1. How do omics technologies foster systems medicine?
In the era of systems medicine, the pathological networks, mutated genes and associated aberrant pathways, and a drug’s actions to rectify the abnormalities, of a disease are integrated across the systems of human biology by using genomics, epigenomics, transcriptomics, and proteomics technologies. Omics technologies are the pillars of systems medicine.
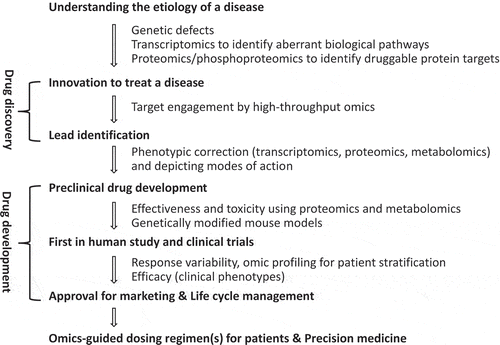
For a disease attributed to a genetic mutation, the specific mutated gene constitutes the biomarker for identifying the patient population for clinical trials and precision medicine at bedside; specific proteins in the associated aberrant pathway are the targets for pharmaceutical research and development (R&D). Omics technologies are vital throughout all phases of pharmaceutical R&D. Synergistic integration of genome and proteome enabled mapping individual clusters of protein kinases to corresponding disease loci [Citation1], contributing to discovery and development of targeted therapy. Chemoproteomics and metabolomics allow illustration of target engagement in a living system [Citation2] and mechanistic determination of biochemical pathways affected by a disease or a drug at the systems level [Citation3], respectively. Transcriptomic profiles provide insight into biological linkages between genomes and proteomes. Indeed, systems medicine has transformed pharmaceutical R&D. Omics has become an integral part of informed pharmaceutical R&D ().
Figure 1. Omics technologies are an integral part of discovery and development of a drug for specific indication(s) throughout its lifecycle. Omics data, including genomics, proteomics, phosphoproteomics and metabolomics, are useful for understanding the etiology of a disease, identifying druggable targets, and demonstrating clinical efficacy and safety. Most importantly, they are useful for managing the quality of a drug throughout its lifecycle.
2. What successes have been accelerated by omics technologies?
Genomics (including epigenomics), transcriptomics, proteomics, and metabolomics technologies make it possible to link genetic mutations and related aberrant molecular pathways to drug targets and to clinical phenotypes of cancers [Citation4] and of genetic diseases[Citation5]. From the number of drugs approved for cancers and genetic diseases, it is clear that omics technologies are, and will continue to be, an important part of pharmaceutical productivity.
2.1. Oncology – from targeting aberrant pathways associated with mutated genes to gene therapy
There have been 18 small molecular drugs and biologics approved for treating various cancers along with corresponding companion diagnostic tests for specific mutated genes [Citation6].
Since approval of imatinib in 2001 for treating chronic myeloid leukemia (CML), approval for marketing of oncology products reached a peak period with 19 products approved in 2012 and 2015 each [Citation5]. Imatinib, a small molecular tyrosine kinase inhibitor (TKI), is the first drug approved for CML by targeting the fusion protein, BCR-ABL. Its approval marked the beginning of successful discovery and development of targeted therapy. Before long, TKI-resistant cancers due to mutations were found to be the leading cause of cancer relapse. More than 50 mutations were found in imatinib-resistant CML patients at various stages of progression [Citation7], leading to subsequent discovery and development of dasatinib, bosutinib, and nilotinib for imatinib-resistant CML [Citation5]. The findings of signal transduction via phosphorylation of kinases [Citation8] along with advances in phospho-proteomics and proteomics reveal the diversity of human kinome [Citation1] and a larger number of kinases as potential drug targets for treating diseases. Over the last 15 years, there have been approximately 30 TKIs approved for treating cancers. Interestingly, though TKIs share a shortcoming of lack of target specificity, their clinical use of treating cancers other than what is indicated in their approved labels has been shown to be ineffective [Citation9]. In brief, TKIs were developed to precisely target specific survival pathways of each individual type of cancer.
The horizon of combating cancers has recently been extended by the first gene therapy (CAR-T cell therapy) approved by Food and Drug Administration for treating B-cell acute lymphoblastic leukemia [Citation10]. The patient’s T cells are genetically engineered to express chimeric antigen receptors on their cell membrane to effectively eradicate cancer cells. The CAR-T therapy may well be the only chance that some patients have, and applications of such technology to treat diseases will likely increase.
2.2. Genetic disorders
Omics technologies have catapulted discovery and development of drug products to treat disorders with specific genetic mutations, most notably over the past five years. Muscular dystrophies (MDs) are a group of muscle disease associated with genetic mutations [Citation11]. In 2016, eteplirsen and nusinersen, two antisense oligonucleotide products, were approved for treating Duchene MD (DMD) and spinal MD, respectively [Citation5]. Eteplirsen, a 30-unit antisense oligonucleotide, increases muscular dystrophin levels in patients with the DMD mutation that is amenable to exon 51 skipping. Nusinersen is a modified antisense for treating patients with mutations leading to deficiency in survival motor neuron-2.
