustainability Institute (cE3c & CHANGES). She has worked on environmental assessment for several years and has done research on the impacts of increased C and N availability in terrestrial ecosystems and greenhouse gas budgets. Her research is focused on plant-bacteria associations and on the effect of soil thermophilic bacteria on C, S, and N cycling and on plant nutrient availability.
Teresa Dias graduated in Plant Biology at the Faculty of Sciences, University of Lisbon (FCUL) in 2002. In 2012, she received a PhD in Biology-Ecophysiology, from the same University. Since 2019, she is a researcher at FCUL and member of the cE3c & CHANGES. Throughout her research, Teresa Dias has always developed work within Plant-Soil Interactions, applying the knowledge acquired in the sphere of ecology and physiology to the resolution of fundamental and applied ecological problems.
Abdul M. Mouazen is a Full Professor and leader of the Precision Soil and Crop Engineering (PreSco) Group. He holds a PhD degree in numerical modelling of soil-tillage tools interaction. He has a background in the application of engineering principles to soil and water management, with specific applications in soil dynamics, tillage, traction, compaction, mechanical weeding, soil remediation and management and precision agriculture.
Cristina Cruz received a PhD in Ecology and Plant Systematics from the University of Lisbon in 1994. She is an Auxiliar Professor at the Faculty of Sciences, University of Lisbon, and Researcher at cE3c & CHANGES. She has a background in ecophysiology of plant nutrition and soil ecology.
Margarida M. Santana received a PhD in Sciences from the University of Paris XI in 1995, and an equivalence of the doctoral degree in Microbiology from the Faculty of Sciences, University of Lisbon (FCUL) in 1996. She is currently a researcher at FCUL and member of the cE3c & CHANGES, where she develops her work in the Plant-Soil Ecology research group. Her research focuses on plant-bacteria associations and on the effect of soil thermophilic bacteria on plant nutrient availability.
References
- Abedini D, Jaupitre S, Bouwmeester H, Dong L. 2021. Metabolic interactions in beneficial microbe recruitment by plants. Curr Opin Biotechnol. 70:241–247.
- Abuelsoud W, Hirschmann F, Papenbrock J. 2016. Sulfur metabolism and drought stress tolerance in plants. In: Hossain M, Wani S, Bhattacharjee S, Burritt D, Tran LS, editors. Drought stress tolerance in plants. Vol. 1. Cham: Springer; p. 227–249. doi: 10.1007/978-3-319-28899-4_9.
- Acosta-Martínez V, Cotton J, Gardner T, Moore-Kucera J, Zak J, Wester D, Cox S. 2014. Predominant bacterial and fungal assemblages in agricultural soils during a record drought/heat wave and linkages to enzyme activities of biogeochemical cycling. Appl Soil Ecol. 84:69–82.
- Adedeji AA, Max A, Häggblomb MM, Babalola OO. 2020. Sustainable agriculture in Africa: plant growth-promoting rhizobacteria (PGPR) to the rescue. Sci African. 9:e00492. doi: 10.1016/j.sciaf.2020.e00492.
- Adesemoye AO, Torbert HA, Kloepper JW. 2009. Plant growth-promoting rhizobacteria allow reduced application rates of chemical fertilizers. Microb Ecol. 58:921–929.
- Akhtar SS, Amby DB, Hegelund JN, Fimognari L, Großkinsky DK, Westergaard JC, Müller R, Moelbak L, Liu F, Roitsch T. 2020. Bacillus licheniformis FMCH001 increases water use efficiency via growth stimulation in both normal and drought conditions. Front Plant Sci. 11:297. doi: 10.3389/fpls.2020.00297.
- Alami Y, Achouak W, Marol C, Heulin T. 2000. Rhizosphere soil aggregation and plant growth promotion of sunflowers by an exopolysaccharide-producing Rhizobium sp. strain isolated from sunflower roots. Appl Environ Microbiol. 66:3393–3398.
- Amaya-Gómez CV, Porcel M, Mesa-Garriga L, Gómez-Álvarez MI. 2020. A framework for the selection of plant growth promoting rhizobacteria based on bacterial competence mechanisms. Appl Environ Microbiol. 86:e00760–20. doi: 10.1128/AEM.00760-20.
- Ansary MH, Rahmani HA, Ardakani MR, Paknejad F, Habibi D, Mafakheri S. 2012. Effect of Pseudomonas fluorescens on proline and phytohormonal status of maize (Zea mays L.) under water deficit stress. Annal Biol Res. 3:1054–1062.
- Armada E, Leite MFA, Medina A, Azcón R, Kuramae EE. 2018. Native bacteria promote plant growth under drought stress condition without impacting the rhizomicrobiome. FEMS Microbiol Ecol. 94(7):1–13. doi: 10.1093/femsec/fiy092.
- Armstrong A, Valverde A, Ramond J-B, Makhalanyane TP, Jansson JK, Hopkins DW, Aspray TJ, Seely M, Trindade MI, Cowan DA. 2016. Temporal dynamics of hot desert microbial communities reveal structural and functional responses to water input. Sci Rep. 6:34434. doi: 10.1038/srep34434.
- Bachar A, Al-Ashhab A, Soares MIM, Sklarz MY, Angel R, Ungar ED, Gillor O. 2010. Soil microbial abundance and diversity along a low precipitation gradient. Microb Ecol. 60:453–461.
- Backer R, Rokem JS, Ilangumaran G, Lamont J, Praslickova D, Ricci E, Subramanian S, Smith DL. 2018. Plant growth-promoting rhizobacteria: context, mechanisms of action, and roadmap to commercialization of biostimulants for sustainable agriculture. Front Plant Sci. 9:1473. doi: 10.3389/fpls.2018.01473.
- Barnard RL, Osborne CA, Firestone MK. 2013. Responses of soil bacterial and fungal communities to extreme desiccation and rewetting. ISME J. 7:2229–2241.
- Barr M, East AK, Leonard M, Mauchline TH, Poole PS. 2008. In vivo expression technology (IVET) selection of genes of Rhizobium leguminosarum biovar viciae A34 expressed in the rhizosphere. FEMS Microbiol Lett. 282:219–227.
- Barret M, Frey-Klett P, Guillerm-Erckelboudt AY, Boutin M, Guernec G, Sarniguet A. 2009. Effect of wheat roots infected with the pathogenic fungus Gaeumannomyces graminis var. tritici on gene expression of the biocontrol bacterium Pseudomonas fluorescens Pf29Arp. Mol Plant Microbe Interact. 22:1611–1623.
