ABSTRACT
In this research, we investigated the role of PIK3R6, a regulatory subunit of PI3Kγ, known for its tumor-promoting properties, in clear cell renal cell carcinoma (CCRCC). Utilizing the UALCAN website, we found PIK3R6 upregulated in CCRCC, correlating with lower survival rates. We compared PIK3R6 expression in CCRCC tumor tissues and adjacent normal tissues using immunohistochemistry. Post RNA interference-induced knockdown of PIK3R6 in 786-O and ACHN cell lines, we performed CCK-8, colony formation, Edu staining, flow cytometry, wound healing, and transwell assays. Results showed that PIK3R6 silencing reduced cell proliferation, migration, and invasion, and induced G0/G1 phase arrest and apoptosis. Molecular analysis revealed decreased CDK4, Cyclin D1, N-cadherin, Vimentin, Bcl-2, p-PI3K and p-AKT, with increased cleaved caspase-3, Bax, and E-cadherin levels in CCRCC cells. Moreover, inhibiting PIK3R6 hindered tumor growth. These findings suggest a significant role for PIK3R6 in CCRCC cell proliferation and metastasis, presenting it as a potential therapeutic target.
Introduction
Renal cell carcinoma (RCC), the predominant form of urinary system malignancy, exhibits an escalating incidence and mortality rate annually [Citation1]. Notably, clear cell RCC (CCRCC) constitutes approximately 80% of all RCC cases, as evidenced by histological analysis [Citation2]. In cases of early-stage and localized RCC, radical nephrectomy has proven efficacious [Citation3]. However, a significant subset of patients present with advanced or metastatic RCC at initial diagnosis [Citation4]. Recent advancements in molecular biology have ushered in neoadjuvant therapies targeting VEGFR, offering new prospects for treating advanced or metastatic RCC [Citation5]. Despite these developments, the 5-year survival rate for advanced-stage patients remains a mere 10%, a figure largely attributed to frequent relapses and a high rate of metastasis [Citation6]. Consequently, there is a critical need for the identification of early diagnostic markers and a deeper understanding of the pathogenesis and progression mechanisms of CCRCC to improve the diagnosis and prognosis of this disease.
The phosphoinositide-3-kinase (PI3K) signaling pathway, a vital mechanism in multicellular organisms, responds to external growth stimuli and is notably linked to the development of most human cancers due to its abnormal activation [Citation7]. PI3K is divided into three primary classes – I, II, and III – each targeting specific substrates and effectors, but universally interacting with the Akt substrate [Citation8]. Particularly, Class I PI3K, which includes the isoforms PI3Kα, β, δ, and γ, is involved in regulating cell growth, movement, and differentiation by generating secondary messengers [Citation9]. Among these, the γ isoform (PI3Kγ) has garnered attention in cancer research. PI3Kγ inhibitors have shown promise in targeting cancer stem cells (CSCs), suggesting potential in cancer therapy [Citation10]. Studies have demonstrated that selective inhibition of PI3Kγ can suppress tumor initiation and growth, as seen in a mouse model of carcinogen-induced and obesity-aggravated hepatocellular carcinoma (HCC) [Citation11]. Furthermore, research by Wang et al. [Citation12] revealed that umbelliferon can induce cell death in RCC cells by reducing the expression of the p110γ catalytic subunit of PI3Kγ. In the context of PI3Kγ, the regulatory subunit PIK3R6 (also known as PI3K-P84) has been investigated in various diseases. For example, overexpressing PIK3R6 in ovarian cancer cells reversed the reduced proliferative and migratory capabilities resulting from RBBP6 knockdown [Citation13]. Additionally, Du et al. [Citation14] reported that PIK3R6 enhances angiogenesis in HCC by activating the STAT3 signaling pathway, thus accelerating the disease’s malignant progression. PIK3R6 has also been implicated in preeclampsia-like conditions through its role in the PIK3R6/p-STAT3 signaling pathway [Citation15]. Despite these findings, the specific biological function of PIK3R6 in human RCC remain to be fully elucidated, underlining the need for further research in this area.
In our research, we analyzed the expression levels of PIK3R6 in a RCC cohort and investigated its relationship with patient prognosis. We also delved into the influence of PIK3R6 on the oncogenic characteristics of RCC cells. The conclusive findings underscored the crucial importance of PIK3R6 in RCC prognosis and confirmed its significant connection with the disease’s development.