Cystic fibrosis (CF) is a fatal disease caused by inheritance of two defective copies from both parents carrying the CF gene encoding the cystic fibrosis transmembrane conductance regulator (CFTR). Two products have been approved for treating CF with ivacaftor approved in 2012 and lumacaftor/ivacaftor in 2015 [Citation5]. Each of them activates specific mutated CFTR to increase transport of chloride ion and normalizes water content in the airway epithelial mucus, thus improving lung function.
3. How do omics technologies promote precision-medicine-by-design?
Late-stage drug attrition is most costly. To increase the late-stage successes of developing drugs for complex diseases, one strategy is developing a right drug for right patients with the right disease. Omics data can be leveraged for discovery of prognostic and predictive efficacy biomarkers for patient stratification and design of clinical trials. With patients’ omics data as predictors and disease severity as a response variable, classification/feature extraction computations can uncover useful biomarkers. The PRECISESADS project [Citation12] funded by Innovative Medicines Initiative [Citation13] has been organized precisely for the goal of reclassifying systemic autoimmune diseases based on their molecular characteristics. The ever-increasing omics-centered, molecular profiling of patients’ tissue/fluid samples is changing disease classification. In autoimmune diseases, high variability is observed among patients with the same disease. Moreover, patients with the same clinical presentation can respond very differently to the same treatment. The en masse profiling of patients can characterize the molecular features that could set patients apart into subgroups, and may eventually lead to a revised disease classification. The outcome of such precision-medicine-by-design will be effective clinical trials in specific patient population that will respond to the drug treatment, and thus reduce late-stage drug attrition. Precision-medicine-by-design means engineering design and workflows are applied throughout the process of discovery and development of drugs to achieve precise and effective treatment in individual patients.
Apart from a right drug for right target(s) in right patients, minimizing a drug’s toxicity is equally important. Predictive models of drug safety can be constructed to reduce drug attrition by leveraging publicly available knowledge bases of clinical incidence of adverse reactions, genomics, protein–protein interactions, and big omics data [Citation14,Citation15].
4. Expert opinions
Omics technologies are an integral part of informed pharmaceutical R&D, and their role in R&D will continue to expand. There are still challenges to be addressed to fully utilize omics technologies, especially for treating complex diseases, such as neurological and autoimmune diseases. Just to name a few. How to establish omics data standards to reduce inter-laboratory variability and to increase confidence in distinctly classifying disease subtypes to aid the design of clinical trials? How to translationally bridge omics data in pathological conditions and clinical phenotypes of individual diseases? How to integrate multiple layers of omics information and phenotypic characterizations, including pathological biomarkers of a disease, pharmacodynamics responses to a drug treatment? How to link omics data to brain images or cognitive scores for neurological or psychological diseases? Computational and statistical methods will undoubtedly continue to play a role. Looking ahead, quantitative systems pharmacology may help bridge the gap. It is anticipated that the molecular networks described by omics data and the physiological/pathological networks presented by clinical tests/diagnosis will be quantitatively integrated to inform R&D.
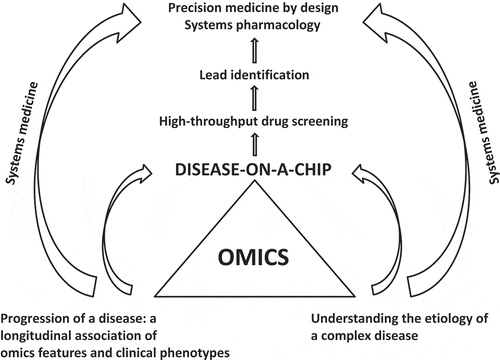
As biomedical technologies evolve, omics technologies need to be integrated with new innovations. The technologies of a disease-on-chip are on the horizon. A disease-on-chip is a novel living disease model that can be constructed to capture the pathological features of a disease throughout the course of its natural history, and to bridge omic profiles and clinical phenotypes. In conjunction with assays for transcriptomics, proteomics, microRNAs, and post-translational modification information, disease-on-chip technologies can uncover the molecular etiology of a disease, identify drug targets, and, most importantly, reveal the responses to a drug across the systems (efficacy and safety). Alzheimer’s disease is an example where a disease-on-chip can be instrumental. Several genetic mutations in the β-amyloid precursor protein gene have been identified in familial Alzheimer’s disease [Citation16]. Conceivably, use of genetic manipulation to induce specific mutations of cells and create specific disease-on-chip might not be far-fetched. In the near future, a 3D Alzheimer’s disease ‘brain-on-chip’ will increase our ability to determine the molecular networks associated with its etiology, and to discover a novel medical product to prevent its onset and/or arrest its progression.
As shown in , omics advances will be at the core of systems medicine-guided precision-medicine-by-design. Tens of thousands of variables (single-nucleotide polymorphisms, proteomes, transcriptomics, microRNAs, etc.) can be measured at different stages of a disease. Harvesting such information in a longitudinal manner, an association can be established between these omics features and corresponding phenotypes of a disease or corresponding responses to a drug treatment. In summary, application of omics technologies to ever-evolving biomedical innovations, such as disease-on-chip technologies, is the path forward for informed pharmaceutical R&D.