- Barret M, Morrissey JP, O’Gara F. 2011. Functional genomics analysis of plant growth-promoting rhizobacterial traits involved in rhizosphere competence. Biol Fertil Soils. 47:729–743.
- Barriuso J, Pereyra MT, García JAL, Megías M, Mañero FJG, Ramos B. 2005. Screening for putative PGPR to improve establishment of the symbiosis Lactarius deliciosus-pinus sp. Microb Ecol. 50:82–89.
- Barriuso J, Solano BR, Lucas JA, Lobo AP, García-Villaraco A, Mañero FJG. 2008. Ecology, genetic diversity and screening strategies of plant growth promoting rhizobacteria (PGPR). In: Ahmad I, Pichtel J, Hayat S, editors. Plant-bacteria interactions:strategies and techniques to promote plant growth. Weinheim: Wiley-VCH Verlag GmbH and Co. KGaA; p. 1–17.doi: 10.1002/9783527621989.ch1.
- Basílio F, Dias T, Santana MM, Melo J, Carvalho L, Correia P, Cruz C. 2022. Multiple modes of action are needed to unlock soil phosphorus fractions unavailable for plants: The example of bacteria- and fungi-based biofertilizers. Appl Soil Ecol. 178:104550. doi: 10.1016/j.apsoil.2022.104550.
- Batool T, Ali S, Seleiman MF, Naveed NH, Ali A, Ahmed K, Abid M, Rizwan M, Shahid MR, Alotaibi M, et al. 2020. Plant growth promoting rhizobacteria alleviates drought stress in potato in response to suppressive oxidative stress and antioxidant enzymes activities. Sci Rep. 10(1):16975. doi: 10.1038/s41598-020-73489-z.
- Baudoin E, Nazaret S, Mougel C, Ranjard L, Moënne-Loccoz Y. 2009. Impact of inoculation with the phytostimulatory PGPR Azospirillum lipoferum CRT1 on the genetic structure of the rhizobacterial community of field-grown maize. Soil Biol Biochem.41:409–413.
- Becker J, Eisenhauer N, Scheu S, Jousset A. 2012. Increasing antagonistic interactions cause bacterial communities to collapse at high diversity. Ecol Lett. 15:468–474.
- Berg G, Kusstatscher P, Abdelfattah A, Cernava T, Smalla K. 2021. Microbiome modulation-toward a better understanding of plant microbiome response to microbial inoculants. Front Microbiol. 12:650610. doi: 10.3389/fmicb.2021.650610.
- Bhaskar PV, Bhosle NB. 2005. Microbial extracellular polymeric substances in marine biogeochemical processes. Curr Sci. 88:45–53.
- Bouasria A, Mustafa T, De Bello F, Zinger L, Lemperiere G, Geremia RA, Choler P. 2012. Changes in root-associated microbial communities are determined by species-specific plant growth responses to stress and disturbance. Eur J Soil Biol. 52:59–66.
- Bouskill NJ, Lim HC, Borglin S, Salve R, Wood TE, Silver WL, Brodie EL. 2013. Pre-exposure to drought increases the resistance of tropical forest soil bacterial communities to extended drought. ISME J 7:384–394.
- Bouskill NJ, Wood TE, Baran R, Ye Z, Bowen BP, Lim H, Zhou Z, Van Nostrand JD, Nico P, Northern TR, et al. 2016. Belowground response to drought in a tropical forest soil. I. changes in microbial functional potential and metabolism. Front. Microbiol. 7:525. doi: 10.3389/fmicb.2016.00525.
- Calvo OC, Franzaring J, Schmid I, Müller M, Brohon N, Fangmeier A. 2016. Atmospheric CO2 enrichment and drought stress modify root exudation of barley. Glob Change Biol. 23:1292–1304.
- Canarini A, Merchant A, Dijkstra FA. 2016. Drought effects on Helianthus annuus and glycine max metabolites: from phloem to root exudates. Rhizosphere. 2:85–97.
- Carlström CI, Field CM, Bortfeld-Miller M, Müller B, Sunagawa S, Vorholt JA. 2019. Synthetic microbiota reveal priority effects and keystone strains in the Arabidopsis phyllosphere. Nat Ecol Evol. 3:1445–1454.
- Chaves MM, Flexas J, Pinheiro C. 2009. Photosynthesis under drought and salt stress: regulation mechanisms from whole plant to cell. Ann Bot. 103:551–560.
- Chen Y, Gozzi K, Yan F, Chai Y. 2015. Acetic acid acts as a volatile signal to stimulate bacterial biofilm formation. mBio. 6(3):e00392. doi: 10.1128/mBio.00392-15.
- Cheng ZY, Duan J, Hao YA, McConkey BJ, Glick BR. 2009. Identification of bacterial proteins mediating the interactions between Pseudomonas putida UW4 and Brassica napus (canola). Mol Plant Microbe Interact. 22:686–694.
- Cherif H, Marasco R, Rolli E, Ferjani R, Fusi M, Soussi A, Mapelli F, Bilou I, Borin S, Boudabous A, et al. 2015. Oasis desert farming selects environment-specific date palm root endophytic communities and cultivable bacteria that promote resistance to drought. Environ Microbiol. Rep. 7:668–678.
- Chiappero J, Cappellari LR, Alderete LGS, Palermo TB, Banchio E. 2019. Plant growth promoting rhizobacteria improve the antioxidant status in Mentha piperita grown under drought stress leading to an enhancement of plant growth and total phenolic content. Ind Crops Prod. 139:111553.
- Cho SM, Kang BR, Han SH, Anderson AJ, Park JY, Lee YH, Cho BH, Yang KY, Ryu CM, Kim YC. 2008. 2R, 3R-butanediol, a bacterial volatile produced by Pseudomonas chlororaphis O6, is involved in induction of systemic tolerance to drought in Arabidopsis thaliana. Mol Plant Microbe Interact. 21:1067–1075.
- Chodak M, Gołebiewski M, Morawska-Płoskonka J, Kuduk K, Niklinska M. 2015. Soil chemical properties affect the reaction of forest ´soil bacteria to drought and rewetting stress. Ann Microbiol. 65:1627–1637.
- Clarke SF, Guy PL, Burritt DJ, Jameson PE. 2002. Changes in the activities of antioxidant enzymes in response to virus infection and hormone treatment. Physiol Plant. 114:157–164.
- Clements LD, Miller BS, Streips UN. 2002. Comparative growth analysis of the facultative anaerobes Bacillus subtilis, Bacillus licheniformis, and Escherichia coli. Syst Appl Microbiol. 25:284–286.
- Cohen AC, Travaglia CN, Bottini R, Piccoli PN. 2009. Participation of abscisic acid and gibberellins produced by endophytic Azospirillum in the alleviation of drought effects in maize. Botany. 87:455–462.
