ABSTRACT
Legumes can contribute to emissions of the potent greenhouse gas nitrous oxide (N2O) directly by some rhizobia species that are able to denitrify under free-living conditions and in symbiotic association with the plant. In this study, the capacity of Phaseolus vulgaris-Rhizobium etli symbiosis to emit N2O in response to nitrate (NO3-) has been demonstrated for the first time. We found that bacteroidal assimilatory nitrate reductase (NarB) as well as nitrite reductase (NirK) and nitric oxide reductase (cNor) denitrifying enzymes contribute to nitric oxide (NO) and N2O formation in nodules. We also show that R. etli NarK is involved in NO2- extrusion and links NO3- reduction by NarB in the cytoplasm with NirK and cNor denitrification activities in the periplasm. The knowledge generated in this work will be instrumental for exploring strategies and sustainable practices in agricultural soil management to increase legume crop yield and mitigate greenhouse gas emissions.
HIGHLIGHTS
Common bean nodules are able to produce N2O in response to nitrate.
Nitrate reduction to nitrite in R. etli bacteroids catalyzed by assimilatory nitrate reductase NarB is involved in N2O emissions from common bean nodules.
R. etli respiratory nitrite reductase NirK and nitric oxide reductase cNor support N2O production in common bean nodules.
R. etli NarB and cNor are involved in NO homeostasis in common bean nodules.
R. etli nitrite exporter NarK links nitrate reduction in the cytoplasm with denitrification activities in the periplasm.
1. Introduction
Nitrous oxide (N2O) is a highly stable greenhouse gas (GHG) and stands alongside carbon dioxide (CO2), methane (CH4) and fluorinated gases as a key driver of global warming. Moreover, the global warming potential for N2O is around 300-fold greater than that from CO2 and has an impact on stratospheric ozone (Müller Citation2021). Agriculture, Forestry and Other Land Use (AFOLU) activities accounted for around 13% of CO2, 44% of CH4, and 81% of N2O emissions from human activities during 2007-2016, representing 23% of total net anthropogenic emissions of GHGs (IPCC Citation2019). Nitrogen (N) in the soil is converted to N2O through two main microbial processes, nitrification and denitrification (Thomson et al. Citation2012; Butterbach-Bahl et al. Citation2013; Pilegaard Citation2013; Torres et al. Citation2016; Sánchez and Minamisawa Citation2019). Under low oxygen conditions some bacteria can produce energy by using nitrate (NO3-) or nitrite (NO2-) as final electron acceptors, via the denitrification pathway. Classical or ‘complete’ denitrification involves four enzymatic steps, which allow sequential conversion of NO3- to NO2-, nitric oxide (NO), N2O and, finally dinitrogen (N2). These four steps are catalysed by the periplasmic nitrate reductase (Nap) or membrane associated nitrate reductase (Nar), nitrite reductases (NirK or NirS), nitric oxide reductases (cNor, qNor or CuANor) and nitrous oxide reductase (Nos) (reviewed by Zumft Citation1997; van Spanning et al. Citation2007; Torres et al. Citation2016; Salas et al. Citation2021). Anthropogenic N2O emissions from agricultural soils are rising primarily due to the application of N fertilizers including inefficiencies such as over-application or poorly synchronization with crop demand timings (IPCC Citation2019; Kudeyarov Citation2020; Lazcano et al. Citation2021). Further to its contribution to N2O emission, excessive usage of NO3--based fertilizers to enhance agricultural production, can also lead runoff causing soil and water contamination. Therefore, it is crucial to rationalize the use of synthetic fertilizers in agriculture (Tian et al. Citation2020). In this context, biological nitrogen fixation (BNF) performed by the rhizobia–legume symbiosis is proposed as an economically and environmentally friendly alternative to N fertilizers, to improve soil fertility and crop yield. Legumes are an important source of non-meat-based protein for humans and animals, and grain legumes such as common bean (Phaseolus vulgaris), account for 27% of primary crop production worldwide (Graham and Vance Citation2003).
These plants have the unique ability to establish a mutualist symbiosis with soil N2-fixing bacteria, termed rhizobia, that leads to the formation of specialized organs called nodules in plant roots. Within the nodules, differentiated rhizobia i.e. bacteroids are able to convert (through the activity of the nitrogenase enzyme) an abundant atmospheric source of N2 that is not normally available to the plant into more bioavailable forms such as ammonia (NH4+) that are readily plant-assimilated. In return, rhizobia obtain a safe niche and stable carbon provision in photosynthates. However, despite clear benefits, bacterial-plant endosymbiosis can be ‘a double-edged sword’ and have negative impacts since it may potentially contribute to N2O emission indirectly through legumes N-rich residue deposition, or directly through some rhizobial species being able to perform denitrification inside nodules where they release free-intermediates (Inaba et al. Citation2009, Citation2012; Hirayama et al. Citation2011; Torres et al. Citation2016). In this context, several reports have demonstrated that the soybean endosymbiont Bradyrhizobium diazoefficiens produces N2O in soybean nodules through the denitrification pathway in response to NO3− and flooding (Tortosa et al. Citation2015; Tortosa et al. Citation2020). However, information concerning the capability of common bean nodules to produce this important GHG and the proteins involved is unknown.
In order to use rhizobia as biofertilizers, it is essential to investigate the mechanisms of legume–rhizobia symbiosis from a holistic perspective including bacterial and/or plant proteins that influence N2O emission. Although denitrification among rhizobia is rare, several of the most agronomical interesting species contain in their genomes some or all the genes required to perform the denitrification pathway (Delgado et al. Citation2007; Torres et al. Citation2016 and references herein). Complete denitrifying rhizobia should be preferred for formulation of bioinoculants, as they possess the NosZ enzyme which is currently the only known enzyme capable of reducing N2O to the ‘safe’ end-product N2 (reviewed by Hein and Simon Citation2019). Rhizobium etli CFN42 is the endosymbiont of common bean, an important globally cultivated grain legume, and does not possess the complete set of denitrification genes. The genome of this bacterium consists of one chromosome and six large plasmids (pRet42a to pRet42f) and contains the nirK and norCBQD genes on plasmid f, which encodes the NirK-type nitrite reductase and c-type nitric oxide reductase (cNor), respectively (Gómez-Hernández et al. Citation2011).
Recently, it has been reported that despite lacking genes encoding the Nap or Nar nitrate reductase enzymes, R. etli produces N2O under microoxic conditions when NO3- is present as the only N source. This ability can be attributed to the activity of the cytoplasmic assimilatory nitrate reductase, NarB (Hidalgo-García et al. Citation2019). Thus, this species of rhizobia can produce N2O from NO3- by coupling nitrate assimilation with the denitrification pathway. Here, NO3- is firstly reduced to NO2- in the cytosol by NarB and the reaction product NO2- is further reduced to NO and then N2O by the sequential action of NirK and cNor, respectively, in the periplasmic space.
In this work, the capacity of the Phaseolus vulgaris-R. etli- symbiosis to emit N2O as well as the contribution of R. etli NarB, NirK and cNor enzymes has been shown for the first time. R. etli connects nitrate reduction in the cytoplasm with nitrite respiration in the periplasm through the nitrite exporter NarK.
2. Material and methods
2.1. Bacterial strains, plasmids and growth conditions
The bacterial strains and plasmids used in this work are listed in . R. etli strains were grown at 30°C in TY rich medium (Tryptone Yeast, Beringer Citation1974) or in Y minimal medium (MMY) with succinate (10 mM) and ammonium chloride (10 mM) as carbon and N sources, respectively (Bravo and Mora Citation1988). To study the growth and N2O production capacity of R. etli from different N sources, 10 mM NH4Cl from MMY medium was replaced by 10 mM KNO3 (this medium was named ‘MMN’) or 1 mM NaNO2. Antibiotics were added to R. etli cultures at the following concentrations (µg/mL): nalidixic acid (Nal) 20, kanamycin (Km) 30, spectinomycin (Sp) 100, streptomycin (Str) 100, tetracycline (Tc) 10. Escherichia coli strains were grown at 37°C in LB medium (Sambrook and Russell Citation2001) with antibiotics added at the following concentrations (µg/mL): Sp 25, Str 25, Km 20, Tc 25 and ampicillin (Ap) 200. For growth under microoxic conditions, 17 mL tubes containing 3 mL of cell cultures were sealed with rubber septa and flushed for 5 min at the start of the incubation with 2% (v/v) O2 and 98% N2 (v/v).
Table 1. Bacterial strains and plasmids.
For determination of growth rates, extracellular NO2- concentration and N2O emission, cells were firstly grown aerobically in TY medium for 24 h, harvested by centrifugation at 8,000 g for 10 min, and washed twice with MMN. Cells were then incubated in minimal medium for another 24 h under the desired oxygen conditions. Initial optical density for cultures (measured at 600 nm, OD600) was around 0.05.
