Abstract
In this study, we conducted a genome-wide association study (GWAS) for five milk production traits in the Valle del Belice sheep. Repeated measurements for milk yield (MY), fat percentage and yield (F% and FY) and protein percentage and yield (P% and PY) on 481 ewes, were available for the analysis. The animals were genotyped using the Illumina Ovine 50k BeadChip. Weighted deregressed breeding values (DEBVw) were used as phenotypes for GWAS analysis. A total of 23 genome-wide significant SNPs were identified: 3 associated with MY, 9 with FY, and 11 with P%. Several SNPs mapped within known candidate genes or previously reported QTL for milk production traits in livestock species. Additional interesting markers were identified on OAR3 for FY and P%. These SNPs supported some previous findings and also added new information useful to understand the genetic mechanisms underlying the milk production and quality traits in dairy sheep.
A total of 23 significant SNPs were detected.
Several SNPs mapped within known candidate genes or previously reported QTL for milk production traits.
These results could provide information to understand the genetic architecture of milk production traits in dairy sheep.
Highlights
Introduction
Dairy sheep represent an important resource for the development of the economies of hill and mountain areas, in which other economic activities are difficult to develop (Scintu and Piredda Citation2007). The Valle del Belice is the main sheep breed reared in Sicily and is used for the production of traditional raw milk cheeses, Pecorino Siciliano and PDO 'Vastedda della Valle del Belice'. Therefore, it appears obvious that milk production traits and their genetic improvement are of great interest in this breed.
Genome-wide association studies (GWAS) represent a powerful tool to identify causal genes associated with important traits in livestock, especially when dealing with complex traits such as milk production traits. GWAS based on high throughput single nucleotide polymorphism (SNP) genotyping technologies opened a broad avenue for exploring candidate genes associated with these traits, making the identification of these genes crucial to understand the genetic architecture of milk production and composition (Goddard and Hayes Citation2009; Wiener et al. Citation2011; Liu et al. Citation2020). GWAS studies in sheep are more recent and less widespread than those of other animal species (Johnston et al. Citation2011; Zhang et al. Citation2013; Mastrangelo et al. Citation2018 ; Sutera, Moscarelli, et al. Citation2021). In particular, lactation performance has been the focal point of the genetic improvement of dairy sheep over the past decades (García-Gámez et al. Citation2012; Di Gerlando et al. Citation2019; Sutera et al. Citation2019; Usai et al. Citation2019; Li et al. Citation2020; Sutera, Tolone, et al. Citation2021). However, so far, little overall consensus has emerged from these studies.
In GWAS of domesticated animals, often, the researchers may have available raw phenotypes of animals and also their estimated breeding values (EBVs) from pedigree-based analyses of historical data. Some animals may have genotypes and EBVs based on information from their relatives, but no direct phenotypes. Although EBVs were used as dependent variables in GWAS (Johnston et al. Citation2011; Becker et al. Citation2013), this approach gave a high false positive rate (Ekine et al. Citation2014). As an alternative, the EBVs can be ‘deregressed’ (DEBVs) and to account for the heterogeneous variance due to differences in breeding value accuracy of individual, the weighting factor and deregressed weighted estimated breeding values (DEBVw) were also proposed (Garrick et al. Citation2009).
The aim of this study was to conduct a GWAS to identify loci associated with milk production traits (i.e. milk yield (MY), fat yield (FY), fat percentage (F%), protein yield (PY) and protein percentage (P%)) in the Valle del Belice dairy sheep using the DEBVw as phenotypes.
Materials and methods
Ethics statement
SNP genotype data were obtained from a previous study (Sutera et al. Citation2019). Therefore, no specific ethical approval was required.
Samples, genotyping data and quality control
Phenotypic data from 481 ewes of Valle del Belice breed were collected by the University of Palermo in four flocks. More details on both animals and phenotypes are provided in a previous study conducted by the same authors (Sutera, Tolone, et al. Citation2021).