- Colaianni NR, Parys K, Lee HS, Conway JM, Kim NH, Edelbacher N, Mucyn TS, Madalinski M, Law TF, Jones CD, et al. 2021. A complex immune response to flagellin epitope variation in commensal communities. Cell Host Microbe. 29(4):635–649. doi: 10.1016/j.chom.2021.02.006.
- Cole BJ, Feltcher ME, Waters RJ, Wetmore KM, Mucyn TS, Ryan EM, Wang G, Ul-Hasan S, McDonald M, Yoshikuni Y, et al. 2017. Genome-wide identification of bacterial plant colonization genes. PLoS Biol. 15(9):e2002860.
- Coleman-Derr D, Desgarennes D, Fonseca-Garcia C, Gross S, Clingenpeel S, Woyke T, North G, Visel A, Partida-Martinez LP, Tringe SG. 2016. Plant compartment and biogeography affect microbiome composition in cultivated and native agave species. New Phytol. 209:798–811.
- Comas LH, Becker SR, Cruz VM, Byrne PF, Dierig DA. 2013. Root traits contributing to plant productivity under drought. Front Plant Sci. 4:442. doi: 10.3389/fpls.2013.00442.
- Conant RT, Ryan MG, Ågren GI, Birge HE, Davidson EA, Eliasson PE, Evans SE, Frey SD, Giardina CP, Hopkins R, et al. 2011. Temperature and soil organic matter decomposition rates – synthesis of current knowledge and a way forward. Glo Chang Biol. 17:3392–4004.
- Cordero I, Balaguer L, Rincón A, Pueyo JJ. 2018. Inoculation of tomato plants with selected PGPR represents a feasible alternative to chemical fertilization under salt stress. J Plant Nutr Soil Sci. 181:694–703.
- Cordier C, Alabouvette C. 2009. Effects of the introduction of a biocontrol strain of Trichoderma atroviride on non target soil micro-organisms. Eur J Soil Biol. 45:267–274.
- da Costa P, Granada CE, Ambrosini A, Moreira F, de Souza R, dos Passos JFM, et al. 2014. A model to explain plant growth promotion traits: a multivariate analysis of 2,211 bacterial isolates. PLoS One. 9:e116020. doi: 10.1371/journal.pone.0116020.
- Datta R, Anand S, Moulick A, Baraniya D, Pathan SI, Rejsek K, Vranova V, Sharma M, Sharma D, Kelkar A, Formanek P. 2017. How enzymes are adsorbed on soil phase and factors limiting its activity: A review. Int Agrophysics. 31:287–302.
- Dawson W, Hör J, Egert M, van Kleunen M, Pester MA. 2017. A small number of low-abundance bacteria dominate plant species-specific responses during rhizosphere colonization. Front Microbiol. 8:975. doi: 10.3389/fmicb.2017.00975.
- Deng S, Caddell DF, Xu G, Dahlen L, Washington L, Yang J, Coleman-Derr D. 2021. Genome wide association study reveals plant loci controlling heritability of the rhizosphere microbiome. ISME J. 15:3181–3194.
- Desgarennes D, Garrido E, Torres-Gomez MJ, Peña-Cabriales JJ, Partida-Martinez LP. 2014. Diazotrophic potential among bacterial communities associated with wild and cultivated agave species. FEMS Microbiol Ecol. 90:844–857.
- Dessaux Y, Faure D. 2018. Quorum sensing and quorum quenching in Agrobacterium: a “go/no go system”? Genes (Basel). 9(4):210. doi:10.3390/genes9040210.
- Dias T, Dukes A, Antunes PM. 2015. Accounting for soil biotic effects on soil health and crop productivity in the design of crop rotations. J. Sci. Food Agric. 95:447–454.
- Duan B, Li L, Chen G, Su-Zhou C, Li Y, Merkeryan H, Liu W, Liu X. 2021. ACC deaminase-producing PGPRs improve drought stress tolerance in grapevine (Vitis vinifera L.). Front Plant Sci. 12:706990. doi: 10.3389/fpls.2021.706990.
- Dubeux JCB, Sollenberger LE. 2020. Nutrient cycling in grazed pastures. In: Rouquette M, Aiken GE, editors. Management strategies for sustainable cattle production in southern pastures. Amsterdam: Academic Press, Elsevier; p. 59–75.
- Edwards PJ. 1998. Sulfur cycling, retention, and mobility in soils: a review. General Technical Report NE-250. U.S. Department of Agriculture, Forest Service, Northeastern Research Station, Radnor, PA.
- Eltlbany N, Baklawa M, Ding GC, Nassal D, Weber N, Kandeler E, Neumann G, Ludewig U, van Overbeek L, Smalla K. 2019. Enhanced tomato plant growth in soil under reduced P supply through microbial inoculants and microbiome shifts. FEMS Microbiol Ecol. 95:fiz124. doi: 10.1093/femsec/fiz124.
- Espinosa-Urgel M, Salido A, Ramos JL. 2000. Genetic analysis of functions involved in adhesion of Pseudomonas putida to seeds. J Bacteriol. 182:2363–2369.
- Feng H, Zhang N, Du W, Zhang H, Liu Y, Fu R, Shao J, Zhang G, Shen Q, Zhang R. 2018. Identification of chemotaxis compounds in root exudates and their sensing chemoreceptors in plant-growth-promoting rhizobacteria Bacillus amyloliquefaciens SQR9. Mol Plant Microbe Interact. 31:995–1005.
- French E, Kaplan I, Iyer-Pascuzzi A, Nakatsu CH, Enders L. 2021. Emerging strategies for precision microbiome management in diverse agroecosystems. Nat Plants. 7:256–267.
- French E, Tran T, Iyer-Pascuzzi A. 2020. Tomato genotype modulates selection and responses to root microbiota. Phytobiomes J. 4:314–326.
- Ghosh D, Sen S, Mohapatra S. 2017. Modulation of proline metabolic gene expression in Arabidopsis thaliana under water-stressed conditions by a drought-mitigating Pseudomonas putida strain. Ann Microbiol. 67:655–668.
- Glick BR. 2014. Bacteria with ACC deaminase can promote plant growth and help to feed the world. Microbiol Res. 169:30–39.
- Gomez EJ, Delgado JA, Gonzalez JM. 2020a. Environmental factors affect the response of microbial extracellular enzyme activity in soils when determined as a function of water availability and temperature. Ecol Evol. 10:10105–10115.