Inoculum for plants was prepared by growing R. etli aerobically in TY rich medium for 24 h, then cells were harvested by centrifugation at 8,000 g for 10 min, washed twice and grown aerobically for another 24 h in MMN. Initial and final OD600 of culture were around 0.3 and 0.6, respectively. The final inoculum was prepared by harvesting cells by centrifugation, washing them with MMN and resuspending them in fresh MMN to a final OD600 of 0.8, corresponding to ∼108 cells/mL.
2.2. Construction and complementation of a R. etli narK mutant
The oligonucleotide primers used in this work were purchased from Sigma (Sigma-Aldrich, Saint Louis, MO, USA) and are listed in Supplementary Table S1. Genomic and plasmid DNA isolation was carried out using the REALPURE Genomic DNA purification Kit (Real) and Qiagen Plasmid Kit (Qiagen), respectively. PCR was performed using the High-Fidelity DNA polymerase Phusion® enzyme (Thermo Fisher Scientific, MA, USA) and DNA digestions were carried out using Fast Digest restriction enzymes (Thermo Fisher Scientific, MA, USA).
To generate a narK mutant, two regions flanking the narK gene (fragments F1 and F2) were amplified by PCR using narK_up_For/narK_up_Rev and narK_down_For/narK_down_Rev primer pairs. Then, fragments F1 and F2 were cloned into the pBluescript KS (pBSKS) vector (Thermo Fisher Scientific, MA, USA) as XbaI/BamHI and BamHI/EcoRI fragments, respectively, to generate plasmid pBSKS_F1F2. The XbaI/EcoRI fragment from pBSKS_F1F2 was cloned into the pK18mobsacB suicide vector (Schäfer et al. Citation1994) yielding plasmid pDR4004 and integrity was confirmed by sequencing. Replacement of the R. etli wild-type narK allele with the truncated mutant allele in plasmid pDR4004 was carried out by double recombination using the sacB sucrose-selectable marker present in plasmid pK18mobsacB. Double recombinants were selected as resistant to 12.5% sucrose and Km sensitive. The derivatives were verified by PCR using primers narK_up-For and narK_down-Rev (Supplementary Table S1), and the narK mutant strain obtained was named DR4004.
The R. etli narK gene was constitutively expressed under control of the lacZ promoter to generate a complemented narK strain. The narK gene was amplified by PCR using narK_compl_For/narK_compl_Rev primers par (Supplementary Table S1) and cloned as a HindIII/BamHI fragment into the broad-host range cloning vector pBBR1MCS-2 (Kovach et al. Citation1994). The plasmid obtained (pDR4005) was sequenced using primers M13_For, M13_Rev and narK_compl_IN (Supplementary Table S1), and then transferred to the R. etli narK + NarB+ strain by bi-parental conjugation using E. coli S17.1. The resulting strain containing pDR4005 (narK + NarB++ NarK) was checked by plasmid isolation and PCR.
2.3. Construction of R. etli strains overexpressing NarB
In previous work (Hidalgo-García et al. Citation2019), it was reported that overexpression of narB led to induction of NR activity and N2O production by free-living R. etli cells. Here, overexpression was performed by introducing plasmid pDR4002 (pBBR1MCS-2 carrying the narB gene) into R. etli WT and narB mutant strains. In this current study we were interested in exploring the role of NarB in N2O metabolism during endosymbiosis, by using the R. etli narB deletion mutant and NarB overexpressing strains. The pTR102 plasmid has been reported to be highly stable in cells of Sinorhizobium meliloti during symbiosis with alfalfa (Weinstein et al. Citation1992) and so, this platform was used to carry the gene to ensure the stability and persistence of the overexpression system over the course of plant experiments. To generate this overexpression platform, the pDR4002 plasmid (pBBR1MCS-2 carrying narB gene) was digested with XbaI/HindIII restriction enzymes, and the resultant fragment carrying narB gene was cloned as an XbaI/HindIII fragment into plasmid pTR102, yielding the pTR102::narB plasmid, where narB gene is under the control of the lacZ promoter.
Once pTR102::narB was constructed, it was introduced into R. etli WT, and narB, nirK, norC and narK mutant strains by conjugation with E. coli S17.1. The derivatives strains containing pTR102::narB (NarB+) were verified by plasmid isolation and PCR. Concurrently, a WT strain containing pTR102 empty vector was obtained (WT-pTR102).
2.4. Extracellular NO2- determination.
Cultures were centrifuged at 8,000 g for 10 min and NO2- concentration was estimated in supernatants by using the Griess-Yallosway diazotization protocol (Nicholas and Nason Citation1957). Briefly, equal volumes of 0.1% (w/v) NNEDA [n-(1-naphthyl)ethylenediamine dihydrochloride] and 1% (w/v) sulfanilamide were added to one volume of supernantant. NO2- was measured colorimetrically at λ = 540 nm after 20 min incubation with the reagents and extrapolating from a standard curve constructed with increasing concentrations of NaNO2 (0, 20, 40, 60, 80 and 100 μM) from a stock solution of 100 μM.
2.5. N2O production by free-living cells
Cells were cultured as indicated above in section 2.1. After 24 h, 1 ml from the headspace was taken using a Hamilton® Gastight syringe, and manually injected into an HP 4890D gas chromatography instrument equipped with an electron capture detector (ECD) (Hewlett Packard, San Jose, CA, USA) as described by Torres et al. (Citation2014).
2.6. Protein concentration measurement
Protein concentration was measured using the Protein Assay Dye Reagent Concentrate (Bio-Rad, CA, USA). Bovine serum albumin (BSA) was used as standard protein for calibration curves.
2.7. Plant inoculation and growth conditions
Seeds of common bean (P. vulgaris, var. Negro jamapa) were surface sterilized as performed by Delgado et al. (Citation1994) with little modifications. Briefly, seeds were immersed in 96% (v/v) ethanol for 30 s, in 30% (v/v) H2O2 for 10 min and then thoroughly washed five times with sterilized distilled water. After incubation for 2 h in sterile distilled water in darkness, the seeds were placed on plates containing 1% water-agar and were allowed to germinate at 30°C for 72 h. Three days after sowing, one germinated seed was transplanted into a sterilized Leonard jar (Trung and Yoshida Citation1983) with a 0.25 L pot filled with vermiculite (N° 3) as growth substrate. Each seedling was inoculated with 1 mL of a single bacterial strain (108 cells/mL). Plants were grown in controlled environmental chambers (16/8 light/dark cycle, 26/22°C, photosynthetic photon flux density of 180 μmol photons/m2 × s, and relative humidity 60–70%). Different sets of plants were watered throughout the experiment with mineral solution (Rigaud and Puppo Citation1975) containing 0, 1, 2 or 4 mM KNO3 or treated with 10 mM KNO3 for one or four days at the end of the experiment. The plants were grown for a total of 25 days following inoculation with R. etli.
2.8. N2O measurement in detached nodules
Plants (10 from each treatment) were harvested from Leonard jars, vermiculite carefully removed from roots and two nodules from each plant transferred into 20 mL headspace vials containing a circle of absorbent paper on its base (20 nodules/vial). Subsequently, 100 µL of mineral solution was added to each vial which were hermetically sealed and incubated at 30°C. After 5, 8 and 24 h of incubation, gas samples were taken from the headspace using luer-lock gas-tight syringes with Mininert® valves and immediately transported to the gas chromatograph for analysis following the protocol described by Tortosa et al. (Citation2015). The emission flux rate [nmol N2O/h x g NFW] was calculated in the interval between 5 and 24 h: Δ N2O molar concentration (24h – 5 h) / Δ incubation time increase (19 h).
2.9. Leghaemoglobin content
Leghaemoglobin (Lb) content was measured as previously described by LaRue and Child (Citation1979). Essentially, nodules (0.3 g) were ground with 6 mL Lb extraction buffer (50 mM Na2HPO4·2H2O/NaH2PO4·H2O, pH 7.4; 0.02% w/v K3Fe(CN)6; 0.1% w/v NaHCO3) supplemented with 0.1 g polyvinylpolypyrrolidone (PVPP). The homogenate was centrifuged at 12,000 g at 4°C for 20 min to retain the supernatant. 50 µL of clear supernatant and 3.15 mL of saturated oxalic acid (66 g/L) were mixed in screw-capped glass tubes, which were sealed and autoclaved for 30 min at 120°C and then allowed to cool to room temperature. The fluorescence of the solutions was measured using a Shimadzu spectrofluorometer (Shimadzu Scientific Instruments, Kyoto, Japan) equipped with a mercury-xenon lamp and a RF-549 red-sensitive photomultiplier. The excitation and the emission wavelengths were 405 and 600 nm, respectively. The difference in fluorescence between heated and unheated samples was proportional to haem protein content.
2.10. Bacteroidal nitrate, nitrite and nitric oxide reductase activities
Bacteroids from detached nodules were extracted according to Mesa et al. (Citation2004). In brief, 1–2 g of fresh nodules were ground with a ceramic mortar and pestle with 7.5 mL of extraction buffer (50 mM Tris-HCl, pH 7.5, 250 mM mannitol). The homogenate was filtered through cheeseclothe and centrifuged at 250 g at 4°C for 5 min to remove nodule debris. The supernatant was recentrifuged at 12,000 g at 4°C for 10 min to pellet the bacteroids, which were washed twice with 50 mM Tris-HCl pH 7.5 for nitrate reductase (NR) and nitrite reductase (NiR) activities or 25 mM sodium phosphate pH 7.4 for nitric oxide reductase (Nor) activity.
