1. Introduction
The goal of data integration in drug discovery is to enable cross-dataset analyses that strengthen or fill gaps in our knowledge of biological, pharmacological, or clinical phenomena. For example, this could be done by testing for consistent statistical associations among similar datasets, as in a meta-analysis, or by chaining together statistical associations across datasets to discover novel associations, as in an enrichment analysis. The potential for data integration to yield valuable insights is high given the vast quantity and variety of data in our field. Drug discovery and development programs generate large amounts of data on genes/proteins, compounds, genomes, variants, patients, animal models, interventions, and more. But these data are heterogeneous, sparse, and high-dimensional, originating from high-throughput sequencing, mass spectrometry, flow cytometry, high-content imaging, high-throughput screening, functional assays, clinical records, pharmacy records, etc. The heterogeneity, sparsity, and high-dimensionality of the data, as well as historically little consideration for data reuse and integration at the time data were generated, make data integration in our field a formidable challenge [Citation1].
Data integration projects have made great progress in the face of these difficulties. There are excellent data integration resources that primarily focus on one data type, such as Gene Expression Omnibus (GEO), UniProt, and ZINC15 [Citation2–Citation4]. There are excellent portals like The European Bioinformatics Institute (EBI) and The National Center for Biotechnology Information (NCBI) that offer a multitude of these resources [Citation5,Citation6]. It has been encouraging to see a new class of resources that integrate data across multiple data types, such as Open Targets, Open PHACTS and Pharos [Citation7–Citation12]. The solution adopted is often to represent each data type in its own column with the ability to query across data types. These may be termed as tabular- or spreadsheet-based resources, which offer ease of use.
Given the great progress in collecting, organizing, and summarizing data, as well as building tools for querying data, our field may be ready for a new paradigm for data integration. We postulate that Artificial Intelligence based representation learning solutions are poised to revolutionize data integration for drug discovery as they offer a flexible, dimensionless way of integrating and converting between disparate data, as we will describe below.
2. Artificial Intelligence and autoencoders
Artificial Intelligence (AI) is a branch of computer science that aims to equip machines with human-like intelligence and the ability to learn from ever-changing environments to successfully achieve their goals. For instance, artificial neural networks (ANN) are mathematical models for data processing, born out of AI, designed to mimic the large array of neurons in the brain that enable humans to parse sensory inputs, learn, and make decisions. A basic ANN consists of an input layer, the hidden layer that is a transformed representation of the input data, and the output layer. Recent advances in computing power and algorithmic innovations have led to the development of deep neural networks from ANN’s, which use multiple hidden layers to learn a hierarchy of increasingly abstract but hopefully more meaningful representations of the data [Citation13]. Deep Learning with these deep neural networks has delivered impressive achievements in the last decade with self-driving cars and language translation [Citation13]. A key concept that has become popular in Deep Learning is that of an Autoencoder.
An Autoencoder is a model that transforms data in some way (encoding) and then attempts to reconstruct the data (decoding) [Citation14]. The output of the encoder (and input to the decoder) is typically a vector of latent variables known as the latent representation, embedding, or code. Autoencoders are typically used for compression, representation learning, and/or visualization of high-dimensional data. Compression can be a preprocessing step or auxiliary task for supervised learning, where the aim is to eliminate noise, collapse correlated variables, and possibly incorporate unlabeled data, making it easier to learn the distribution of the data and hopefully improve generalization [Citation14]. Principal Components Analysis (PCA) is likely the best-known and simplest Autoencoder. In PCA, the encoder and decoder functions are linear transformations, and the parameters of the transformation matrices can be determined analytically [Citation15].
Recently, deep neural network Autoencoders such as the Variational Autoencoder have become popular [Citation14,Citation16]. In these models, the encoder and decoder functions are deep neural networks, and their parameters are determined by minimizing the reconstruction error on a set of training data. Deep neural networks allow a hierarchy of increasingly abstract and ideally more meaningful features of the input data to be learned, such as patterns of input variables corresponding to pathways, regulatory motifs, or binding domains [Citation16]. Deep neural networks are also extremely flexible. Their architectures can be customized to process data in a way that takes advantage of intrinsic characteristics of a data type, such as the regular 2-D arrangement of pixels in an image. Many data type specific architectures already exist that can be plugged in as encoders and decoders. The greater flexibility and expressive power of deep neural networks over other machine learning models comes at the cost of increased model engineering time, increased computational demand, and the need for large amounts of training data.
Large amounts of diverse but sparse data is produced from bespoke projects in the process of drug discovery. Data has historically been integrated by specific types, and only recently across multiple data types (Open Targets, Pharos). Artificial Intelligence (Autoencoders, in particular) can provide compact encodings that span data types. This promises a dimensionless way of integrating drug discovery data, ushering in the next generation of data integration that can use measurements on the system to infer its hidden state. This hidden state discovery will enable machine learning based discovery of predictive biomarkers and allow us to create virtual patient cohorts for trials, thus providing a data driven approach for decision making in drug discovery.