- Gomez EJ, Delgado JA, Gonzalez JM. 2020b. Persistence of microbial extracellular enzymes in soils under different temperatures and water availabilities. Ecol Evol. 10:10167–10176.
- Gomez EJ, Delgado JA, Gonzalez JM. 2021. Influence of water availability and temperature on estimates of microbial extracellular enzyme activity. PeerJ. 9:e10994. doi: 10.7717/peerj.10994.
- Gorlach-Lira K, Coutinho HDM. 2007. Population dynamics and extracellular enzymes activity of mesophilic and thermophilic bacteria isolated from semi-arid soil of northeastern Brazil. Braz J Microbiol. 38:135–141.
- Gowtham HG, Brijesh S, Murali M, Shilpa N, Prasad M, Aiyaz M, Amruthesh KN, Niranjana SR. 2020. Induction of drought tolerance in tomato upon the application of ACC deaminase producing plant growth promoting rhizobacterium Bacillus subtilis Rhizo SF 48. Microbiol Res. 234:126422. doi: 10.1016/j.micres.2020.126422.
- Gu S, Wei Z, Shao Z, Friman VP, Cao K, Yang T, Kramer J, Wang X, Li M, Mei X, et al. 2020. Competition for iron drives phytopathogen control by natural rhizosphere microbiomes. Nat Microbiol. 5:1002–1010.
- Guerrero A, De Neve F, Mouazen AM. 2021. Data fusion approach for map-based variable-rate nitrogen fertilization in barley and wheat. Soil Tillage Res. 205:104789. doi: 10.1016/j.still.2020.104789.
- Hartmann M, Brunner I, Hagedorn F, Bardgett RD, Stierli B, Herzog C, Chen X, Zingg A, Graf-Pannatier E, Rigling A, Frey B. 2017. A decade of irrigation transforms the soil microbiome of a semi-arid pine forest. Mol Ecol. 26:1190–1206.
- He M, Dijkstra FA. 2014. Drought effect on plant nitrogen and phosphorus: a meta-analysis. New Phytol. 204:924–931.
- Hemkemeyer M, Dohrmann AB, Christensen BT, Tebbe CC. 2018. Bacterial preferences for specific soil particle size fractions revealed by community analyses. Front Microbiol. 9:149. doi: 10.3389/fmicb.2018.00149.
- Herrmann L, Lesueur D. 2013. Challenges of formulation and quality of biofertilizers for successful inoculation. Appl Microbiol Biotechnol. 97:8859–8873.
- Hol WH, Garbeva P, Hordijk C, Hundscheid PJ, Gunnewiek PJ, Van Agtmaal M, Kuramae EE, De Boer W. 2015. Non-random species loss in bacterial communities reduces antifungal volatile production. Ecology. 96:2042–2048.
- Jacoby RP, Kopriva S. 2019. Metabolic niches in the rhizosphere microbiome: new tools and approaches to analyse metabolic mechanisms of plant–microbe nutrient exchange. J Exp Bot. 70:1087–1094.
- Jaleel CA, Manivannan P, Wahid A, Farooq M, Al-Juburi HJ, Somasundaram R, Panneerselvam R. 2009. Drought stress in plants: a review on morphological characteristics and pigments composition. Int J Agric Biol. 11:100–105.
- Jiao X, Takishita Y, Zhou G, Smith DL. 2021. Plant associated rhizobacteria for biocontrol and plant growth enhancement. Front Plant Sci. 12:634796. doi: 10.3389/fpls.2021.634796.
- Jochum MD, McWilliams KL, Borrego EJ, Kolomiets MV, Niu G, Pierson EA, Jo Y-K. 2019. Bioprospecting plant growth-promoting rhizobacteria that mitigate drought stress in grasses. Front Microbiol. 10:2106. doi: 10.3389/fmicb.2019.02106.
- Jousset A, Bienhold C, Chatzinotas A, Gallien L, Gobet A, Kurm V, Küsel K, Rillig MC, Rivett DW, Salles JF, et al. 2017. Where less may be more: how the rare biosphere pulls ecosystems strings. ISME J. 11:853–862.
- Kanchiswamy CN, Malnoy M, Maffei ME. 2015. Chemical diversity of microbial volatiles and their potential for plant growth and productivity. Front. Plant Sci. 6:151. doi: 10.3389/fpls.2015.00151.
- Kaplan D, Maymon M, Agapakis CM, Lee A, Wang A, Prigge BA, Volkogon M, Hirsch AM. 2013. A survey of the microbial community in the rhizosphere of two dominant shrubs of the negev desert highlands, Zygophyllum dumosum (Zygophyllaceae) and Atriplex halimus (Amaranthaceae), using cultivation-dependent and cultivation-independent methods. Am J Bot. 100:1713–1725.
- Kaplan I, Bokulich NA, Caporaso JG, Enders LS, Ghanem W, Ingerslew KS. 2020. Phylogenetic farming: Can evolutionary history predict crop rotation via the soil microbiome? Evol Appl. 13:1984–1999.
- Kasim WA, Osman ME, Omar MN, Abd El-Daim IA, Bejai S, Meijer J. 2013. Control of drought stress in wheat using plant growth promoting bacteria. J Plant Growth Regul. 32:122–130.
- Kaushal M, Wani SP. 2016. Plant-growth-promoting rhizobacteria: drought stress alleviators to ameliorate crop production in drylands. Ann Microbiol. 66:35–42.
- Kehe J, Kulesa A, Ortiz A, Ackerman CM, Thakku SG, Sellers D, Kuehn S, Gore J, Friedman J, Blainey PC. 2019. Massively parallel screening of synthetic microbial communities. Proc Natl Acad Sci USA. 116:12804–12809.
- Khan N, Bano A. 2019. Exopolysaccharide producing rhizobacteria and their impact on growth and drought tolerance of wheat grown under rainfed conditions. PLoS One. 14(9):e0222302. doi: 10.1371/journal.pone.0222302.
- Khan N, Bano A, Ali S, Babar Md A. 2020. Crosstalk amongst phytohormones from planta and PGPR under biotic and abiotic stresses. Plant Growth Regul. 90:189–203.
- Kim N, Zabaloy MC, Guan K, Villamil MB. 2020. Do cover crops beneft soil microbiome? A meta-analysis of current research. Soil Biol Biochem. 142:107701.
- Kochakinezhad H, Peyvast G-A, Kashi A-K, Olfati J-A, Asadi A. 2012. A comparison of organic and chemical fertilizers for tomato production. J Org Syst. 7:14–25.
- Konstantinidis KT, Tiedje JM. 2004. Trends between gene content and genome size in prokaryotic species with larger genomes. Proc Natl Acad Sci USA. 101:3160–3165.