For NR activity, bacteroids were disrupted using a French pressure cell (SLM Aminco, Jessup, MD, USA) at about 120 MPa. The resultant cell extract was used to measure methyl viologen-dependent (MV)-NR activity as described by Delgado et al. (Citation2003). Briefly, equal volumes of reaction mixture (10 mM KNO3 and 200 μM MV) were added to bacteroid cell extract aliquots containing 0.2–0.4 mg of protein and mixed gently. Enzymatic reactions started by addition of freshly prepared sodium dithionite (144 mM) dissolved in NaHCO3 (300 mM). After incubation for 30–60 min at 30°C, the reaction was stopped by vigorous shaking until the samples lost their blue color. Controls were done in parallel by oxidizing sodium dithionite at the start of the reaction (by shaking). Nitrite production was estimated after diazotization as described previously in section 2.4. Results are expressed as nmol NO2- produced/h x mg protein.
For MV-dependent NiR activity measurements, equal volumes of reaction mixture (0.1 mM NaNO2 and 200 μM MV) were added to bacteroid aliquots containing 0.1–0.3 mg of protein and thereafter the procedure was followed as previously described above for NR activity in reactions carried out for 15–30 min. Results were expressed as nmol NO2- consumed/h x mg protein.
Bacteroidal NO consumption rate (Nor activity) was determined by using an ISONOP NO electrode APOLLO 4000® (World Precision Instruments, Friedberg, Germany). The reaction chamber (2 mL) was temperature-controlled, magnetically stirred, and contained 760 μL of 25 mM Na2HPO4/NaH2PO4 buffer (pH 7.4) and 900 μL of cell suspension (1–2 mg protein). To generate an anoxic atmosphere, the method described in Cabrera et al. (Citation2016) was used. Once a steady base line was obtained, 15 μL of 2 mM NO were added to the chamber and NO consumption was recorded. Results were expressed as nmol NO consumed/h x mg protein.
2.11. Bacteroidal N2O emission
Bacteroids from detached nodules were extracted as described above in section 2.10. Once bacteroids were isolated, they were resuspended in a sterile solution of 50 mM Tris-HCl (pH 7.5), 10 mM KNO3, 10 mM sodium succinate dibasic hexa-hydrate. Then, 3mL of the suspension was placed in 17mL tubes and treated as free-living cells to obtain microoxic conditions (see section 2.1.). These were then incubated at 30°C at 170 rpm for 24 h. The initial and final OD600 of the suspensions was 2.5. N2O concentration in the headspace was measured as specified in section 2.5.
2.12. Detection of LbNO complexes in whole nodules by Electron Paramagnetic Resonance (EPR) spectroscopy
Continuous wave X-band EPR spectroscopy was performed on whole-nodule samples as described previously with minor modifications (Sánchez et al. Citation2010). Individual frozen and intact root nodules with a size between 2.5- and 2.8-mm diameter were selected and transferred under liquid nitrogen to pre-cooled 20-cm Wilmad® quartz (CFQ) EPR sample tubes with 3 mm internal diameter. Each sample contained intact nodules closely packed to a sample depth of 25 mm in the tubes (sample biomass was 79 ± 8 mg wet wt.). To improve signal to noise during data collection, a 10-cm length of ethylene-tetrafluoroethylene tubing (GE Healthcare, Piscataway, NJ, USA) was inserted into the tube and was secured with parafilm at the tube entrance to prevent nodule movement and allow liquid gas boil-off. Control experiments confirmed that neither the tubes nor tubing gave rise to any EPR signals in the absence of nodules.
EPR spectra were recorded using a Bruker ELEXSYS 500 spectrometer with an ER049X SuperX microwave bridge and a super high Q cavity (Bruker Analytische Messtechnik GmBH, Karlsruhe, Germany). Low temperature experiments were performed using an Oxford Instruments ESR-900 helium flow cryostat and an ITC3 temperature controller (Oxford Instruments, Oxfordshire, UK). Microwave frequency was 9.45 GHz with 2 mW power and 0.3 mT (3 Gauss) modulation amplitude applied. Spectra were recorded at 40 ± 1 K and are the average of three scans.
2.13. Statistical analyzes
For each assay, the total number of replicates is shown in each figure and table. Descriptive statistical analyzes (mean and standard deviation) for each parameter were calculated and data were checked for normal distribution and homoscedasticity. The analyzes of variance (ANOVA) within treatments were performed using the post hoc HSD Tukey test (P ≤ 0.05). Statistical software used for these analyzes was IBM SPSS Statistics v.26.
3. Results
3.1. Rhizobium etli RHE_CH01783 encodes a NarK-type nitrate/nitrite transporter
The gene RHE_CH01783 from R. etli CFN42 lies upstream of nirB (RHE_CH01782), nirD (RHE_CH01781) and narB (RHE_CH01780) genes (). According to bioinformatics predictions, nirB and nirD have been proposed to encode the large and small subunits of the sirohaem-ferredoxin nitrite reductase (NirB, NirD), respectively. The narB gene encodes the large subunit of the Mo[MGD]2 and iron-sulfur-binding nitrate reductase (NarB) that has been demonstrated in previous work to be involved in assimilatory nitrate reduction (Hidalgo-García et al. Citation2019). Upstream of RHE_CH01783, two other genes are located, RHE_CH01784 and RHE_CH01785, which have been annotated as a NO3--ABC transporter, ATP-binding protein (NrtCch), and a two-component response regulator protein (NasT), respectively (). Although RHE_CH01784 is annotated as nrtCch, this gene is more likely to be nasS given its proximity to nasT (RHE_CH01785). The alignment of the NrtCch amino acids sequence revealed between 60 and 50% similarity to NasS proteins from B. diazoefficiens USDA 110 and Paracoccus denitrificans PD1222, respectively (Source: NCBI Blast, data not shown). Consequently, these two genes are likely to encode the NasS-NasT two component regulatory system for NO3-/NO2- perception and control of genes involved in NO3-/NO2- assimilation that has been previously characterized in the α-proteobacteria P. denitrificans (Luque-Almagro et al. Citation2013) and B. diazoefficiens (Cabrera et al. Citation2016).
Figure 1. Genomic context of RHE_CH01783 gene. Source: https://www.ncbi.nlm.nih.gov/gene.
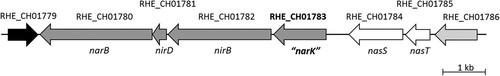
Concerning RHE_CH01783, this gene has been predicted to encode a probable NO3--transporter belonging to the major facilitator superfamily (MFS)-type transporter (Source: https://www.genome.jp/kegg/genes.html). A sequence alignment (Supplementary Figure S1A) showed that RHE_CH01783 protein shares from 79% to 38% amino acid similarity with NarK-like transporters (a subgroup of MFS proteins) from P. denitrificans, Thermus thermophilus, E. coli and B. diazoefficiens (Supplementary Figure S1B). Interestingly, among rhizobia, B. diazoefficiens NarK, whose translated sequence has 40% similarity with the predicted primary sequence of R. etli NarK, has been reported to be involved in NO2- export (Cabrera et al. Citation2016).
Since proline residues usually have a crucial role in dynamic processes involving transmembrane α-helices, we looked at the R. etli NarK sequence (Supplementary Figure S1A) and we found six prolines located within the residues that conform the α-helices (P58, P157, P240, P302, P364 and P375; Supplementary Figure S1A). Among them, four are highly conserved within NarK1 homologous (P58, P157, P302 and P375), and three are highly conserved within NarK2 homologous (P58, P157 and P375). Particularly, P302 has been reported to be essential for NarK1 function, meanwhile P58, P157 and P375 were shown to be essential for NarK2 function in P. denitrificans (Goddard et al. Citation2017).
As shown in Supplementary Table S2, NarK homologous are present in many rhizobia species, however is only in B. diazoefficiens (Cabrera et al. Citation2016) and Sinorhizobium meliloti (Ruiz et al. Citation2022) where a connection between assimilatory and denitrification pathways has been reported.