Materials and methods
Tissue specimens
The study involved 20 cases of CCRCC patients from Chongqing Ninth People’s Hospital (Chongqing, China), collected between January and June 2021. All samples, including tumor tissues and adjacent non-cancerous tissues, were treated with formalin, embedded in paraffin, and snap-frozen in liquid nitrogen for preservation at −80°C. These patients were specifically selected for not having other renal or systemic diseases, other tumors, or prior surgical interventions before their radical nephrectomies. The research adhered to strict medical ethical standards and received approval from the Institutional Ethics Committee of Chongqing Ninth People’s Hospital (Approval No. CNP20210356A). All patients provided their written informed consent for participation in the study.
Immunohistochemistry
Thin sections (5-μm) of paraffin-embedded tissues were first dewaxed using xylene and graded ethanol solutions, then rehydrated with distilled water. These sections were incubated with a rabbit anti-PIK3R6 antibody (PA5–54689, Thermo Fisher Scientific, Waltham, MA, USA) at 4°C overnight, followed by labeling with biotinylated goat anti-rabbit serum and streptavidin-peroxidase for 15 min, and visualized using diaminobenzidine (DAB). Blinded evaluations of the staining were conducted by two independent pathologists using a microscope. PIK3R6 positive cell distribution was scored as 0, 1, 2, and 3 for 0%, 0–20%, 20–60%, 60–100% distributions, respectively. Staining intensities, categorized as light yellow, yellow, brown-yellow, and reddish-brown, received corresponding scores. The total score, combining distribution and intensity, was classified into four grades: negative (0), weak (1), medium (2–4), and strong (5–6).
Cell culture and transfection
Two CCRCC cell lines (786-O and ACHN) were acquired from China Cell Bank (Shanghai, China), accompanied by STR identification certificates. They were cultured in DMEM (HyClone, Logan, UT, USA) with 10% FBS (Gibco, NY, USA) and 1% penicillin/streptomycin (Gibco) at 37°C with 5% CO2. Post reaching the logarithmic phase in six-well plates, these cells underwent transfection using siRNAs targeting PIK3R6 (si-PIK3R6) and a negative control (si-NC) via Lipofectamine 2000 (Thermo Fisher Scientific). The siRNA sequences were si-PIK3R6#1: 5'-GCTGCACACTGTAATGTACGT-3“; si-PIK3R6#2: 5”-GGCCACAGGAATCACAGAAGA-3“; and si-NC: 5”-GCTCTATCAGGTCGACGTAAT-3'. Cells were harvested for analysis 48 h post-transfection.
Cell proliferation assays
To assess cell proliferation, three methods were employed: Cell Counting Kit-8 (CCK-8), colony formation, and 5-ethynyl-2'-deoxyuridine (EdU) assays. For the CCK-8 assay, cells were seeded at 1 × 10 [Citation3] cells/well in 96-well plates overnight, followed by a 2-h incubation with 10 µl of CCK-8 solution (Beyotime Biotechnology, Jiangsu, China) at 37°C. Viability was determined by absorbance at 450 nm at 0, 24, 48, and 72 h. In the colony formation assay, 600 cells/well were cultured in six-well plates for two weeks at 37°C, then fixed with 4% paraformaldehyde, stained with 0.3% crystal violet, and colonies were counted. The EdU assay utilized a BeyoClick™ EdU Imaging Detection Kit (Beyotime), involving cell fixation with 4% paraformaldehyde, permeabilization with Triton X-100 (Sigma-Aldrich), and a 30-min incubation with the Click-iT EdU reaction cocktail. Cell nuclei were stained with DAPI, and the cells were observed and photographed using a fluorescence microscope (Olympus, Tokyo, Japan), with Image J software quantifying stained cells.
Flow cytometry assays
In the cell cycle assay, cells underwent trypsinization and were then centrifuged for 10 min at 1500 rpm. Following three PBS washes, they were resuspended in PBS and treated with a mixture of propidium iodide (PI, 40 µg/mL) and RNase (100 µg/mL), followed by an overnight incubation at 4°C in darkness. For detecting cell apoptosis, a dual staining method was used with Annexin V-FITC/PI, as per the guidelines of the Annexin V-FITC/PI Apoptosis Kit (KeyGEN Biotech, Nanjing, China). The FACS Calibur flow cytometer (Beckman Coulter, CA, USA) was utilized to assess the variations in the proportion of stained cells.