- Kurm V, van der Putten WH, de Boer W, Naus-Wiezer S, Hol WH. 2017. Low abundant soil bacteria can be metabolically versatile and fast growing. Ecology. 98:555–564. doi: 10.1002/ecy.1670.
- Lau JA, Lennon JT. 2012. Rapid responses of soil microorganisms improve plant fitness in novel environments. Proc Natl Acad Sci USA. 109:14058–14062.
- Le TA, Pék Z, Takács S, Neményi A, Daood HG, Helyes L. 2018. The effect of plant growth promoting rhizobacteria on the water-yield relationship and carotenoid production of processing tomatoes. HortScience. 53:816–822.
- Lebeis SL, Paredes SH, Lundberg DS, Breakfield N, Gehring J, McDonald M, Malfatti S, Glavina del Rio T, Jones CD, Tringe SG, Dangl JL. 2015. Plant microbiome. salicylic acid modulates colonization of the root microbiome by specific bacterial taxa. Science. 349:860–864.
- Li D, Mou W, Van de Poel B, Chang C. 2022. Something old, something new: conservation of the ethylene precursor 1-amino-cyclopropane-1-carboxylic acid as a signaling molecule. Curr Opin Plant Biol. 65:102116. doi: 10.1016/j.pbi.2021.102116.
- Li X, Rui J, Xiong J, Li J, He Z, Zhou J, Yanaarell AC, Mackie RI. 2014. Functional potential of soil microbial communities in the maize rhizosphere. PLoS One. 9:e112609. doi: 10.1371/journal.pone.0112609.
- Liu XM, Zhang H. 2015. The effects of bacterial volatile emissions on plant abiotic stress tolerance. Front Plant Sci. 6:774. doi: 10.3389/fpls.2015.00774.
- Liu Y, Chen L, Wu G, Feng H, Zhang G, Shen Q. 2017. Identification of root-secreted compounds involved in the communication between cucumber, the beneficial Bacillus amyloliquefaciens, and the soil-borne pathogen Fusarium oxysporum. Mol Plant-Microbe Interact. 30:53–62.
- López-Bellido RJ, Lal R, Danneberger TK, Street JR. 2010. Street plant growth regulator and nitrogen fertilizer effects on soil organic carbon sequestration in creeping bentgrass fairway turf. Plant Soil. 332:247–255.
- Lundberg DS, Lebeis SL, Paredes SH, Yourstone S, Gehring J, Malfatti S, Tremblay J, Engelbrektson A, Kunin V, Del Rio TG, et al. 2012. Defining the core Arabidopsis thaliana root microbiome. Nature. 488:86–90.
- Mahmoudi N, Caeiro MF, Mahdhi M, Tenreiro R, Ulm F, Mars M, Cruz C, Dias T. 2021. Arbuscular mycorrhizal traits are good indicators of soil multifunctionality in drylands. Geoderma. 397:115099. doi: 10.1016/j.geoderma.2021.115099.
- Mahmud K, Missaoui A, Lee K, Ghimire B, Presley HW, Makaju S. 2021. Rhizosphere microbiome manipulation for sustainable crop production. Curr Plant Biol. 27:100210. doi: 10.1016/j.cpb.2021.100210.
- Mallon CA, Le Roux X, Van Doorn GS, Dini-Andreote F, Poly F, Salles JF. 2018. The impact of failure: unsuccessful bacterial invasions steer the soil microbial community away from the invader’s niche. ISME J. 12:728–741.
- Marasco R, Rolli E, Ettoumi B, Vigani G, Mapelli F, Borin S, Abou-Hadid AF, El-Behairy UA, Sorlini C, Cherif A, et al. 2012. A drought resistance-promoting microbiome is selected by root system under desert farming. Plos one. 7(10):e48479. doi: 10.1371/journal.pone.0048479.
- Marchant R, Banat IM, Rahman TJ, Berzano M. 2002a. The frequency and characteristics of highly thermophilic bacteria in cool soil environments. Environ Microbiol. 4:595–602.
- Marchant R, Banat IM, Rahman TJ, Berzano M. 2002b. What are high temperature bacteria doing in cold environments? Trends Microbiol. 10:120–121.
- Marchant R, Franzetti A, Pavlostathis SG, Tas DO, Erdbrugger I, Unyayar A, Mazmanci MA, Banat IM. 2008. Thermophilic bacteria in cool temperate soils: are they metabolically active or continually added by global atmospheric transport? Appl Microbiol Biotechnol. 78:841–852.
- Marschner P, Grierson PF, Rengel Z. 2005. Microbial community composition and functioning in the rhizosphere of three Banksia species in native woodland in western Australia. Appl Soil Ecol. 28:191–201.
- Martinez-Gil M, Yousef-Coronado F, Espinosa-Urgel M. 2010. Lapf, the second largest Pseudomonas putida protein, contributes to plant root colonization and determines biofilm architecture. Mol Microbiol. 77:549–561.
- Mawarda PC, Le Roux X, Van Elsas JD, Salles JF. 2020. Deliberate introduction of invisible invaders: a critical appraisal of the impact of microbial inoculants on soil microbial communities. Soil Biol Biochem. 148:107874. doi: 10.1016/j.soilbio.2020.107874.
- Mayak S, Tirosh T, Glick BR. 2004. Plant growth-promoting bacteria that confer resistance to water stress in tomatoes and peppers. Plant Sci. 166:525–530.
- Meena KK, Sorty AM, Bitla UM, Choudhary K, Gupta P, Pareek A, Singh DP, Prabha R, Sahu PK, Gupta VK, et al. 2017. Abiotic stress responses and microbe-mediated mitigation in plants: the omics strategies. Front Plant Sci. 8:172. doi: 10.3389/fpls.2017.00172.
- Meldau S, Hoang LH, Underberg S, Wunsche H, Baldwin IT. 2013. Dimethyl disulfide produced by the naturally associated bacterium Bacillus sp B55 promotes Nicotiana attenuata growth by enhancing sulfur nutrition. Plant Cell. 25:2731–2747.
- Miche L, Belkin S, Rozen R, Balandreau J. 2003. Rice seedling whole exudates and extracted alkylresorcinols induce stress-response in Escherichia coli biosensors. Environ Microbiol. 5:403–411.
- Mishra SK, Khan MH, Misra S, Dixit VK, Khare P, Srivastava S, Chauhan PS. 2017. Characterisation of Pseudomonas spp. and Ochrobactrum sp. isolated from volcanic soil. Antonie Leeuwenhoek. 110:253–270.