3.2. Involvement of R. etli NarK in nitrite extrusion and nitrous oxide production by free-living cells
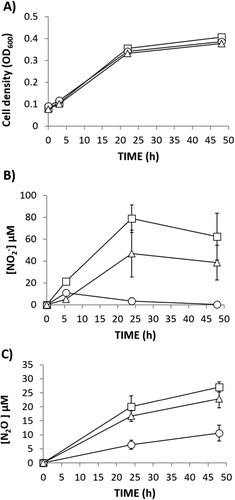
To investigate the role of R. etli NarK in NO2- export and N2O production, a narK knockout mutant was constructed. The narK mutant strain was able to grow with NO3- as the sole N source at the same rate as the WT cells under oxic, as well as microoxic, conditions (A,C, respectively). This demonstrated that NO3- assimilation was not impaired by mutation of narK gene. However, extracellular NO2- accumulation was significantly lower in the growth medium of the narK mutant compared to the WT strain (B) under oxic conditions indicating a NarK role in nitrite extrusion. When cultured microoxically, extracellular NO2- was not detected in the growth medium of any strains (D). This observation suggests that, as previously reported (Hidalgo-García et al. Citation2019), under microoxic conditions, NO2- exported from the cytoplasm might be subsequently reduced to NO and N2O in the periplasm by NirK and cNor denitrification enzymes, respectively. However, we can assume that NO2- export is also affected under microoxic conditions since N2O production is notably decreased in the headspace of the narK cultures compared to that from the WT cultures (E). In order to confirm that the decrease of N2O production in the narK mutant was due to an impaired NO2- transport from cytoplasm to periplasm, we performed the same experiment but using NO2- as the sole N source (). Under these conditions the WT and narK mutant grew (A) and emit N2O (B) at similar rates. Hidalgo-García et al. (Citation2019) reported that R. etli CE3 overexpressing NarB shows increased levels of extracellular NO2- and headspace N2O under microoxic conditions with NO3- as the sole N source. To further confirm the NarK involvement in NO2- extrusion and N2O production, WT and narK strains overexpressing NarB were used (). As previously reported by Hidalgo-García et al. (Citation2019), extracellular NO2- accumulation in growth medium (80 µM NO2- after 25 h incubation) for a WT + NarB+ strain was observed during microoxic culture with NO3-. However, when narB was overexpressed in the narK mutant, NO2- was practically undetectable in the medium (B). The WT + NarB+ and narK + NarB+ strains showed similar growth (A), however, a noticeable difference was N2O production that was much lower in the narK mutant compared to that observed in the WT, both overexpressing NarB (C). Collectively, these in vivo results suggest that NarK is involved in NO2- transport from the cytoplasm to the periplasm and, therefore in N2O emission, since the NO2- supplied to the periplasm by NarK is the substrate for NirK and cNor denitrification enzymes that reduce it to NO and N2O, respectively. Complementation of the narK + NarB+ mutant with an expression plasmid carrying the narK gene under the control of a constitutive promotor, restored the extracellular NO2- and N2O production to levels comparable with those observed for the WT + NarB+ strain (B,C, respectively).
Figure 2. Nitrate-dependent growth (A, C), extracellular nitrite (NO2-) concentration (B, D) and N2O accumulation (E) of R. etli CE3 (WT) (▪) and narK (●). Cells were cultured with NO3- as the sole N source under oxic (A, B) or microoxic (C, D, E) conditions. Data are means with standard error bars from at least two independent cultures assayed in triplicate.
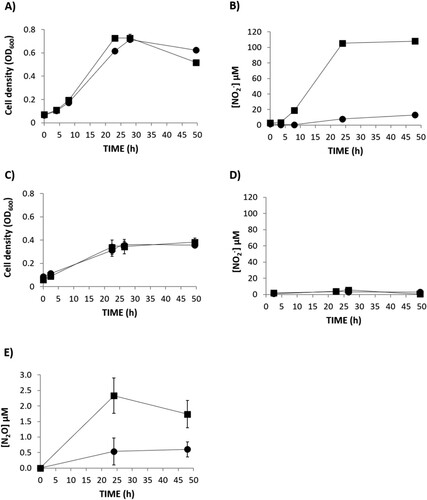
Figure 3. Growth (A) and N2O accumulation (B) by R.etli CE3 (WT) (▪) and narK(●), cultured microoxically with NO2- as the sole N source. Data are means with standard error bars from at least two independent cultures assayed in triplicate.
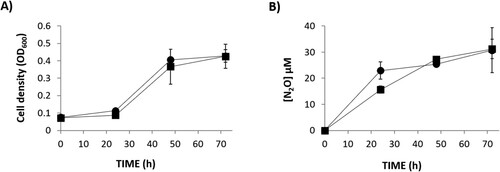
Figure 4. Nitrate-dependent growth (A) extracellular NO2- concentration (B) and N2O accumulation (C) by R.etli WT + NarB+ (□), narK + NarB+ (○) and narK + NarB++NarK+ (Δ), cultured microoxically with NO3− as the sole N source. Data are means with standard error bars from at least two independent cultures assayed in triplicate.
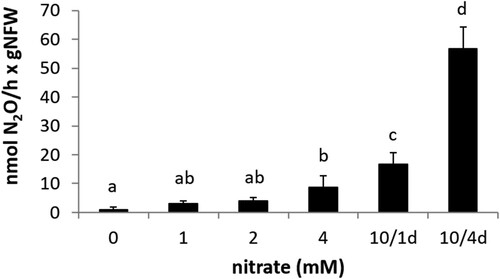
3.3. Nitrate induces N2O emission by common bean nodules
The capacity of common bean nodules to emit N2O was explored in plants inoculated with R. etli CE3 and N2O emission was measured in detached nodules by gas chromatography. Several treatments were tested to determine which plant growth conditions could support rhizobia denitrification and, therefore, N2O production by common bean nodules. shows a positive correlation between 1, 2, and 4 mM KNO3 concentration in the mineral solution applied to irrigate the plants and N2O emission. Treatment of 10 mM NO3- for one day before nodules harvesting slightly increases N2O production by nodules compared to 1, 2 or 4 mM treatment during the entire plant growth period. However, the highest N2O emission rate was reached after the application of 10 mM KNO3 four days before plant harvesting (). Although this treatment significantly induced N2O production by the nodules, it had an adverse effect in nodule biomass and fitness since decreased nodules fresh weight as well as leghaemoglobin content of nodules was observed in 10 mM KNO3 treated plants compared to plants watered with a N-free mineral solution throughout the experiment (Supplementary Figure S2).
Figure 5. N2O emission by detached nodules from plants inoculated with R. etli CE3 and watered with mineral solution containing 0, 1, 2 or 4 mM KNO3 throughout the growth period or with 10 mM KNO3 for one day (10/1d) or four days (10/4d) before plant harvesting. Data are expressed as the mean value and standard deviation error bars of two independent experiments. In each experiment, 5 replicates collected from ten plants were assayed. Lower-case letters indicate comparisons between nitrate treatments. Same lower-case letters are not statistically significant according to HSD Tukey test at p ≤ 0.05. NFW, nodule fresh weight.
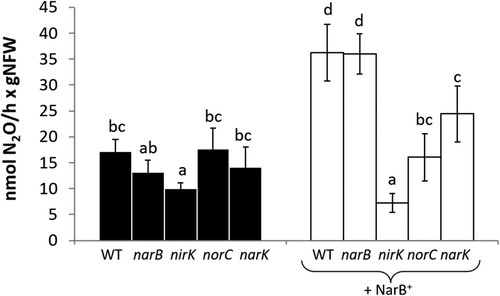
3.4. R. etli NarB, NirK, NorC and NarK proteins are involved in N2O emission from common bean nodules
Common bean plants were inoculated with R. etli CE3 (WT) and a set of mutant strains individually lacking the narB, nirK, norC or narK genes. Plants were watered with N-free mineral solution and 4 days before plant harvesting, they were treated with 10 mM KNO3. N2O production was detected in nodules from common bean plants regardless of the R. etli strain used as inoculum (). We found that only nodules formed in plants inoculated with the nirK mutant emitted 41% less N2O than those from plants inoculated with the WT strain (). A parallel experiment was performed using the WT and the same set of mutants overexpressing NarB (WT + NarB+, narB + NarB+, nirK + NarB+, norC + NarB+ and narK + NarB+). Overexpression of the narB gene in WT and narB strains resulted in about 2-fold increase of N2O emission. Interestingly, nodules formed by nirK + NarB+, norC + NarB+ and narK + NarB+ mutants emitted significantly less N2O than the WT + NarB+ (). In fact, nirK + NarB+ nodules produced about 77% less N2O than WT + NarB+ nodules. In the case of norC + NarB+ nodules, they emitted around 56% less N2O than WT + NarB+ nodules (). Finally, N2O production capacity of the nodules induced by narK + NarB+ mutant was only decreased by about 25% compared to WT + NarB+ nodules ().
Figure 6. N2O emission by detached nodules from plants inoculated with R. etli CE3 (WT), narB, nirK, norC and narK (black bars) or R. etli WT + NarB+, narB + NarB+, nirK + NarB+, norC + NarB+ and narK + NarB+ (white bars). Plants were treated with 10 mM KNO3 for four days before plant harvesting. Data are expressed as the mean value and standard deviation error bars of two independent experiments. In each experiment, 5 replicates collected from ten plants were assayed. Lower-case letters indicate comparisons between nitrate treatments. Same lower-case letters are not statistically significant according to HSD Tukey test at p ≤ 0.05. NFW, nodule fresh weight.