Wound healing assay
The migration capabilities of cells were analyzed using a wound healing assay. In this assay, a scratch was created on a layer of confluent 786-O and ACHN cells, which had been seeded in 24-well plates, using a micropipette tip. After creating the scratch, the cells were allowed to migrate for 24 h in a serum-free medium. The migration progress was documented using a Zeiss microscope (Germany) at the start (0 h) and end (24 h) of the migration period. The relative wound healing rate was calculated with the formula: (wound area at 0 h-wound area at 48 h)/wound area at 0 h × 100%. This procedure was independently repeated three times for consistency.
Transwell assay
The invasion assay utilized 24-well Transwell inserts with 8-μm pores (Corning, Costar®, Washington, DC, USA), which were coated with 0.1 ml of Matrigel (50 µg/ml, BD Biosciences). 786-O and ACHN cells, post-transfection, were placed in the upper chamber in serum-free medium, while the lower chamber was filled with 600 µl of medium containing 10% FBS. Following a 24-h incubation at 37°C, cells that invaded through the membrane were fixed using 4% paraformaldehyde, stained with 0.3% crystal violet, and then imaged using a Zeiss microscope (Germany).
Animal experiments
Male BALB/c athymic nude mice, aged five weeks, were acquired from Beijing Vital River Laboratory Animal Technology Co., Ltd (Beijing, China). To create a subcutaneous tumor model, 786-O cells (1 × 10 [Citation6], transfected with either si-PIK3R6#1 or si-NC, were suspended in 200 µl of PBS and injected subcutaneously into the lower right flank region of the mice. The dimensions of the subcutaneous tumors, specifically width (W) and length (L), were measured at five-day intervals. Tumor volume (V) was calculated using the formula: V = (W [Citation2] × L/2). After 25 days, the mice were euthanized, and the tumors were excised for weight recording, photographing, and further analysis. All animal studies followed the ARRIVE guidelines (https://arriveguidelines.org/) and were performed in accordance with the Chongqing Medical University’s Guidelines for the Care and Use of Laboratory Animals.
Quantitative real-time PCR
RNA was isolated from both cultured cells and tissue specimens using Trizol reagent (Gibco). For cDNA synthesis, Oligo (dT) primer and M-MLV Reverse Transcriptase (Promega) were utilized. The quantitative real-time PCR analysis was conducted using the BioRad connect Real-Time PCR platform (Bio-Rad, Hercules, CA, USA) and SYBR Green Master Mix Kits (TaKaRa, Tokyo, Japan). The amount of total RNA and cDNA which were used for the reverse transcription (RT) and PCR reactions were 2 μg total RNA and 5 µl cDNA (30 ng/µl) respectively. The detailed PCR procedure was initially denatured at 95°C for 1 min, and 40 cycles of denaturation at 95°C for 5 s, and extension at 60°C for 20 s. The absorbance value was read in the extension stage. The primers for PIK3R6 were 5'-CGCACCCTGGAGCACTATTT-3' (forward) and 5'-GAGCACCAGTTCCTTCCAGA-3' (reverse); for GAPDH, they were 5'-GGTGAAGGTCGGAGTCAACG-3' (forward) and 5'-GCATCGCCCCACTTGATTTT-3' (reverse). The 2−ΔΔCt method was used to calculate the relative levels of PIK3R6, with normalization to GAPDH.
Western blot analysis
Protein samples were extracted using RIPA buffer (Beyotime, Shanghai, China) and their concentrations were determined using the BCA method (Beyotime). These proteins were separated by 10% SDS-PAGE and then transferred to polyvinylidene fluoride (PVDF) membranes (Millipore). The PVDF membranes were subsequently blocked with 5% nonfat milk in TBST for 1 h at room temperature. Afterward, they were incubated overnight with primary antibodies: PIK3R6 (ab192540, Abcam), CDK4 (ab226474, Abcam), Cyclin D1 (60186–1-1 g, Proteintech), Bax (ab53154, Abcam), E-cadherin (ab238099, Abcam), N-cadherin (ab76059, Abcam), Vimentin (ab137321, Abcam), PI3K/p-PI3K (ab154598, Abcam), AKT/p-AKT (ab38449, Abcam) and GAPDH (10494–1-AP, Proteintech). This was followed by a 2-h incubation with horseradish peroxidase-conjugated goat anti-rabbit IgG antibody (SC-2054, Santa Cruz) at room temperature. Protein signals were visualized using enhanced chemiluminescence (Thermo Fisher Scientific), with GAPDH serving as the internal control.