3. Expert opinion
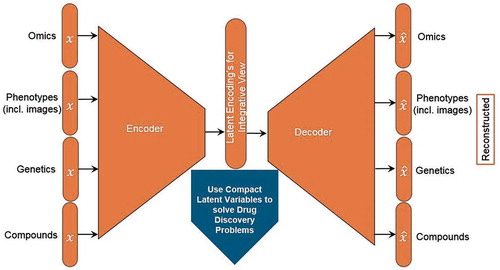
A useful way to interpret an Autoencoder’s latent variables is to consider them master dials that completely define the ‘state’ of a system, entity, or object. From this point-of-view, if we know a system’s latent variables, then we know its state, and we should be able to predict almost everything that we could possibly observe about that system. For example, we could conceive of patients as having a set of latent variables that define their health state, and clinical records, prescriptions, and lab results would all be observations determined by that health state. This point-of-view leads to an approach to data integration that is conceptually straightforward, general purpose, and data-driven. If different types of data about a system are simply different types of observations of the same latent state, then we should be able to use an Autoencoder (or set of Autoencoders) to infer a single set of latent variables from which we can explain all our observations (). More importantly, the encoder and decoder functions would provide mappings between the different types of data, which we could use for cross-dataset predictive analytics.
Figure 1. Autoencoders: a natural framework for data integration that may learn the hidden state of the biological system.
Autoencoders consist of encoder and decoder functions that can compress high-dimensional data into a latent representation and then reconstruct the output, bringing light to hidden states that may exist in the data from disparate sources. Drug discovery problems include target identification, biomarker selection, compound prioritization, and patient stratification.
As an example, imagine we have oncology clinical trial data on 100 patients with 30 responders and 70 non-responders. We would like to find a response biomarker. For each patient at baseline, we have anthropomorphic and lifestyle traits, clinical data, lab values, blood cytokine levels, tissue mRNA expression, immunohistochemistry, and biopsy images. We do not have complete data for any single patient. Given the small sample size and high dimensionality of the data, it is unlikely that we will be able to find a response predictor using conventional statistical or machine learning approaches [Citation17,Citation18]. This raises a troubling issue, having a large amount of data on these patients does not seem to help us in this situation. Rather, it seems to make the problem more difficult by increasing the probability of false discovery. This is strange; why does having more data seem to provide no benefit?
We argue the latent state approach to data integration provides a way to unlock the benefits of high-dimensional, multi-modal data, with Autoencoders being a key enabling tool. If we knew our patients’ latent states, we should be able to predict their response. For the response prediction task, the Autoencoder provides a reduced set of candidate predictors (the latent variables) that also explain the other patient observations. We expect these candidate predictors to generalize better than those found by a purely supervised approach, which need not contain any coherent information about the patient.
Several challenges must be resolved for Autoencoders to be used successfully for data integration. For example, a significant amount of neural network architecture engineering will be required, with different data types needing different encoder/decoder architectures (e.g. convolutional neural networks for images, recurrent neural networks for sequences, and graph convolutional neural networks for compounds). Also, experimentation with the training objective will be required. Reconstruction error may need to be computed differently for different data types (e.g. squared error loss for normally distributed data and cross entropy loss for binary data), data types may need weights, with lower quality data receiving less weight, and data types may need to be randomly masked at the input during training to force the model to produce reconstructions given only partial observations. Also, validation will depend on use case and each use case may require fine-tuning of the model, e.g. training an Autoencoder to integrate patient data and then fine-tuning the encoder to predict response to treatment. Finally, a strategy to collect the same types of data across clinical trials or across preclinical projects would be required to have a sufficient number of samples to fit the models. Autoencoders could be used to integrate data across patients, targets, compounds, diseases, and more. Autoencoders could also be used to integrate data of only one of the aforementioned data types from multiple resources accounting for batch-effects. Some early examples of using Autoencoders for data integration have emerged [Citation19–Citation22], and we anticipate research on this topic will grow. Recent research has shown that Autoencoders can be used to select features and reproduce virtual patient images, thus producing virtual cohorts [Citation23]. The emergence of affordable data generation methods has led to huge amounts of labeled omic, genetic, compound and clinical data. The large-scale data generation coinciding with advances in computing hardware and deep learning, suggests that this is the right time for integration across drug discovery data.
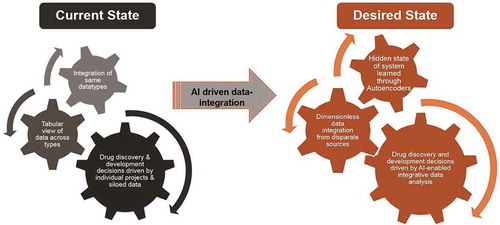
Data integration has promised to revolutionize drug discovery for the past two decades. Though resources around single data types continue to flourish, advances have come from assembling disparate pieces of data together in large tables and databases, with convenient access provided through web interfaces and APIs [Citation10]. Representation Learning developments in Artificial Intelligence, such as Autoencoders, will provide the new paradigm for data integration (). For example, the ability of an Autoencoder to learn the latent master dials to completely define the state of the system may provide a solution to the problem of combining very different aspects and measurements of a system in a dimensionless way.