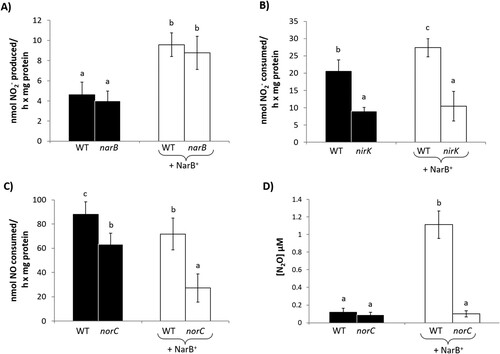
To better understand the involvement of NarB, NirK and cNor enzymes in N2O emission from common bean nodules, NR, NiR and Nor activities, as well as N2O production, were analyzed in isolated bacteroids from nodules of plants inoculated with the WT strain or the narB, nirK and norC mutants. With respect to NR, very low levels of activity were observed in bacteroids of the WT and narB mutant (A). However, the overexpression of NarB protein in the WT and narB strains resulted in a significant induction of NR activity by more than double compared to the strains that did not overexpress narB gene (A). NiR activity was clearly impaired in bacteroids from nodules recovered from plants inoculated with the nirK mutant strain compared to those of WT strain in both scenarios: with or without overexpression of NarB protein (B). Bacteroids produced by the norC mutant consumed around 25% less NO than WT bacteroids. This difference was more notable between WT and norC strains overexpressing NarB protein, where the rate of NO consumption of norC + NarB+ bacteroids decreased around 58% compared to WT + NarB+ bacteroids (C). Finally, we analyzed N2O production by bacteroids induced by WT, norC, WT + NarB+ or norC + NarB+ strains (D). Interestingly, NarB overexpression in the WT strain increased the bacteroid N2O emission by around 10-fold (D). By contrast, very low levels of N2O were produced by norC and norC + NarB+ bacteroids compared to those produced by WT + NarB+ bacteroids (D).
Figure 7. Nitrate reductase (A), nitrite reductase (B), nitric oxide reductase (C) activities and N2O production (D) of bacteroids from nodules of common bean plants inoculated with R. etli CE3 (WT), narB, nirK or norC (black bars) or WT + NarB+, narB + NarB+, nirK + NarB+, or norC + NarB+ (white bars). Data are expressed as the mean value and standard deviation error bars of two independent experiments. In each experiment, bacteroids from 3 replicates collected from ten plants were assayed. Lower-case letters indicate comparisons between strains. Same lower-case letters are not statistically significant according to HSD Tukey test at p ≤ 0.05.
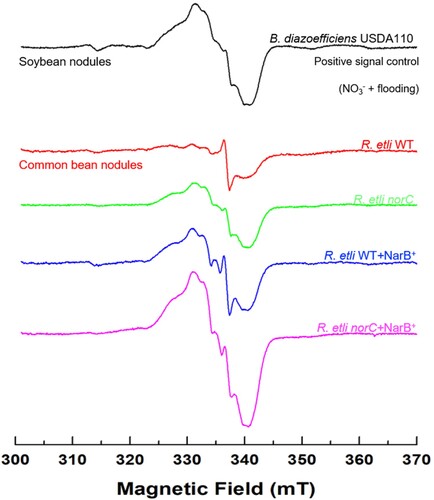
3.5. R. etli NarB and cNor proteins are involved in NO homeostasis in common bean nodules
Common bean plants were inoculated with WT, norC, WT + NarB+ and norC + NarB+ strains of R. etli and treated with 10 mM KNO3 four days before harvesting. Electron paramagnetic resonance (EPR) spectroscopy was used to detect NO bound to Lb (LbNO complexes) within intact frozen nodules recovered from the plant root systems. It has been previously reported that soybean nodules from plants inoculated with B. diazoefficiens, watered with 4 mM KNO3 and subjected to hypoxia produced an EPR signal specific for LbNO complexes (Meakin et al. Citation2007; Sánchez et al. Citation2010). Therefore, in this current study, nodules from soybean plants treated with nitrate and flooding were used as a positive control for detection of EPR signals from LbNO complexes (). Interestingly, overexpression of the narB gene in the WT strain gave rise to clear formation of these complexes in common bean nodules (). Furthermore, the magnitude of the specific EPR signal was greater in nodules from plants inoculated with the R. etli norC or norC + NarB+ strains compared to nodules recovered from plants inoculated with either WT or WT + NarB+ strains, indicating a higher accumulation of LbNO complexes in nodules deficient in the bacteroidal NO reductase. Notably, nodules from plants inoculated with the norC + NarB+ strain gave the largest EPR signal for LbNO ().
Figure 8. Leghaemoglobin-NO (LbNO) complexes detected by electron paramagnetic resonance (EPR) from whole intact nodules of common bean plants inoculated with R. etli CE3 (WT), norC, WT + NarB+ and norC + NarB + . Nodule EPR signal from soybean plants subjected to NO3− and flooding was used as positive control. For each strain, a representative EPR spectrum observed from triplicate nodule samples from three different nodule harvests is presented.
4. Discussion
4.1. Rhizobium etli NarK is involved in nitrite extrusion and N2O emission by free-living cells
R. etli is a N2-fixing bacterium able to live freely in soil or symbiotically associated with common bean plants. It is considered an incomplete denitrifier since its genome only contains the nirK and nor denitrification genes, which code for the nitrite and nitric oxide reductases, respectively, but lacks genes that encode for the respiratory nitrate reductases (Nar or Nap) and nitrous oxide reductase (Nos). In a previous work (Hidalgo-García et al. Citation2019), it was demonstrated that R. etli is able to produce NO and N2O when cultivated under microoxic conditions with NO3- as the sole N source, due to the action of an assimilatory nitrate reductase, NarB. In the cytosol, NO3- is reduced to NO2- via NarB, then NO2- can be further reduced to NH4+ or exported outside the cell since this molecule can be toxic at certain concentrations. However, once NO2- passes into the periplasmic space, it can be subsequently reduced to NO and N2O by NirK and cNor denitrification enzymes, respectively. Until now, it was unknown which transporter(s) were involved in NO2- export to the periplasmic space in R. etli, acting as a nexus between the nitrate assimilatory and denitrification pathways. Bioinformatic analyses showed that, in the R. etli genome, upstream nirB, nirD, and narB genes, a gene (RHE_CH01783) was predicted to encode a putative NarK-like NO3--transporter belonging to the MFS-type transporters superfamily. Using a R. etli narK knockout mutant, we have demonstrated that NarK performs a role in NO2- export to the periplasm and supports N2O production under microoxic conditions. Consistent with a NO2- extrusion role for NarK, when the narK mutant was cultured with NO2- as the sole N source under microoxic conditions, N2O emission was similar to that observed for WT cultures. In this case, NO2- is externally provided and its export from cytosol to periplasm is not necessary for N2O formation in the periplasmic space.
In bacteria, there are two classes of MFS transporters involved in NO3- transport: NarK1 are predicted to be NO3-/proton symporters (Moir and Wood Citation2001), whereas NarK2 are believed to be NO3-/ NO2- antiporters (Moir and Wood Citation2001; Goddard et al. Citation2008). In this context, it has been also reported that in P. denitrificans, NarK1 and NarK2 are fuzed into a single protein where the NarK1-like domain functions primarily as a NO3- transporter, while the NarK2-like domain is more specialized in NO3-/NO2- antiport (Goddard et al. Citation2017). R. etli NarK shows higher amino acid sequence similarity with NarK1 protein from P. denitrificans, however our results suggest a NO2- export role for this protein. Accordingly, among rhizobia, B. diazoefficiens NarK has also been previously reported to be involved in NO2- export (Cabrera et al. Citation2016). The capacity of the narK mutant to grow on NO3- or NO2- suggests that NarK is not implicated, or at least is not essential, for the import of such substrates to the cytoplasm in R. etli. This suggests that other unknown importers that are encoded by genes remote to the NO3- or NO2- assimilatory genes are involved in the transport of these molecules to the cytosol. In this context, it has been reported that free nitrous acid (HNO2) diffusion might be a way of NO2- transport across the inner membrane (Gates et al. Citation2011).
4.2. N2O is emitted by common bean nodules by the coupling, through NarK transporter, of R. etli nitrate assimilation and denitrification routes
Our next goal was to explore the role of R. etli NarB, NarK, NirK and cNor in N2O emission by common bean root nodules. It is well known that soybean nodules produce N2O in response to nitrate and flooding (Tortosa et al. Citation2015; Tortosa et al. Citation2020). However, due to the high sensitivity of common bean to flooding (Hidalgo-García et al. Citation2016), this standard laboratory experimental conditions could not be used to study N2O emissions by common bean root nodules. Instead, plants were exposed to different NO3− treatments to investigate the capacity of the nodules to produce N2O. The application of 10 mM KNO3 for four days resulted in the highest N2O production in nodules. This treatment also reduced the fitness of the nodules as determined by the analysis of Lb content. As observed in our work, it has been previously reported that 10 mM KNO3 treatment of common bean or soybean for 4 or 6 days, respectively, affected nodules functionality by inducing their premature senescence (Calvo-Begueria et al. Citation2018). The nitrate treatment (10 mM KNO3 four days before plant harvesting) used in this work to induce N2O emissions is quite improbable under natural conditions and it is detrimental to symbiosis and nitrogen fixation. However, those conditions allowed us to demonstrate N2O emissions from common bean nodules as well as the involvement of R. etli NarB, NarK, NirK and cNor proteins under high nitrate fertilization when nodulation is established.