Animals were genotyped with the OvineSNP50 BeadChip (Illumina Inc, San Diego, CA, USA). The quality control was performed using GenABEL package v3.4 (Aulchenko et al. Citation2007) in R environment (http://www.r-project.org). Only SNPs located on autosomes were considered. Animals and markers that fulfilled the following criteria were kept in the analysis: (i) call rate per individuals and per SNPs >95%, (ii) minor allele frequency >2% and (iii) no extreme deviation from Hardy-Weinberg equilibrium (p < .001). A total of 37,228 SNPs and 469 individuals were retained for further analyses. The phenotypic data from 469 ewes included 5,328 test-day records for MY, FY, F%, PY and P%.
Genome-wide association analyses
The EBVs were estimated fitting the same effects reported in Sutera, Tolone, et al. (2021), using the pedigree rather than the SNPs to build the relationship matrix. In addition, EBVs were first deregressed (DEBVs) according to Garrick et al. (Citation2009), and then weighted to obtain DEBVw, using ASReml v3.0 (Gilmour et al. Citation2009), fitting the genomic relationship matrix (GRM).
The association analysis was carried out using GenABEL package (Aulchenko et al. Citation2007) and DEBVw as phenotype. No polygenic step to account for relatedness among the individuals was needed, as this was already accounted when estimating the DEBVw (i.e. by fitting the GRM). Association was therefore tested using a qtscore function (Aulchenko et al. Citation2007). After Bonferroni correction, significant thresholds were p < 1.34 × 10−6 for genome-wide (p < 0.05) and p < 2.69 × 10−5 for suggestive (i.e. one false positive for genome scan) levels, corresponding to -log10(P) of 5.87 and 4.57, respectively. Quantile-quantile (Q–Q) plots were used to compare the observed and expected distribution of p-values under the null hypothesis of no association. Population substructure was explored using multidimensional scaling (MDS) in order to verify the genetic homogeneity of the sample before analysis.
Annotation
To investigate if the significant SNPs detected in this study laid within genes or previously reported quantitative trait loci (QTL) for milk production traits, we searched them in NCBI Genome Data Viewer for Ovis aries v4.0 genome assembly (https://www.ncbi.nlm.nih.gov/genome/gdv/browser/?context=genome&acc=GCF_000298735.2) and in the SheepQTLdb (https://www.animalgenome.org/cgi-bin/QTLdb/OA/index).
Results and discussion
Previous GWAS have been conducted for milk production traits in the Valle del Belice breed, using SNP data and repeated measures (Sutera et al. Citation2019; Sutera, Tolone, et al. 2021), and Copy Number Variants and DEBV (Di Gerlando et al. Citation2019). Here, we report the results from a GWAS for five most commonly evaluated milk production traits using the DEBVw. The choice to use DEBVw as response variable in our analysis was guided by the fact that GenABEL cannot handle multiple records (i.e. it only allows to fit a polygenic effect as random, making it not possible to account for the permanent environmental effects due to the repeated measurements). It is not uncommon to use EBVs as dependent variables in GWAS (Johnston et al. Citation2011; Becker et al. Citation2013). However, it was demonstrated that this approach has a high false positive rate (Ekine et al. Citation2014), whereas the use of DEBVs and DEBVw as dependent variables can improve the power of GWAS (Sell-Kubiak et al. Citation2015; Sevillano et al. Citation2015).
A total of 23 genome-wide significant SNPs were detected, with 43 reaching the suggestive significance threshold (by the definition we used), across all five traits (Figure and for the Q–Q plots). As SNP rs406975522 is associated with both FY and PY, the total number of distinct identified SNPs was 65. The details of the identified SNPs, including the trait they are associated to, positions on Ovis aries v4.0 genome assembly, p-values and the nearest known genes they laid within, are given in Table .
Figure 1. Genome-wide plots of -log10(p-values) for association of SNPs with milk yield (MY), fat percentage (F%), fat yield (FY), protein percentage (P%) and protein yield (PY). Genome-wide and suggestive thresholds are also shown.