Figure 2. The future of drug discovery with AI-driven data integration.
The current state of drug discovery has been achieved by integrating data of single types from individual projects and querying large databases that were built to collate data from disparate sources. AI and Autoencoders can learn any hidden states present among high-dimensional, sparse data from disparate source, thus building a platform for data-driven decision making in drug discovery.
This will enable us to create virtual clinical subject cohorts and simulate their response in silico to experimental therapeutic interventions prior to any human clinical intervention study [Citation23]. It could also help stratify patients based on these master dials that will incorporate anthropomorphic, genetics, genomics and drug treatment data, leading to better biomarkers and diagnoses. Integrating data across the pipeline using Autoencoders will provide a data driven approach, which would mark the new paradigm for decision making throughout the drug discovery and development pipeline.
Declaration of Interest
All authors are employees of GlaxoSmithKline. The authors have no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript apart from those disclosed.
Reviewer Disclosures
Peer reviewers on this manuscript have no relevant financial or other relationships to disclose.
Additional information
Funding
References
- Searls DB. Data integration: challenges for drug discovery. Nat Rev Drug Discov. 2005 Jan 01;4:45. online.
- Barrett T, Wilhite SE, Ledoux P, et al. NCBI GEO: archive for functional genomics data sets—update. Nucleic Acids Res. 2013;41(D1):D991–D95.
- The UniProt Consortium. UniProt: the universal protein knowledgebase. Nucleic Acids Res. 2017;45(D1):D158–D69.
- Sterling T, Irwin JJ. ZINC 15 – ligand discovery for everyone. J Chem Inf Model. 2015;55(11):2324–2337.
- European bioinformatics institute. [cited 2019]. Available from: https://www.ebi.ac.uk/
- National center for biotechnology information (NCBI). Bethesda (MD): National Library of Medicine (US), National Center for Biotechnology Information. 1988 [cited 2019]. Available from: https://www.ncbi.nlm.nih.gov/
- Nguyen D-T, Mathias S, Bologa C, et al. Pharos: collating protein information to shed light on the druggable genome. Nucleic Acids Res. 2017;45(Databaseissue):D995–D1002.
- Koscielny G, An P, Carvalho-Silva D, et al. Open targets: a platform for therapeutic target identification and validation. Nucleic Acids Res. 2017;45(Databaseissue):D985–D94.
- Khaladkar M, Koscielny G, Hasan S, et al. Uncovering novel repositioning opportunities using the open targets platform. Drug Discov Today. 2017;22(12):1800–1807.
- Reisdorf WC, Chhugani N, Sanseau P, et al. Harnessing public domain data to discover and validate therapeutic targets. Expert Opin Drug Discov. 2017;12(7):687–693.
- Williams AJ, Harland L, Groth P, et al. Open PHACTS: semantic interoperability for drug discovery. Drug Discov Today. 2012 Nov;17(21–22):1188–1198.
- Ajami A, Buvailo A. 36 web resources for target hunting in drug discovery; 2017 [ cited 2019]. Available from: https://www.biopharmatrend.com/post/45-27-web-resources-for-target-hunting-in-drug-discovery/
- LeCun Y, Bengio Y, Hinton G. Deep learning. Nature. 2015;521:436.
- Goodfellow I, Bengio Y, Courville A. Deep learning; 2016 [cited 2019]. Available from: http://www.deeplearningbook.org/
- Trapnell C. Defining cell types and states with single-cell genomics. Genome Res. 2015;25(10):1491–1498.
- Chen H, Engkvist O, Wang Y, et al. The rise of deep learning in drug discovery. Drug Discov Today. 2018;23(6):1241–1250.
- Rubingh CM, Bijlsma S, Derks EP, et al. Assessing the performance of statistical validation tools for megavariate metabolomics data. Metabolomics. 2006;2(2):53–61.
- Christin C, Hoefsloot HC, Smilde AK, et al. A critical assessment of feature selection methods for biomarker discovery in clinical proteomics. Mol Cell Proteomics. 2013 Jan;12(1):263–276.
- Gligorijevic V, Barot M, Bonneau R. deepNF: deep network fusion for protein function prediction. Bioinformatics. 2018 Jun 1.
- Rohart F, Gautier B, Singh A, et al. mixOmics: an R package for ‘omics feature selection and multiple data integration. PLoS Comput Biol. 2017 Nov;13(11):e1005752.
- Eser U, Churchman LS. FIDDLE: an integrative deep learning framework for functional genomic data inference. bioRxiv. 2016.
- Ngiam J, Khosla A, Kim M, et al. Multimodal deep learning. Proceedings of the 28 th International Conference on Machine Learning; Bellevue, Washington, USA. 2011.
- Chen J, Xie Y, Wang K, et al. Generative invertible networks (GIN): pathophysiology-interpretable feature mapping and virtual patient generation. CoRR. 2018. abs/1808.04495.