Next, the contribution of R. etli NarB, NirK, cNor, and NarK proteins in N2O emissions by common bean nodules was assessed. In contrast to free-living conditions where NarB is required for N2O production (Hidalgo-García et al. Citation2019), mutation of narB did not significantly affect N2O emission by common bean nodules. Similar to our observations, it has been shown in Medicago truncatula nodules that S. meliloti nitrate assimilation pathway encoded by nirBDnarB did not play any detectable role in the production of NO, the N2O precursor (Ruiz et al. Citation2022). Despite this, we have demonstrated the involvement of NarB in bacteroidal NR activity as well as in nodule and bacteroidal N2O emissions by overexpressing narB. Our results also reveal an important contribution of bacteroidal NirK to N2O emissions by common bean nodules. This was supported with nirK and nirK + NarB+ nodules emitting significantly less N2O than WT and WT + NarB+ nodules, respectively, presumably due to the impaired bacteroidal Nir activity. The involvement of cNor in N2O production was only observed in nodules produced by a norC mutant additionally overexpressing narB. However, norC + NarB+ still produced significant levels of N2O (about 44%). These results suggest that, in addition to cNor, other plant or bacterium proteins may be involved in N2O formation. Since, norC or norC + NarB+ bacteroids were defective in N2O production, a plant protein may provide an additional source of N2O in common bean nodules. Supporting this hypothesis, it has been reported that N2O can be produced from NO in plant mitochondria (Timilsina et al. Citation2020 and references herein).
Although the norC mutation resulted in a lower capacity of bacteroids to consume NO compared to WT, the NO consumption activity remained higher than that observed under free-living conditions, where NO consumption by the norC mutant was almost abolished compared to WT cells (Hidalgo-García et al. Citation2019). This discrepancy between free-living and symbiotic lifestyles might indicate the existence of other systems to detoxify NO that are induced specifically under symbiotic conditions in R. etli bacteroids. In this context, in addition to cNor, a combined role for S. meliloti flavohaemoglobin (Hmp), and NnrS1 and NnrS2 proteins in NO degradation has been reported in Medicago truncatula nodules (Blanquet et al. Citation2015). Furthermore, in soybean nodules, the B. diazoefficiens hemoglobin, Bjgb, indirectly contributes to NO levels modulation in the nodules (Salas et al. Citation2020). Finally, R. etli NarK also plays an important role in N2O formation by contributing to NO2− export from the bacteroidal cytosol to the periplasmic space. However, in contrast to free-living conditions, narK nodules still produce significant levels of N2O. These results suggest the involvement of additional specific NO2- transport systems in the bacteroids or alternatively the participation of plant proteins in N2O formation.
4.3. R. etli NarB and cNor proteins modulate NO levels in common bean nodules
In addition to being a precursor to N2O, NO is a very important signaling molecule in plants and is involved in many processes including vegetative development, defence against pathogens, abiotic stress signalling among others. Interestingly, NO also plays a role at different stages of the rhizobia-legume symbiotic relationship, from the early steps of infection to nodule senescence (reviewed by Berger et al. Citation2019). Moreover, NO inhibits nitrogenase activity (Sánchez et al. Citation2010). Consequently, NO concentration has to be finely controlled inside nodules to maintain viability. It has been previously reported that denitrification performed by B. diazoefficiens or S. meliloti is an important source of NO in soybean or M. truncatula nodules (Horchani et al. Citation2010; Sánchez et al. Citation2010; Tortosa et al. Citation2015). This molecule can bind to leghaemoglobin yielding LbNO complexes which can be detected readily by EPR (Sánchez et al. Citation2010). In order to investigate the contribution of R. etli NarB and cNor to NO levels in common bean nodules, LbNO complexes were analysed by EPR in intact nodules. Overexpression of R. etli narB in the WT strain gave rise to clear LbNO complex signals in intact nodules suggesting that NarB contributes to NO formation in common bean nodules. Similar to what has been reported for soybean nodules by using a B. diazoefficiens norC mutant (Sánchez et al. Citation2010; Calvo-Begueria et al. Citation2018), mutation of R. etli norC gene also resulted in higher levels of LbNO complexes demonstrating the involvement of R. etli NarB and NorC in NO modulation in common bean nodules.
5. Conclusions
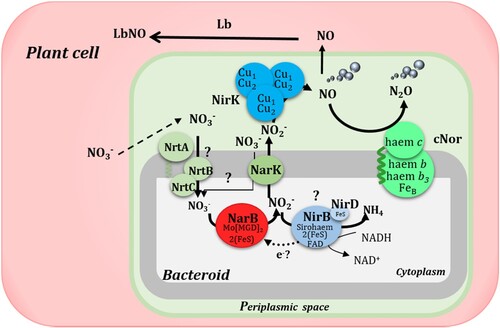
Legumes contribute to N2O emissions by providing N-rich residues for decomposition or directly by some rhizobia that can denitrify under free-living and symbiotic conditions. In the case of common beans, its endosymbiont, Rhizobium etli CFN42, is an incomplete denitrifier since it lacks the nar or nap genes encoding the respiratory nitrate reductases (Nap or Nar) as well as the nos genes encoding the nitrous oxide reductase enzyme. In this work, we have demonstrated the capacity of common bean nodules to produce N2O by coupling NO3- assimilation and denitrification in the bacteroids (). In this pathway, NO2- formed by the R. etli assimilatory nitrate reductase NarB is exported by NarK from the cytosol to the periplasm. Then, the denitrifying enzymes NirK and NorC reduce NO2- to NO and N2O, respectively (). In conclusion, this work highlights the importance of rhizobia NO3- assimilation and denitrification in emission of NO and N2O by common bean nodules under high nitrate fertilization when the nodulation is established. The knowledge generated in this work including the identification of proteins involved in NO and N2O production by legume root nodules will contribute to development of new solutions to improve legume crop production while mitigating greenhouse gas emissions.
Figure 9. Schematic representation of the contribution of NarB, NarK, NirK and cNor to NO and N2O production in common bean nodules. In this model, NO3− provided in the nutrient solution enters the nodule and can be transported to the cytoplasm of the bacteroid where, by the action of NarB it would be reduced to NO2−. Then, NO2− can be exported to the periplasmic space through NarK and there the denitrifying enzymes NirK and cNor would reduce it to NO and N2O, respectively. In the plant cell, NO produced by the bacteroids would be bound to Lb to form LbNO complexes. Question marks denote mechanisms still unknown. Adapted from Salas et al. Citation2021.
Supplemental Material
Download MS Word (2.2 MB)Acknowledgements
We thank Dr Fraser Macmillan and Dr Myles Cheesman from the School of Chemistry at the University of East Anglia for their help with assistance with acquisition of EPR spectra and useful discussions and Marisa Rodríguez from the Centro de Ciencias Genómicas, Universidad Nacional Autónoma de México (UNAM) for technical assistance. We also acknowledge the support of the publication fee by the CSIC Open Access Publication Support Initiative through its Unit of Information Resources for Research (URICI). M.J.D, M.J.T. and A.H-G conceived and designed the study. A.H-G., G.T., P.J.P., and A.J.G. performed the experiments. A.H-G., A.J.G, M.J.T., and M.J.D. analysed the results and wrote the manuscript. A.H-G., and A.J.G prepared the figures and tables. A.J.G., D.J.R., E.J.B., L.G., M.J.T., and M.J.D. critically revised the manuscript. All authors read and approved the final version of the manuscript.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Additional information
Funding
Notes on contributors
Alba Hidalgo-García
Alba Hidalgo-García is a postdoctoral researcher in the Nitrogen Metabolism in Rhizospheric Bacteria (NITRORHIZ) group at the Estación Experimental del Zaidín (EEZ-CSIC), Granada, Spain. At the moment she is developing a Project about the implication of bacterial proteins in nitric and nitrous oxide emission by the Rhizobium etli - Phaseolus vulgaris symbiosis.
Germán Tortosa
Germán Tortosa is a research technician in the Nitrogen Metabolism in Rhizospheric Bacteria (NITRORHIZ) group at the Estación Experimental del Zaidín (EEZ-CSIC), Granada, Spain. His research primarily focuses on how soil microorganisms contribute to plant growth and how they can recycle organic waste to produce compost. He possesses expertise in composting, organic and biological fertilizers, biostimulants, rhizobia, legumes, nitrous oxide (N2O) emissions, the denitrification process, and environmental nitrate pollution.
Pedro J. Pacheco
Pedro J. Pacheco is a postdoctoral researcher at the Department of Soil and Plant Microbiology, in the Estación Experimental del Zaidín (EEZ-CSIC), Granada, Spain. His research has been focused on the study of the environmental factors and molecular mechanisms involved in nitrous oxide emissions from endosymbiotic bacteria either under free-living conditions or in symbiosis with their host legume, with special emphasis on the role of copper in denitrification.