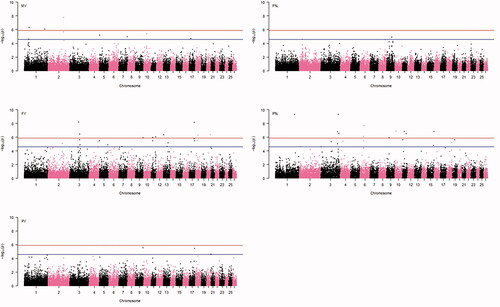
Table 1. SNPs significantly associated with milk production traits at genome-wide (p < 1.34 × 10−6) and suggestive (p < 2.69 × 10−5) thresholds.
Out of 10 SNPs associated with MY, four were mapped within known genes or previously reported QTL for the milk yield or other milk production traits (Table ). In particular, SNPs located between 177.26 and 178.47 Mb on OAR2 (rs419432879 and rs429723758, in bold in Table ), laid into the region containing a QTL (ID = 13992) for milk and lactose yield in sheep (Raadsma et al. Citation2009). The SNP rs419432879 mapped within the NCKAP5 (Nck—associated protein 5) gene, which has been associated with F% in Holstein cattle (Jiang et al. Citation2019). The SNP rs160014503 on OAR5 was located in the intronic region of SLC27A1 (Solute Carrier Family 27 member 1) gene, which shows enzymatic activity similar to acyl-CoA synthetase (Hall et al. Citation2003). In cattle, SLC27A1 was proposed as candidate gene for milk traits (Kulig et al. Citation2013; Nafikov et al. Citation2013) and polymorphisms at this gene have potential effects on milk yield trait (Lv et al. Citation2011). Finally, the SNP rs424648601 (located at 19.38 Mb on OAR17) was within a QTL (ID = 57756) previously identified by García-Gámez et al. (Citation2013) for MY in the Churra sheep breed, using two different approaches.
Only one SNP was associated with F% and it is located on OAR9, within the EXT1 (Exostosin 1) gene. On the other hand, 30 SNPs (located on 14 different chromosomes) were found associated with FY; some of these markers are associated to this trait in other sheep breeds (e.g. SNP rs403120738 on OAR2 (García-Gámez et al. Citation2013); SNP rs406975522 on OAR9 (Jonas et al. Citation2011)). The highest number of significant SNPs was located between 97.58 and 120.23 Mb on OAR3. Sutera, Tolone, et al. (2021) identified significant genomic regions on OAR 3 for F% and P%; however, none of our SNPs laid into this region. Several SNPs associated with FY mapped within known genes. The SNP rs423430191 on OAR5 was located in the intronic region of MAPK9 (Mitogen-Activated Protein Kinase 9), that is involved in immune system pathway in sheep (Yang et al. Citation2015) and is one of the key signalling nodes in mammary gland development in humans (Whyte et al. Citation2009). Zhou et al. (Citation2019) examining the transcriptome and proteome profiles of liver tissue samples from Chinese Holstein cows, identified MAPK9 as candidate gene that regulates milk fat synthesis, transport, and metabolism. The SNP rs407468867 on OAR11 mapped within the RNF157 (Ring Finger Protein 157), a candidate gene involved in milk production traits in sheep (Saridaki et al. Citation2019 ). The SNP rs424842019 on OAR18 was located within the ADAMTSL3 (A Disintegrin-Like And Metalloprotease Domain With Thrombospondin Type I Motifs-Like) gene. Mateescu and Thonney (Citation2010) reported two QTLs (ID = 14149 and ID = 14150) for MY in the same region.