- Mitter EK, Tosi M, Obregon D, Dunfield KE, Germida JJ. 2021. Rethinking crop nutrition in times of modern microbiology: innovative biofertilizer technologies. Front. Sustain. Food Syst. 5:1–2. doi: 10.3389/fsufs.2021.606815.
- Morcillo RJL, Manzanera M. 2021. The effects of plant-associated bacterial exopolysaccharides on plant abiotic stress tolerance. Metabolites. 11(6):337. doi: 10.3390/metabo11060337.
- Morella NM, Weng FC-H, Joubert PM, Metcalf CJE, Lindow S. 2020. Successive passaging of a plant-associated microbiome reveals robust habitat and host genotype-dependent selection. Proc Natl Acad Sci USA. 117:1148–1159.
- Mosqueira MJ, Marasco R, Fusi M, Michoud G, Merlino G, Cherif A, Daffonchio D. 2019. Consistent bacterial selection by date palm root system across heterogeneous desert oasis agroecosystems. Sci Rep. 9(1):4033. doi: 10.1038/s41598-019-40551-4.
- Mueller UG, Sachs JL. 2015. Engineering microbiomes to improve plant and animal health. Trends Microbiol. 23(10):606–617. doi: 10.1016/j.tim.2015.07.009.
- Mulvaney RL, Khan SA, Ellsworth TR. 2009. Synthetic nitrogen fertilizers deplete soil nitrogen: a global dilemma for sustainable cereal production. J Environ Qual. 38:2295–2314.
- Munnaf MA, Haesaert G, Mouazen AM. 2021. Map-based site-specific seeding of seed potato production by fusion of proximal and remote sensing data. Soil Tillage Res. 205:104801. doi: 10.1016/j.still.2020.104801.
- Naseem H, Bano A. 2014. Role of plant growth-promoting rhizobacteria and their exopolysaccharide in drought tolerance of maize. J Plant Interact. 9:689–701.
- Nautiyal CS, Dion P. 2008. Molecular mechanisms of plant and microbe coexistence. Berlin: Springer. doi: 10.1007/978-3-540-75575-3.
- Naylor D, Coleman-Derr D. 2018. Drought stress and root-associated bacterial communities. Front Plant Sci. 8:2223. doi: 10.3389/fpls.2017.02223.
- Naylor D, DeGraaf S, Purdom E, Coleman-Derr D. 2017. Drought and host selection influence bacterial community dynamics in the grass root microbiome. ISME J. 11:2691–2704.
- Nazina T N, Tourova TP, Poltaraus AB, Novikova EV, Grigoryan AA, Ivanova AE, Lysenko AM, Petrunyaka VV, Osipov GA, Belyaev SS, Ivanov MV. 2001. Taxonomic study of aerobic thermophilic bacilli: descriptions of Geobacillus subterraneus gen. nov., sp. nov. and Geobacillus uzenensis sp. nov. from petroleum reservoirs and transfer of Bacillus stearothermophilus, Bacillus thermocatenulatus, Bacillus thermoleovorans, Bacillus kaustophilus, Bacillus thermodenitrificans to Geobacillus as the new combinations G. stearothermophilus, G. th. Int J Syst Evol Microbiol. 51(Pt 2):433–446.
- Neal AL, Ahmad S, Gordon-Weeks R, Ton J. 2012. Benzoxazinoids in root exudates of maize attract Pseudomonas putida to the rhizosphere. PLoS One. 7(4):e35498. doi: 10.1371/journal.pone.0035498.
- Niu B, Paulson JN, Zheng X, Kolter R. 2017. Simplified and representative bacterial community of maize roots. Proc Natl Acad Sci U S A. 114(12):E2450–E2459.
- Niu XG, Song LC, Xiao YN, Ge WD. 2018. Drought-tolerant plant growth-promoting rhizobacteria associated with foxtail millet in a semi-arid agroecosystem and their potential in alleviating drought stress. Front. Microbiol. 8:2580. doi: 10.3389/fmicb.2017.02580.
- Nuccio EE, Anderson-Furgeson J, Estera KY, Pett-Ridge J, De Valpine P, Brodie EL, Firestone MK. 2016. Climate and edaphic controllers influence rhizosphere community assembly for a wild annual grass. Ecology. 97:307–1318.
- Oku S, Komatsu A, Tajima T, Nakashimada Y, Kato J. 2012. Identification of chemotaxis sensory proteins for amino acids in Pseudomonas fluorescens Pf0-1 and their involvement in chemotaxis to tomato root exudate and root colonization. Microbes Environ. 27:462–469.
- Or D, Smets BF, Wraith JM, Dechesn A, Friedman SP. 2007. Physical constraints affecting bacterial habitats and activity in unsaturated porous media – a review. Adv Water Resour. 30:1505–1527.
- Pacheco I, Ferreira R, Correia P, Carvalho L, Dias T, Cruz C. 2021. Microbial consortium increases maize productivity and reduces grain phosphorus concentration under field conditions. Saudi J Biol Sci 28:232–237.
- Pascault N, Ranjard L, Kaisermann A, Bachar D, Christen R, Terrat S, Mathieu O, Lévêque J, Mougel C, Henault C, et al. 2013. Stimulation of different functional groups of bacteria by various plant residues as a driver of soil priming effect. Ecosystems. 16:810–822.
- Paul D, Nair S. 2008. Stress adaptations in a plant growth promoting rhizobacterium (PGPR) with increasing salinity in the coastal agricultural soils. J Basic Microbiol. 48:378–384.
- Pawar RR, Borkar SG. 2018. Isolation and characterization of thermophilic bacteria from different habitats and their assessment for antagonism against soil-borne fungal plant pathogens. Afr J Microbiol Res. 12:556–566.
- Pérez CA, Kim M, Aravena JC, Silva W. 2022. Diazotrophic activity and denitrification in two long-term chronosequences of maritime Antarctica. Sci Total Environ. 809:152234. doi: 10.1016/j.scitotenv.2021.152234.
- Portillo MC, Santana M, Gonzalez JM. 2012. Presence and potential role of thermophilic bacteria in temperate terrestrial environments. Naturwissenschaften. 99:43–53.
- Renoud S, Abrouk D, Prigent-Combaret C, Wisniewski-Dyé F, Legendre L, Moënne-Loccoz Y, Muller D. 2022. Effect of inoculation level on the impact of the PGPR Azospirillum lipoferum CRT1 on selected microbial functional groups in the rhizosphere of field maize. Microorganisms. 10(2):325. doi: 10.3390/microorganisms10020325.
- Rhodes D, Hanson AD. 1993. Quaternary ammonium and tertiary sulfonium compounds in higher plants. Annu Rev Plant Physiol Plant Mol Biol. 44:357–384.