Andrew J. Gates
Andrew Gates is an Associate Professor of Bacterial Bioenergetics at the University of East Anglia, Norwich, UK. His research focusses on investigating the role of proteins and regulatory nucleic acid secondary structures in controlling nitrate, nitrite, nitric oxide and nitrous oxide metabolism in denitrifying microorganisms.
David J. Richardson
David Richardson is a Professor of Microbial Biochemistry at the University of East Anglia, Norwich, UK. He has used a range of approaches to investigate respiratory processes of anaerobic bacteria from a wide range of environments. His work on the nitrogen cycle as well as the mechanisms by which bacteria use insoluble extracellular minerals as respiratory electron acceptors has made major contributions to environmental and climate science, industrial biotechnology and synthetic biology.
Eulogio J. Bedmar
Eulogio J Bedmar is Professor Emeritus in the Nitrogen Metabolism in Rhizospheric Bacteria (NITRORHIZ) group at the Estación Experimental del Zaidín (EEZ-CSIC), Granada, Spain. His research has been focused on the biodiversity of soil microorganisms associated with plants. He has also expertise in rhizobia, legumes, nitrous oxide (N2O) emissions, the denitrification process, and environmental nitrate pollution.
Lourdes Girard
Lourdes Girard is an Associated Professor in the Center for Genome Sciences at the National Autonomous University of Mexico, Cuernavaca, México. She performs research within the field of Molecular Genetics in bacteria, focusing primarily on nitrogen-fixing bacteria known as Rhizobia, which establish symbiosis with legume plants. Her main contributions are within the transcriptional regulation and the analysis of the functional networks that are essential to control biological nitrogen fixation in response to environmental stimuli such as oxygen and abiotic stresses. Her research combines genetics, molecular biology and genomic approaches
María J. Torres
María J Torres is a postdoctoral researcher in the Department of Biochemistry and Molecular Biology at the University of Córdoba, Spain. She is studying how microalgae-bacteria consortia can be used to bioremediate rural wastewater, and also the mechanisms implicated in the N2O gas emission from soil microorganisms.
María J. Delgado
María J Delgado is a senior researcher and the head of the Nitrogen Metabolism in Rhizospheric Bacteria (NITRORHIZ) group at the Estación Experimental del Zaidín (EEZ-CSIC), Granada, Spain. Her current research interest is related to sustainable legume production and mitigation of greenhouse gas emissions. She has been coordinator of several national and international projects focused on rhizobia, legumes, nitrous oxide (N2O) emissions, and the denitrification process.
References
- Berger A, Boscari A, Frendo P, Brouquisse R. 2019. Nitric oxide signaling, metabolism and toxicity in nitrogen-fixing symbiosis. J Exp Bot. 70:4505–4520. doi:10.1093/jxb/erz159.
- Beringer JE. 1974. R factor transfer in Rhizobium leguminosarum. J Gen Microbiol. 84:188–198.
- Blanquet P, Silva L, Catrice O, Bruand C, Carvalho H, Meilhoc E. 2015. Sinorhizobium meliloti controls nitric oxide–mediated post-translational modification of a Medicago truncatula nodule protein. Mol Plant-Microbe Interact. 28:1353–1363. doi:10.1094/MPMI-05-15-0118-R.
- Bravo A, Mora J. 1988. Ammonium assimilation in Rhizobium phaseoli by the glutamine synthetase-glutamate synthase pathway. J Bacteriol. 170:980–984. doi:10.1128/jb.170.2.980-984.1988.
- Butterbach-Bahl K, Baggs EM, Dannenmann M, Kiese R, Zechmeister-Boltenstern S. 2013. Nitrous oxide emissions from soils: how well do we understand the processes and their controls? Philos Trans R Soc Lond, B Biol Sci. 368:1–13. doi:10.1098/rstb.2013.0122.
- Cabrera JJ, Salas A, Torres MJ, Bedmar EJ, Richardson DJ, Gates AJ, Delgado MJ. 2016. An integrated biochemical system for nitrate assimilation and nitric oxide detoxification in Bradyrhizobium japonicum. Biochem J. 473:297–309. doi:10.1042/BJ20150880.
- Calvo-Begueria L, Rubio MC, Martínez JI, Pérez-Rontomé C, Delgado MJ, Bedmar EJ, Becana M. 2018. Redefining nitric oxide production in legume nodules through complementary insights from electron paramagnetic resonance spectroscopy and specific fluorescent probes. J Exp Bot. 69:3703–3714. doi:10.1093/jxb/ery159.
- Delgado MJ, Bonnard N, Tresierra-Ayala A, Bedmar EJ, Muller P. 2003. The Bradyrhizobium japonicum napEDABC genes encoding the periplasmic nitrate reductase are essential for nitrate respiration. Microbiology. 149:3395–3403. doi:10.1099/mic.0.26620-0.
- Delgado MJ, Casella S, Bedmar EJ. 2007. Denitrification in rhizobia-legume symbiosis. In: Bothe H, Ferguson SJ, Newton WE, editor. Biology of the nitrogen cycle. Amsterdam: Elsevier Science; p. 57–66.
- Delgado MJ, Ligero F, Lluch C. 1994. Effect of salt stress on growth and nitrogen fixation by pea, faba bean, common bean and soybean plants. Soil Biol Biochem. 26:371–376. doi:10.1016/0038-0717(94)90286-0.
- Gates AJ, Luque-Almagro VM, Goddard AD, Ferguson SJ, Roldan MD, Richardson DJ. 2011. A composite biochemical system for bacterial nitrate and nitrite assimilation as exemplified by Paracoccus denitrificans. Biochem J. 435:743–753. doi:10.1042/BJ20101920.
- Goddard AD, Bali S, Mavridou DA, Luque-Almagro VM, Gates AJ, Roldán MD, Newstead S, Richardson DJ, Ferguson SJ. 2017. The Paracoccus denitrificans NarK-like nitrate and nitrite transporters probing nitrate uptake and nitrate/nitrite exchange mechanisms. Mol Microbiol. 103:117–133. doi:10.1111/mmi.13546.
- Goddard AD, Moir JW, Richardson DJ, Ferguson SJ. 2008. Interdependence of two NarK domains in a fused nitrate/nitrite transporter. Mol Microbiol. 70:667–681. doi:10.1111/j.1365-2958.2008.06436.x.
- Gómez-Hernández N, Reyes-González A, Sánchez C, Mora Y, Delgado MJ, Girard L. 2011. Regulation and symbiotic role of nirK and norC expression in Rhizobium etli. Mol Plant-Microbe Interact. 24:233–245. doi:10.1094/MPMI-07-10-0173.
- Graham PH, Vance CP. 2003. Legume Importance and Constraints to Greater Use. Plant Physiol. 131:872–877. doi:10.1104/pp.017004.
- Hein S, Simon J. 2019. Bacterial nitrous oxide respiration: electron transport chains and copper transfer reactions. Adv Microb Physiol. 75:137–175. doi:10.1016/bs.ampbs.2019.07.001.
- Hidalgo-García A, Torres MJ, Salas A, Bedmar EJ, Girard L, Delgado MJ. 2019. Rhizobium etli produces nitrous oxide by coupling the assimilatory and denitrification pathways. Front Microbiol. 10:1–11. doi:10.3389/fmicb.2019.00980.
- Hidalgo-García A, Tortosa G, Bedmar EJ, Delgado MJ. 2016. Efecto del nitrato y el encharcamiento en la emisión del gas invernadero N2O por la simbiosis Phaseolus vulgaris-Rhizobium etli. In: Clemente A, De Ron AM, editors. 7ª acta de la asociación española de leguminosas. Granada: Editorial Atrio S.L.; p. 51–61.
- Hirayama J, Eda S, Mitsui H, Minamisawa K. 2011. Nitrate dependent N2O emission from intact soybean nodules via denitrification by Bradyrhizobium japonicum bacteroids. Appl Environ Microbiol. 77:8787–8790. doi:10.1128/AEM.06262-11.
- Horchani F, Prévot M, Boscari A, Evangelisti E, Meilhoc E, Bruand C, Raymond P, Boncompagni E, Aschi-Smiti S, Puppo A, Brouquisse R. 2010. Both plant and bacterial nitrate reductases contribute to nitric oxide production in Medicago truncatula nitrogen-fixing nodules. Plant Physiol. 155:1023–1036. doi:10.1104/pp.110.166140.
- Inaba S, Ikenishi F, Itakura M, Kikuchi M, Eda S, Chiba N, Katsuyama C, Suwa Y, Mitsui H, Minamisawa K. 2012. N2O emission from degraded soybean nodules depends on denitrification by Bradyrhizobium japonicum and other microbes in the rhizosphere. Microbes Environ. 27:470–476. doi:10.1264/jsme2.ME12100.
- Inaba S, Tanabe K, Eda S, Ikeda S, Higashitani A, Mitsui H, Minamisawa K. 2009. Nitrous oxide emission and microbial community in the rhizosphere of nodulated soybeans during the late growth period. Microbes Environ. 24:64–67. doi:10.1264/jsme2.ME08544.