For the protein traits (i.e. P% and PY), 22 and 3 SNPs were identified, respectively. Only the significant SNP for P% (rs42391986) on OAR2 mapped within a QTL (ID = 57738) for the considered trait (García-Gámez et al. Citation2013). The SNP rs424276469 on OAR3 was located within MED27 (Mediator Complex Subunit 27), a candidate gene for milk yield in water Buffalo (Du et al. Citation2019). Significant SNPs were also found between 41.70 and 46.45 Mb on OAR6. The OAR6 has received wide attention because harbours, among others, the well-known casein clusters, generally accepted as major genes affecting milk production traits (Moioli et al. Citation2007). A significant marker identified in this study on this chromosome mapped within SEL1L3 gene, which has been reported as candidate gene for milk protein composition and cheese-making properties (Sanchez et al. Citation2019). Of the three SNPs associated with PY, the SNP rs406975522 on OAR9 (reported as significant also for FY) mapped within MMP16 (Matrix Metallopeptidase 16) gene. This SNP could be an example of cross-phenotype association, by which variants are associated with multiple traits that are often relevant to pleiotropy (Solovieff et al. Citation2013), although this hypothesis will have to be confirmed by further statistical analysis.
It is worth to highlight that no common regions were identified when comparing these results with those previously obtained in other GWAS on this dataset (Sutera et al. Citation2019; Sutera, Tolone, et al. Citation2021), in which a different definition of the traits and different methodologies were used. This might be partially due to the different approaches used and to the small sample size, which can be a possible cause of power reduction to detect significant regions. On the other hand, it might also be a confirmation of the complex nature of milk production traits, with many variants with small effect involved. We are therefore hypothesising that using different approaches and definition of the traits could be considered as complementary approaches, as they allow identifying different variants worth exploring.
Conclusion
A total of 23 genome-wide significant SNPs associated with milk production traits were identified in this study, with several SNPs mapped within known candidate genes or previously reported QTL. These SNPs supported some previous findings and also added new information useful to understand the genetic mechanisms underlying the milk production and quality traits in dairy sheep. As no common regions were identified when comparing these results with those previously obtained in other GWAS on this dataset, we are hypothesising that using different approaches and definition of the traits could be considered as complementary approaches, as they allow identifying different variants.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The data have been submitted to the repository at: https://ww.animalgenome.org/repository/pub/UPIT2018.0801/
Additional information
Funding
References
- Aulchenko YS, de Koning DJ, Haley C. 2007. Genomewide rapid association using mixed model and regression: a fast and simple method for genomewide pedigree-based quantitative trait loci association analysis. Genetics. 177(1):577–585.
- Becker DK, Wimmers H, Luther A, Hofer T, Leeb A. 2013. A genome-wide association study to detect QTL for commercially important traits in Swiss Large White boars. PLOS One. 8(2):e55951.
- Di Gerlando R, Sutera AM, Mastrangelo S, Tolone M, Portolano B, Sottile G, Bagnato A, Strillacci MP, Sardina MT. 2019. Genome-wide association study between CNVs and milk production traits in Valle del Belice sheep. PLOS One. 14(4):e0215204.
- Du C, Deng T, Zhou Y, Ye T, Zhou Z, Zhang S, Shao B, Wei P, Sun H, Khan FA, et al. 2019. Systematic analyses for candidate genes of milk production traits in water buffalo (Bubalus Bubalis). Anim Genet. 50(3):207–216.
- Ekine CC, Rowe SJ, Bishop SC, de Koning DJ. 2014. Why breeding values estimated using familial data should not be used for genome-wide association studies. G3. 4(2):341–347.
- García-Gámez E, Gutiérrez-Gil B, Sahana G, Sánchez JP, Bayón Y, Arranz JJ. 2012. GWA analysis for milk production traits in dairy sheep and genetic support for a QTN influencing milk protein percentage in the LALBA gene. PLoS One. 7(10):e47782.
- García-Gámez E, Gutiérrez-Gil B, Suarez-Vega A, de la Fuente LF, Arranz JJ. 2013. Identification of quantitative trait loci underlying milk traits in Spanish dairy sheep using linkage plus combined linkage disequilibrium and linkage analysis approaches. J Dairy Sci. 96(9):6059–6069.
- Garrick DJ, Taylor JF, Fernando RL. 2009. Deregressing estimated breeding values and weighting information for genomic regression analyses. Genet Sel Evol. 41:55.