Figure 2. Omics technologies play a central role in the era of precision medicine-by-design now and upon availability of a disease-on-a-chip. Systems medicine/pharmacology and omics technologies will be applied together to accelerate the pipelines of drug discovery and development. Precision-medicine-by-design means the concept of engineering design and workflows are applied throughout the process of discovery and development of drugs to achieve precise targeting in individual patients. Considering interactions among systems in the hierarchy of human biology, systems medicine is the compass to achieve precision-medicine-by-design.
Declaration of Interest
IN Melas is an employee of UCB Celltech. The views expressed are those of the authors and do not necessarily represent the position of, nor imply endorsement from, the US Food and Drug Administration or the US government. The authors have no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript apart from those disclosed. Peer reviewers on this manuscript have no relevant financial or other relationships to disclose.
Additional information
Funding
References
- Manning G, Whyte DB, Martinez R, et al. The protein kinase complement of the human genome. Science. 2002 Dec 06;298(5600):1912–1934. PubMed PMID: 12471243. DOI:10.1126/science.1075762
- Simon GM, Niphakis MJ, Cravatt BF. Determining target engagement in living systems. Nat Chem Biol. 2013 Apr;9(4):200–205. PubMed PMID: 23508173; PubMed Central PMCID: PMCPMC4004587. DOI:10.1038/nchembio.1211
- Johnson CH, Ivanisevic J, Siuzdak G. Metabolomics: beyond biomarkers and towards mechanisms. Nat Rev Mol Cell Biol. 2016 Jul;17(7):451–459. PubMed PMID: 26979502. DOI:10.1038/nrm.2016.25
- Vucic EA, Thu KL, Robison K, et al. Translating cancer ‘omics’ to improved outcomes. Genome Res. 2012 Feb;22(2):188–195. PubMed PMID: 22301133; PubMed Central PMCID: PMCPMC3266027. DOI:10.1101/gr.124354.111
- Drugs@FDA: FDA Approved Drug Products. [ cited 2017 July]. Available from: https://wwwaccessdatafdagov/scripts/cder/daf/
- Food and Drug Administration. List of cleared or approved companion diagnostic devices (In Vitro and Imaging Tools. [ Cited 2017 October]. Available from: https://wwwfdagov/MedicalDevices/ProductsandMedicalProcedures/InVitroDiagnostics/ucm301431htm.
- Apperley JF. Part I: mechanisms of resistance to imatinib in chronic myeloid leukaemia. Lancet Oncol. 2007 Nov;8(11):1018–1029. PubMed PMID: 17976612. DOI:10.1016/S1470-2045(07)70342-X
- Hunter T. Signaling–2000 and beyond. Cell. 2000 Jan 07;100(1):113–127. PubMed PMID: 10647936.
- Le TC, Delord JP, Goncalves A, et al. Molecularly targeted therapy based on tumour molecular profiling versus conventional therapy for advanced cancer (SHIVA): a multicentre, open-label, proof-of-concept, randomised, controlled phase 2 trial. Lancet Oncol. 2015 Oct;16(13):1324–1334. PubMed PMID: 26342236. DOI:10.1016/S1470-2045(15)00188-6
- Food and Drug Administration. FDA approval brings first gene theerapy to the United States. [ cited 2017 August 30]. Available from: https://wwwfdagov/NewsEvents/Newsroom/PressAnnouncements/ucm574058htm
- Shieh PB. Muscular dystrophies and other genetic myopathies. Neurol Clin. 2013 Nov;31(4):1009–1029. PubMed PMID: 24176421. DOI:10.1016/j.ncl.2013.04.004
- Hofmann-Apitius M, Alarcon-Riquelme ME, Chamberlain C, et al. Towards the taxonomy of human disease. Nat Rev Drug Discov. 2015 Feb;14(2):75–76. PubMed PMID: 25633780.
- PRECISESADS. [ cited 2017 Oct 17]. Available from: http://www.precisesads.eu.
- Melas IN, Sakellaropoulos T, Iorio F, et al. Identification of drug-specific pathways based on gene expression data: application to drug induced lung injury. Integr Biol (Camb). 2015 Aug;7(8):904–920. PubMed PMID: 25932872. DOI:10.1039/c4ib00294f
- Hur J, Guo AY, Loh WY, et al. Integrated systems pharmacology analysis of clinical drug-induced peripheral neuropathy. CPT Pharmacometrics Syst Pharmacol. 2014 May 14;3:e114. PubMed PMID: 24827872; PubMed Central PMCID: PMCPMC4051377. DOI:10.1038/psp.2014.11
- Goate A, Hardy J. Twenty years of Alzheimer’s disease-causing mutations. J Neurochem. 2012 Jan;120(Suppl 1):3–8. PubMed PMID: 22122678. DOI:10.1111/j.1471-4159.2011.07575.x