- Ribas-Agustí A, Seda M, Sarraga C, Montero J, Castellari M, Muñoz P. 2017. Municipal solid waste composting: application as a tomato fertilizer and its effect on crop yield, fruit quality and phenolic content. Renew Agric and Food Syst. 32:358–365.
- Rodriguez-Salazar J, Suarez R, Caballero-Mellado J, Itturiaga G. 2009. Trehalose accumulation in Azospirillum brasilense improves drought tolerance and biomass in maize plants. FEMS Microbiol Lett. 296:52–59.
- Rouwenhorst KHR, Krzywda PM, Benes NE, Mul G, Lefferts L. 2021. Ammonia production technologies. In: Valera-Medina A, Banares-Alcantara R, editors. Techno-economic challenges of green ammonia as an energy vector. Amsterdam: Academic Press, Elsevier; p. 41–83, doi: 10.1016/B978-0-12-820560-0.00004-7.
- Ryu CM, Farag MA, Hu CH, Reddy MS, Kloepper JW, Paré PW. 2004. Bacterial volatiles induce systemic resistance in Arabidopsis. Plant Physiol. 134:1017–1026.
- Ryu CM, Farag MA, Hu CH, Reddy MS, Wei HX, Paré PW, Kloepper JW. 2003. Bacterial volatiles promote growth in Arabidopsis. Proc Natl Acad Sci USA. 100:4927–4932.
- Sakamoto A, Murata N. 2002. The role of glycine betaine in the protection of plants from stress, clues from transgenic plants. Plant Cell Environ. 25:163–171.
- Salah Ud-Din AIM, Roujeinikova A. 2017. Methyl-accepting chemotaxis proteins: a core sensing element in prokaryotes and archaea. Cell Mol Life Sci. 74:3293–3303.
- Santana MM, Carvalho L, Melo J, Araújo ME, Cruz C. 2020. Unveiling the hidden interaction between thermophiles and plant crops: wheat and soil thermophilic bacteria. J Plant Interact. 15:127–138.
- Santana MM, Dias T, Gonzalez JM, Cruz C. 2021. Transformation of organic and inorganic sulfur– adding perspectives to new players in soil and rhizosphere. Soil Biol. Biochem. 160:108306. doi: 10.1016/j.soilbio.2021.108306.
- Santana MM, Gonzalez JM. 2015. High temperature microbial activity in upper soil layers. FEMS Microbiol Lett. 362(22):fnv182. doi: 10.1093/femsle/fnv182.
- Santana MM, Portillo MC, Gonzalez JM, Clara MIE. 2013. Characterization of new soil thermophilic bacteria potentially involved in soil fertilization. J Plant Nutr Soil Sci. 176:47–56.
- Schalk IJ, Hannauer M, Braud A. 2011. New roles for bacterial siderophores in metal transport and tolerance. Environ Microbiol. 13:2844–2854.
- Schimel J, Balser TC, Wallenstein M. 2007. Microbial stress-response physiology and its implications for ecosystem function. Ecology. 88:1386–1394.
- Schreiter S, Sandmann M, Smalla K, Grosch R. 2014. Soil type dependent rhizosphere competence and biocontrol of two bacterial inoculant strains and their effects on the rhizosphere microbial community of field-grown lettuce. Plos one. 9:e103726. doi: 10.1371/journal.pone.0103726.
- Searcy SW. 1997. Precision farming:a new approach to crop management. Texas agriculture extension service. The Texas A&M University System, College station, Texas, 4. http://lubbock.tamu.edu/files/2011/10/precisionfarm_1.pdf.
- Sessitsch A, Pfafenbichler N, Mitter B. 2019. Microbiome applications from lab to field: facing complexity. Trends Plant Sci. 24:194–198.
- Shade A. 2017. Diversity is the question, not the answer. ISME J. 11:1–6.
- Shrivastava P, Kumar R. 2015. Soil salinity: a serious environmental issue and plant growth promoting bacteria as one of the tools for its alleviation. Saudi J Biol Sci. 22:123–131.
- Signorelli S, Tarkowski ŁP, O’Leary B, Tabares-da Rosa S, Borsani O, Monza J. 2021. GABA and proline metabolism in response to stress. In: Gupta DK, Corpas FJ, editors. Hormones and plant response. Plant in challenging environments. Vol 2. Cham: Springer; p. 291–314. doi: 10.1007/978-3-030-77477-6_12.
- Song F, Han X, Zhu X, Herbert SJ. 2012. Response to water stress of soil enzymes and root exudates from drought and non-drought tolerant corn hybrids at different growth stages. Can. J. Soil Sci. 92:501–507.
- Teixeira PJP, Colaianni NR, Fitzpatrick CR, Dangl JL. 2019. Beyond pathogens:microbiota interactions with the plant immune system. Curr Opin Microbiol. 49:7–17.
- Terhorst CP, Lennon JT, Lau JA. 2014. The relative importance of rapid evolution for plant-microbe interactions depends on ecological context. Proc R Soc B Biol Sci. 281(1785):20140028. doi: 10.1098/rspb.2014.0028.
- Timmusk S, Abd El-Daim IA, Copolovici L, Tanilas T, Kännaste A, Behers L, Nevo E, Seisenbaeva G, Stemström E, Niinemets Ü. 2014. Drought-tolerance of wheat improved by rhizosphere bacteria from harsh environments: enhanced biomass production and reduced emissions of stress volatiles. PLoS One. 9:e96086. doi: 10.1371/journal.pone.0096086.
- Tiwari S, Lata C, Chauhan PS, Nautiyal CS. 2016. Pseudomonas putida attunes morphophysiological, biochemical and molecular responses in Cicer arietinum L. during drought stress and recovery. Plant Physiol Biochem. 99:108–117.
- Tocheva EI, Ortega DR, Jensen GJ. 2016. Sporulation, bacterial cell envelopes and the origin of life. Nat Rev Microbiol. 14:535–542.
- Totsche KU, Amelung W, Gerzabek MH, Guggenberger G, Klumpp E, Knief C, Lehndorff E, Mikutta R, Peth S, Prechtel A, et al. 2018. Microaggregates in soils. J Plant Nutr Soil Sci. 181:104–136.
- Treseder KK, Kivlin SN, Hawkes CV. 2011. Evolutionary trade-offs among decomposers determine responses to nitrogen enrichment: evolutionary trade-offs among decomposers. Ecol Lett. 14:933–938.
- Tringe SG, von Mering C, Kobayashi A, Salamov AA, Chen K, Chang HW, Podar M, Short JM, Mathur EJ, Detter JC, et al. 2005. Comparative metagenomics of microbial communities. Science. 308:554–557.