- Gilmour AR, Gogel BJ, Cullis BR, Thompson R. 2009. ASReml user guide release 3.0VSN International Ltd, Hempstead, HP1 1ES UK.
- Goddard ME, Hayes BJ. 2009. Mapping genes for complex traits in domestic animals and their use in breeding programmes. Nat Rev Genet. 10(6):381–391.
- Hall AM, Smith AJ, Bernlohr DA. 2003. Characterization of the Acyl-CoA synthetase activity of purified murine fatty acid transport protein 1. J Biol Chem. 278(44):43008–43013.
- Jiang J, Ma L, Prakapenka D, VanRaden PM, Cole JB, Da Y. 2019. A large-scale genome-wide association study in US Holstein cattle. Front Genet. 10:412.
- Johnston SE, McEwan JC, Pickering NK, Kijas JW, Beraldi D, Pilkington JG, Pemberton JM, Slate J. 2011. Genome-wide association mapping identifies the genetic basis of discrete and quantitative variation in sexual weaponry in a wild sheep population. Mol Ecol. 20(12):2555–2566.
- Jonas E, Thomson PC, Hall EJ, McGill D, Lam MK, Raadsma HW. 2011. Mapping quantitative trait loci (QTL) in sheep. IV. Analysis of lactation persistency and extended lactation traits in sheep. Genet Sel Evol. 43:1–10.
- Kulig H, Kowalewska-Łucza KŻ, Żukowski K, Kunicka M, Kruszyński W. 2013. SLC27A1 SNPs in relation to breeding value of milk production traits in Polish Holstein-Friesian cows. Anim Sci Pap Rep. 31:273–279.
- Li H, Wu XL, Tait RG, Bauck S, Thomas DL, Murphy TW, Rosa GJM. 2020. Genome‐wide association study of milk production traits in a crossbred dairy sheep population using three statistical models. Anim Genet. 51(4):624–628.
- Liu L, Zhou J, Chen CJ, Zhang J, Wen W, Tian J, Zhang Z, Gu Y. 2020. Gwas-based identification of new loci for milk yield, fat, and protein in Holstein cattle. Animals. 10(11):2048.
- Lv Y, Wei C, Zhang L, Lu G, Liu K, Du L. 2011. Association between polymorphisms in the SLC27A1 gene and milk produc-tion traits in Chinese Holstein cattle. Anim. Biotechnol. 22(1):1–6.
- Mastrangelo S, Sottile G, Sutera AM, Di Gerlando R, Tolone M, Moscarelli A, Sardina MT, Portolano B. 2018. Genome-wide association study reveals the locus responsible for microtia in Valle del Belice sheep breed. Anim Genet. 49(6):636–640.
- Mateescu RG, Thonney ML. 2010. Genetic mapping of quantitative trait loci for milk production in sheep. Anim Genet. 41(5):460–466.
- Moioli B, D’Andrea M, Pilla F. 2007. Candidate genes affecting sheep and goat milk quality. Small Rumin Res. 68(1–2):179–192.
- Nafikov RA, Schoonmaker JP, Korn KT, Noack K, Garrick DJ, Koehler K, Minick-Bormann J, Reecy JM, Spur-Lock D, Beitz DC. 2013. Association of polymorphisms in solute carrier family 27, isoform A6 (SLC27A6) and fatty acid-binding protein-3 and fatty acid-binding protein-4 (FABP3 and FABP4) with fatty acid composition of bovine milk. J Dairy Sci. 96(9):6007–6021.
- Raadsma HW, Jonas E, McGill D, Hobbs M, Lam Thomson PC. 2009. Mapping quantitative trait loci (QTL) in sheep. II. Meta-assembly and identification of novel QTL for milk production traits in sheep. Genet Sel Evol. 41:45.