- IPCC. 2019. Summary for policymakers. In: Shukla PR, Skea J, Calvo Buendía E, Masson-Delmotte V, Pörtner H-O, Roberts DC, Zhai P, Slade R, Connors S, van Diemen R., et al., editors. Climate change and land: An IPCC special report on climate change, desertification, land degradation, sustainable land management, food security, and greenhouse Gas fluxes in terrestrial ecosystems. Geneva: Intergovernmental panel on climate change; p. 1–34.
- Kovach ME, Phillips RW, Elzer PH, Roop RM, Peterson KM. 1994. pBBR1MCS: a broad-host-range cloning vector. Biotechniques. 16:800–802.
- Kudeyarov VN. 2020. Nitrous oxide emission from fertilized soils: an analytical review. Eurasian Soil Science. 53(10):1396–1407. doi:10.1134/S1064229320100105.
- LaRue TA, Child JJ. 1979. Sensitive fluorometric assay for leghemoglobin. Anal Biochem. 92:11–15. doi:10.1016/0003-2697(79)90618-3.
- Lazcano C, Zhu-Barker X, Decock C. 2021. Effects of organic fertilizers on the soil microorganisms responsible for N2O emissions: a review. Microorganisms. 9:983. doi:10.3390/microorganisms9050983.
- Luque-Almagro VM, Lyall VJ, Ferguson SJ, Roldan MD, Richardson DJ, Gates AJ. 2013. Nitrogen oxyanion-dependent dissociation of a two-component complex that regulates bacterial nitrate assimilation. J Biol Chem. 288:29692–29702. doi:10.1074/jbc.M113.459032.
- Meakin GE, Bueno E, Jepson B, Bedmar EJ, Richardson DJ, Delgado MJ. 2007. The contribution of bacteroidal nitrate and nitrite reduction to the formation of nitrosylleghaemoglobin complexes in soybean root nodules. Microbiology. 153:411–419. doi:10.1099/mic.0.2006/000059-0.
- Mesa S, Alché JD, Bedmar E, Delgado MJ. 2004. Expression of nir, nor and nos denitrification genes from Bradyrhizobium japonicum in soybean root nodules. Physiol Plant. 120:205–211. doi:10.1111/j.0031-9317.2004.0211.x.
- Moir JW, Wood NJ. 2001. Nitrate and nitrite transport in bacteria. Cell Mol Life Sci. 58:215–224. doi:10.1007/PL00000849.
- Müller R. 2021. The impact of the rise in atmospheric nitrous oxide on stratospheric ozone. Ambio. 50:35–39. doi:10.1007/s13280-020-01428-3.
- Nicholas DJ, Nason A. 1957. Determination of nitrate and nitrite. Meth Enzymol. 3:981–984. doi:10.1016/S0076-6879(57)03489-8.
- Noel K D, Sanchez A, Fernandez L, Leemans J, Cevallos M A. 1984. Rhizobium phaseoli symbiotic mutants with transposon Tn5 insertions. Journal of Bacteriology. 158(1):148–155. http://doi.org/10.1128/jb.158.1.148-155.1984.
- Pilegaard K. 2013. Processes regulating nitric oxide emissions from soils. Philos Trans R Soc Lond B Biol Sci. 368(1621):20130126. doi:10.1098/rstb.2013.0126.
- Rigaud J, Puppo A. 1975. Indole-3-acetic acid catabolism by soybean bacteroids. J Gen Microbiol. 88:223–228. doi:10.1099/00221287-88-2-223.
- Ruiz B, Sauviac L, Brouquisse R, Bruand C, Meilhoc E. 2022. Role of nitric oxide of bacterial origin in the Medicago truncatula-Sinorhizobium meliloti symbiosis. Mol Plant Microbe Interact. 35:887–892. doi:10.1094/MPMI-05-22-0118-SC.
- Salas A, Cabrera JJ, Jiménez-Leiva A, Mesa S, Bedmar EJ, Richardson DJ, Gates AJ, Delgado MJ. 2021. Bacterial nitric oxide metabolism: recent insights in rhizobia. Adv Microb Physiol. 78:259–315. doi:10.1016/bs.ampbs.2021.05.001.
- Salas A, Tortosa G, Hidalgo-García A, Delgado A, Bedmar EJ, Richardson DJ, Gates AJ, Delgado MJ. 2020. The Hemoglobin Bjgb from Bradyrhizobium diazoefficiens controls NO homeostasis in soybean nodules to protect symbiotic nitrogen fixation. Front Microbiol. 10:2915. doi:10.3389/fmicb.2019.02915.
- Sambrook J, Fritsch EF, Maniatics T. 1989. Molecular cloning: A laboratory manual. New York: Cold Spring Harbor Laboratory, Cold Spring Harbor.
- Sambrook J, Russell D. 2001. Molecular cloning: a laboratory manual, 3rd ed. New York: Cold Spring Harbor Laboratory, Cold Spring Harbor.
- Sánchez C, Gates AJ, Meakin GE, Uchiumi T, Girard L, Richardson DJ, Bedmar EJ, Delgado MJ. 2010. Production of nitric oxide and nitrosylleghaemoglobin complexes in soybean nodules in response to flooding. Mol Plant Microbe Interact. 23:702–711. doi:10.1094/MPMI-23-5-0702.
- Sánchez C, Minamisawa K. 2019. Nitrogen cycling in soybean rhizosphere: sources and sinks of nitrous oxide (N2O). Front Microbiol. 10(1943):1–7. doi:10.3389/fmicb.2019.01943.
- Schäfer A, Tauch A, Jäger W, Kalinowski J, Thierbach G, Pühler A. 1994. Small mobilizable multi-purpose cloning vectors derived from the Escherichia coli plasmids pK18 and pK19: selection of defined deletions in the chromosome of Corynebacterium glutamicum. Gene. 145:69–73. doi:10.1016/0378-1119(94)90324-7.
- Simon R, Priefer U, Pühler A. 1983. Molecular Genetics of the Bacteria-Plant Interaction. Heidelberg: Springer.
- Thomson AJ, Giannopulos G, Pretty J, Baggs EM, Richardson DJ. 2012. Biological sources and sinks of nitrous oxide and strategies to mitigate emissions. Philos Trans R Soc Lond, B Biol Sci. 367:1157–1168. doi:10.1098/rstb.2011.0415.
- Tian H, Xu R, Canadell JG, Thompson RL, Winiwarter W, Suntharalingam P, Davidson EA, Ciais P, Jackson RB, Janssens-Maenhout G, et al. 2020. A comprehensive quantification of global nitrous oxide sources and sinks. Nature. 586:248–256. doi:10.1038/s41586-020-2780-0.
- Timilsina A, Zhang C, Pandey B, Bizimana F, Dong X, Hu C. 2020. Potential pathway of nitrous oxide formation in plants. Front Plant Sci. 11:1177. doi:10.3389/fpls.2020.01177.
- Torres MJ, Rubia MI, de la Peña T, Pueyo JJ, Bedmar EJ, Delgado MJ. 2014. Genetic basis for denitrification in Ensifer meliloti. BMC Microbiol. 14:142–151. doi:10.1186/1471-2180-14-142.
- Torres MJ, Simon J, Rowley G, Bedmar EJ, Richardson DJ, Gates AJ, Delgado MJ. 2016. Nitrous oxide metabolism in nitrate-reducing bacteria: physiology and regulatory mechanisms. Adv Microb Physiol. 68:353–432. doi:10.1016/bs.ampbs.2016.02.007.
- Tortosa G, Hidalgo A, Salas A, Bedmar EJ, Mesa S, Delgado MJ. 2015. Nitrate and flooding induce N2O emissions from soybean nodules. Symbiosis. 67:125–133. doi:10.1007/s13199-015-0341-3.
- Tortosa G, Pacheco PJ, Hidalgo-García A, Granados A, Delgado A, Mesa S, Bedmar EJ, Delgado MJ. 2020. Copper modulates nitrous oxide emissions from soybean root nodules. Environ Exp Bot. 180:104262. doi:10.1016/j.envexpbot.2020.104262.
- Trung BC, Yoshida S. 1983. Improvement of Leonard jar assembly for screening of effective rhizobium. Soil Sci Plant Nutr. 29:97–100. doi:10.1080/00380768.1983.10432410.
- van Spanning RJ, Richardson DJ, Ferguson SJ. 2007. Introduction to the biochemistry and molecular biology of denitrification. In: Bothe H, Ferguson SJ, Newton WE, editor. Biology of the nitrogen cycle. Amsterdam: Elsevier; p. 3–20.
- Weinstein M, Roberts RC, Helinski DR. 1992. A region of the broad-host-range plasmid RK2 causes stable in planta inheritance of plasmids in Rhizobium meliloti cells isolated from alfalfa root nodules. J Bacteriol. 174:7486–7489. doi:10.1128/jb.174.22.7486-7489.1992.
- Zumft WG. 1997. Cell biology and molecular basis of denitrification. Microbiol Mol Biol Rev. 61:533–616.