Statistical analysis
Three biological replicates’ data were analyzed using GraphPad Prism 8.0, presented as mean ± standard deviation (SD). The Wilcoxon signed rank test assessed PIK3R6 expression differences between CCRCC tissues and their corresponding non-cancerous counterparts. For si-NC vs. si-PIK3R6 groups, the student’s t-test was used, with a p-value <.05 indicating significance.
Results
Elevated PIK3R6 expression in CCRCC linked to adverse progression
Analysis of the UALCAN website (https://ualcan.path.uab.edu) revealed that PIK3R6 mRNA levels were significantly elevated in tumor tissues (n = 533) compared to normal tissues (n = 72) in CCRCC patients (). Additionally, heightened PIK3R6 expression was notably associated with various aspects of CCRCC such as tumor stage (), grade (), subtype (), and metastasis states (). Furthermore, increased PIK3R6 expression showed a correlation with poorer survival outcomes in CCRCC patients (). To validate PIK3R6 overexpression, immunohistochemistry assays were conducted to measure PIK3R6 levels in human CCRCC and adjacent normal kidney tissues. The results, represented in , indicated a significant elevation of PIK3R6 in CCRCC tissues compared to para-carcinoma normal kidney tissues (), with the statistical difference being highly significant (p < .001).
Figure 1. Elevated PIK3R6 expression in CCRCC linked to adverse progression. (a–e) the difference of PIK3R6 mRNA expression among CCRCC tissues and adjacent tissues was analyzed based on sample types, tumor stage, tumor grade, tumor subtype, and tumor metastasis states. (f) TCGA database analysis on UALCAN website showed the relationship between PIK3R6 expression and the patients’ survival rate.
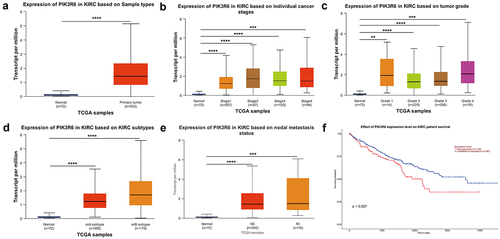
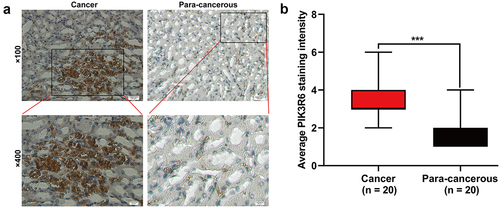
Figure 2. PIK3R6 protein level was upregulated in CCRCC. (a) Representative immunohistochemical images are presented about PIK3R6 expression in CCRCC and paired para-carcinoma tissues. (b) PIK3R6 expression is significantly higher in the CCRCC tissues than that in the paired para-carcinoma tissues (n = 20; ***p < .001).
PIK3R6 silencing suppressed the proliferation of CCRCC cells in vitro
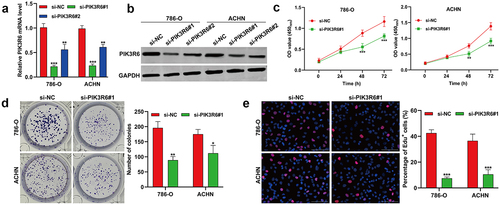
In response to the heightened expression of PIK3R6 in CCRCC, we undertook in vitro loss-of-function experiments to determine its specific role. This involved transfecting 786-O and ACHN cells with si-PIK3R6#1 and si-PIK3R6#2 to knock down PIK3R6. We evaluated the transfection’s success using quantitative real-time PCR () and western blot analysis (), which revealed a significant reduction in both mRNA and protein levels of PIK3R6. Notably, si-PIK3R6#1 exhibited a more pronounced inhibitory effect on PIK3R6 and was thus chosen for further studies. The CCK-8 assay revealed that si-PIK3R6#1 transfection markedly reduced cell viability in both 786-O and ACHN cells (). This finding was supported by colony formation () and Edu assays (), which consistently indicated that PIK3R6 knockdown notably inhibited the proliferation of these cell lines.
Figure 3. PIK3R6 silencing suppressed the proliferation of CCRCC cells in vitro. 786-O and ACHN cells were transfected with si-PIK3R6#1, si-PIK3R6#2 or si-NC for 48 h. (a) Quantitative real time PCR and (b) western blot analysis showed efficient knockdown of PIK3R6 in 786-O and ACHN cells. (c) CCK-8 assay showed the cell viability of the si-PIK3R6#1 or si-NC group in 786-O and ACHN cells after 24 h, 48 h and 72 h incubation. The effects of PIK3R6 knockdown on cell proliferation were assessed by colony formation assay (d) and edu staining (e) in 786-O and ACHN cells. The data were shown as the mean ± SD. *p < .05, **p < .01, ***p < .001, compared with si-NC.