- Sanchez M-P, Ramayo-Caldas Y, Wolf V, Laithier C, El Jabri M, Michenet A, Boussaha M, Taussat S, Fritz S, Delacroix-Buchet A, et al. 2019. Sequence-based GWAS, network and pathway analyses reveal genes co-associated with milk cheese-making properties and milk composition in Montbéliarde cows. Genet Sel Evol. 51(1):34.
- Saridaki A, Antonakos G, Hager-Theodorides AL, Zoidis E, Tsiamis G, Bourtzis K, Kominakis A. 2019. Combined haplotype blocks regression and multi-locus mixed model analysis reveals novel candidate genes associated with milk traits in dairy sheep. Livest Sci. 220:8–16.
- Scintu MF, Piredda G. 2007. Typicity and biodiversity of goat and sheep milk products. Small Rumin Res. 68(1–2):221–231.
- Sell-Kubiak E, Duijvesteijn N, Lopes MS, Janss LLG, Knol EF, Bijma P, Mulder HA. 2015. Genome-wide association study reveals novel loci for litter size and its variability in a large white pig population. BMC Genomics. 16:1049.
- Sevillano CA, Lopes MS, Harlizius B, Hanenberg EH, Knol EF, Bastiaansen JVM. 2015. Genome-wide association study using deregressed breeding values for cryptorchidism and scrotal/inguinal hernia in two pig lines. Genet Sel Evol. 47:18.
- Solovieff N, Cotsapas C, Lee PH, Purcell SM, Smoller JW. 2013. Pleiotropy in complex traits: challenges and strategies. Nat Rev Genet. 14(7):483–495.
- Sutera AM, Riggio V, Mastrangelo S, Di Gerlando R, Sardina MT, Pong‐Wong R, Tolone M, Portolano B. 2019. Genome-wide association studies for milk production traits in Valle del Belice sheep using repeated measures. Anim Genet. 50(3):311–314.
- Sutera AM, Moscarelli A, Mastrangelo S, Sardina MT, Di Gerlando R, Portolano B, Tolone M. 2021. Genome-wide association study identifies new candidate markers for somatic cells score in a local dairy sheep. Front Genet. 12:643531.
- Sutera AM, Tolone M, Mastrangelo S, Di Gerlando R, Sardina MT, Portolano B, Pong-Wong R, Riggio V. 2021. Detection of genomic regions underlying milk production traits in Valle del Belice dairy sheep using regional heritability mapping. J Anim Breed Genet.0:1–10
- Usai MG, Casu S, Sechi T, Salaris SL, Miari S, Sechi S, Carta P, Carta A. 2019. Mapping genomic regions affecting milk traits in Sarda sheep by using the OvineSNP50 Beadchip and principal components to perform combined linkage and linkage disequilibrium analysis. Genet Sel Evol. 51(1):65.
- Whyte J, Bergin O, Bianchi A, McNally S, Martin F. 2009. Key signalling nodes in mammary gland development and cancer. Mitogen-activated protein kinase signalling in experimental models of breast cancer progression and in mammary gland development. Breast Cancer Res. 11(5):209.
- Wiener P, Edriss MA, Williams JL, Waddington D, Law A, Woolliams JA, Gutiérrez–Gil B. 2011. Information content in genome-wide scans: concordance between patterns of genetic differentiation and linkage mapping associations. BMC Genomics. 12:65.
- Yang Y, Zhou QJ, Chen XQ, Yan BL, Guo XL, Zhang HL, Du AF. 2015. Profiling of differentially expressed genes in sheep T lymphocytes response to an artificial primary Haemonchus contortus infection. Parasit Vectors. 8:235.
- Zhang L, Liu J, Zhao F, Ren H, Xu L, Lu J, Zhang S, Zhang X, Wei C, Lu G, et al. 2013. Genome-wide association studies for growth and meat production traits in sheep. PLoS One. 8(6):e66569.
- Zhou C, Shen D, Li C, Cai W, Liu S, Yin H, Shi S, Cao M, Zhang S. 2019. Comparative transcriptomic and proteomic analyses identify key genes associated with milk fat traits in Chinese Holstein cows. Front Genet. 10:672.