- Turner TR, Ramakrishnan K, Walshaw J, Heavens D, Alston M, Swarbreck D, Osbourn A, Grant PS. 2013. Comparative metatranscriptomics reveals kingdom level changes in the rhizosphere microbiome of plants. ISME J. 7:2248–2258.
- Ulm F, David A, Hobson P, Penha-Lopes G, Dias T, Máguas C, Cruz C. 2019. Sustainable urban agriculture using compost and an open-pollinated maize variety. J. Clean. Prod. 212:622–629.
- Uroz S, Buée M, Murat C, Frey-Klett P, Martin F. 2010. Pyrosequencing reveals a contrasted bacterial diversity between oak rhizosphere and surrounding soil. Environ Microbiol. 2:281–288.
- Valente J, Gerin F, Le Gouis J, Moënne-Loccoz Y, Prigent-Combaret C. 2020. Ancient wheat varieties have a higher ability to interact with plant growth-promoting rhizobacteria. Plant Cell Environ. 43:246–260.
- Vendruscolo ECG, Schuster I, Pileggi M, Scapim CA, Molinari HBC, Marur CJ, Vieira LGE. 2007. Stress-induced synthesis of proline confers tolerance to water deficit in transgenic wheat. J Plant Physiol. 164:1367–1376.
- Venturi V, Keel C. 2016. Signaling in the rhizosphere. Trends Plant Sci. 21:187–198.
- Veresoglou SD, Menexes G. 2010. Impact of inoculation with Azospirillum spp. on growth properties and seed yield of wheat: a meta-analysis of studies in the ISI Web of science from 1981 to 2008. Plant Soil. 337:469–480.
- Vurukonda SS, Vardharajula S, Shrivastava M, SkZ A. 2016. Enhancement of drought stress tolerance in crops by plant growth promoting rhizobacteria. Microbiol Res. 184:13–24.
- Walters WA, Jin Z, Youngblut N, Wallace JG, Sutter J, Zhang W, González-Peña A, Peiffer J, Koren O, Shi Q, et al. 2018. Large-scale replicated feld study of maize rhizosphere identifes heritable microbes. Proc Natl Acad Sci USA. 115:7368–7373.
- Wang L, Tsuda K, Sato M, Cohen JD, Katagiri F, Glazebrook J. 2009. Arabidopsis CaM binding protein CBP60 g contributes to MAMP-induced SA accumulation and is involved in disease resistance against Pseudomonas syringae. PLoS Pathog. 5(2):e1000301. doi: 10.1371/journal.ppat.1000301.
- Wang S, Ouyang L, Ju X, Zhang L, Zhang Q, Li Y. 2014. Survey of plant drought-resistance promoting bacteria from Populus euphratica tree living in arid area. Indian J. Microbiol. 54:419–426.
- Weng J, Wang Y, Li J, Shen Q, Zhang R. 2013. Enhanced root colonization and biocontrol activity of Bacillus amyloliquefaciens SQR9 by abrB gene disruption. Appl. Microbiol. Biotechnol. 97:8823–8830.
- Wigley P, George C, Turner S. 2017. Accelerated directed evolution of microbial consortia for the development of desirable plant phenotypic traits. Patent US-9150851-B2. https://pubchem.ncbi.nlm.nih.gov/patent/US-9260713-B2.
- Wintermans PCA, Bakker PAHM, Pieterse CMJ. 2016. Natural genetic variation in Arabidopsis for responsiveness to plant growth-promoting rhizobacteria. Plant Mol. Biol. 90:623–634.
- Xie J, Dawwam GE, Sehim AE, Li X, Wu J, Chen S, Zhang D. 2021. Drought stress triggers shifts in the root microbial community and alters functional categories in the microbial gene pool. Front Microbiol. 744897(12):744897. doi: 10.3389/fmicb.2021.
- Xun W, Shao J, Shen Q, Zhang R. 2021. Rhizosphere microbiome: functional compensatory assembly for plant fitness. Comput Struct Biotechnol J. 19:5487–5493.
- Yamaguchi-Shinozaki K, Shinozaki K. 1994. A novel cis-acting element in an arabidopsis gene is involved in responsiveness to drought, low-temperature, or high-salt stress. Plant Cell. 6:251–264.
- Yan Y, Kuramae EE, de Hollander M, Klinkhamer PG, van Veen JA. 2017. Functional traits dominate the diversity-related selection of bacterial communities in the rhizosphere. ISME J. 11:56–66.
- Yancey PH. 2001. Water stress, osmolytes and proteins. Am Zool. 41:699–709.
- Yin D, Wang N, Xia F, Li Q, Wang W. 2013. Impact of biocontrol agents Pseudomonas fluorescens 2P24 and CPF10 on the bacterial community in the cucumber rhizosphere. Eur J Soil Biol. 59:36–42.
- Zengler K, Hofmockel K, Baliga NS, Behie SW, Bernstein HC, Brown JB, Dinneny JR, Floge SA, Forry SP, Hess M, et al. 2019. EcoFABs: advancing microbiome science through standardized fabricated ecosystems. Nat Methods. 16:567–571.
- Zhalnina K, Louie KB, Hao Z, Mansoori N, da Rocha UN, Shi S, Cho H, Karaoz U, Loqué D, Bowen BP, et al. 2018. Dynamic root exudate chemistry and microbial substrate preferences drive patterns in rhizosphere microbial community assembly. Nat Microbiol. 3:470–480.
- Zhang H, Murzello C, Sun Y, Kim MS, Xie X, Jeter RM, Zak JC, Dowd SE, Paré PW. 2010. Choline and osmotic-stress tolerance induced in Arabidopsis by the soil microbe Bacillus subtilis (GB03). Mol Plant Microbe Interact. 23:1097–1104.
- Zhang J, Guerrero A, Mouazen AM. 2021. Map-based variable-rate manure application in wheat using a data fusion approach. Soil Tillage Res. 207:104846. doi: 10.1016/j.still.2020.104846.
- Zhang J, Liu YX, Zhang N, Hu B, Jin T, Xu H, Qin Y, Yan P, Zhang X, Guo X, et al. 2019b. NRT1.1B is associated with root microbiota composition and nitrogen use in feld-grown rice. Nat Biotechnol. 37:676–684.
- Zhang Y, Gao X, Shen Z, Zhu C, Jiao Z, Li R, Shen Q. 2019a. Pre-colonization of PGPR triggers rhizosphere microbiota succession associated with crop yield enhancement. Plant Soil. 439:553–567.