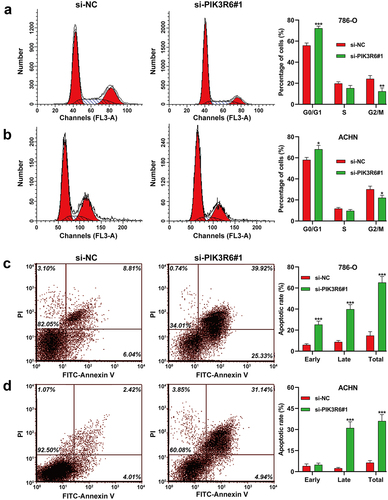
PIK3R6 silencing induced G0/G1 phase arrest and apoptosis in CCRCC cells
This investigation further examined the effects of PIK3R6 downregulation on cell cycle dynamics and apoptosis in CCRCC cell lines. Flow cytometry analysis () demonstrated that PIK3R6 silencing notably augmented the proportion of 786-O and ACHN cells in the G0/G1 phase (786-O cells: 72.25% ± 1.96 vs. 55.90% ± 2.31, p < .001; ACHN cells: 68.17% ± 3.65 vs. 58.09% ± 2.36, p < .05), while concurrently diminishing the fraction in the G2/M phase (786-O cells: 12.38% ± 3.16 vs. 24.24% ± 3.05, p < .01; ACHN cells: 22.13% ± 2.32 vs. 30.16% ± 2.96, p < .05). Moreover, apoptotic assays indicated a significant elevation in the apoptotic rate of both 786-O () and ACHN () cells post-transfection with si-PIK3R6#1, as compared to the si-NC transfection group. These findings collectively suggest that silencing of PIK3R6 disrupts normal cell cycle progression, primarily by inducing G0/G1 arrest, and enhances apoptosis, highlighting its potential as a therapeutic target in CCRCC treatment.
Figure 4. PIK3R6 silencing induced G0/G1 phase arrest and apoptosis in CCRCC cells. 786–O and ACHN cells were transfected with si-PIK3R6#1 or si-NC for 48 h. (a–b) the effects of si–PIK3R6#1 transfection on cell cycle distribution were determined in 786–O and ACHN cells. The representative results of cell cycle were shown in left panel and corresponding quantification of cell cycle distribution was depicted in right panel. (c-d) the effects of si-PIK3R6#1 transfection on apoptotic rate were determined in 786-O and ACHN cells. The representative results of cell apoptosis were shown in left panel and corresponding quantification of cell cycle distribution was depicted in right panel. The data were shown as the mean ± SD. *p < .05, **p < .01, ***p < .001, compared with si–NC.
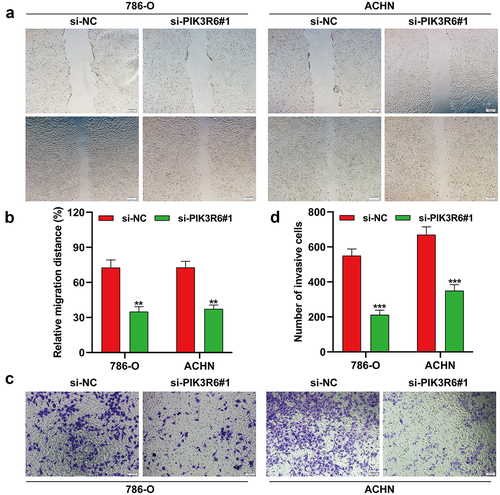
PIK3R6 silencing inhibited the migratory and invasive ability of CCRCC cells
Acknowledging the critical influence of metastasis in cancer progression, we explored how PIK3R6 knockdown affects the metastatic abilities of CCRCC cells through wound healing and transwell assays. illustrated that si-PIK3R6#1 transfection notably slowed wound closure in both 786-O and ACHN cells relative to controls. Detailed analysis showed a marked decrease in migration distance for the si-PIK3R6#1 group compared to the si-NC group in both cell types (). Aligning with these results, the transwell assays also indicated a significant reduction in the number of invasive cells following PIK3R6 knockdown, as compared to the control (). These findings collectively indicate that silencing PIK3R6 effectively inhibits the migration and invasion of CCRCC cells, underscoring its role in metastasis.
Figure 5. PIK3R6 silencing inhibited the migratory and invasive ability of CCRCC cells. 786–O and ACHN cells were transfected with si-PIK3R6#1 or si–NC for 48 h. (a–b) Cell migration was determined in transfected 786-O and ACHN cells by wound healing assay. (c–d) Cell invasion was determined in transfected 786-O and ACHN cells by transwell invasion assay. The data were shown as the mean ± SD. **p < .01, ***p < .001, compared with si-NC.
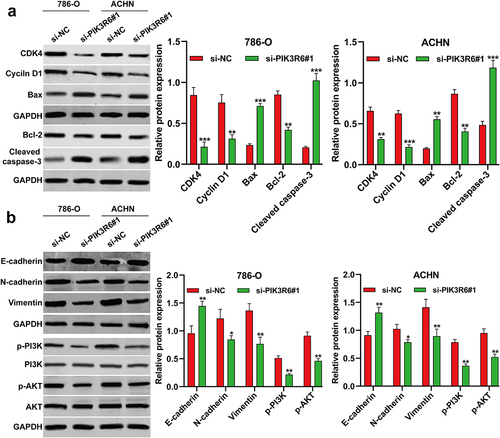
PIK3R6 silencing modulated growth and metastasis indicators in CCRCC cells
To delve deeper into the mechanisms driving G0/G1 phase arrest, apoptosis, and metastasis in CCRCC cells, we examined various related markers via western blot analysis. revealed that post-si-PIK3R6#1 transfection, there was a noticeable decrease in the expression of CDK1 and Cyclin D1, which are crucial for the G1-S phase transition. Concurrently, an increase in pro-apoptotic markers (Bax and cleaved caspase-3) and a decreased in anti-apoptotic marker Bcl-2, were observed in both 786-O and ACHN cells. Additionally, our examination of epithelial-mesenchymal transition (EMT) and PI3K/AKT signaling markers found that PIK3R6 knockdown significantly raised E-cadherin levels, while reducing N-cadherin, vimentin, p-PI3K and p-AKT levels in these cells ().
Figure 6. PIK3R6 silencing modulated growth and metastasis indicators in CCRCC cells. 786–O and ACHN cells were transfected with si-PIK3R6#1 or si–NC for 48 h. (a) The protein levels of CDK4, cyclin D1, Bax, Bcl-2 and cleaved caspase-3 were detected in transfected 786–O and ACHN cells. (b) The protein levels of E-cadherin, N-cadherin, Vimentin, p–PI3K, PI3K, p–AKT and AKT were detected in transfected 786–O and ACHN cells. The data were shown as the mean ± SD. *p < .05, **p < .01, ***p < .001, compared with si-NC.
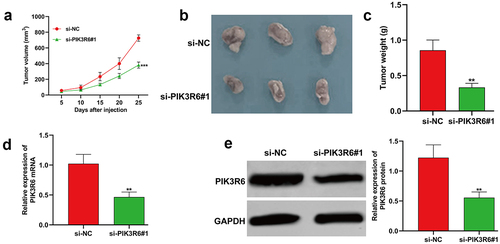
PIK3R6 silencing repressed the tumor growth of CCRCC cells in vivo
To explore the contribution of PIK3R6 to CCRCC tumorigenesis in vivo, we injected nude mice with 786-O cells that had been stably transfected with si-PIK3R6#1. Notably, the tumors in the PIK3R6-inhibited group demonstrated a significant reduction in volume compared to the si-NC group (). Upon removal of the xenograft tumors, it was clear that both the size () and weight () of the tumors in the si-PIK3R6#1 group were substantially lower than those in the si-NC group. Additionally, the levels of PIK3R6 mRNA () and protein () were markedly decreased in the si-PIK3R6#1 group relative to the si-NC group. These in vivo findings corroborate our in vitro results, further affirming the role of PIK3R6 as a facilitator in the progression of CCRCC tumors.
Figure 7. PIK3R6 silencing repressed the tumor growth of CCRCC cells in vivo. 7786-O cells grew as subcutaneous tumors in si-PIK3R6#1 and si-NC groups of BALB/c athymic nude mice (representative 3 mice per group) for 25 days. (a) Tumor volume, (b) xenograft tumors, and quantitative results (c) of tumor weight from two groups. (d-e) the expression levels of PIK3R6 mRNA and protein were determined in tumor tissues was determined. The data were shown as the mean ± SD. **p < .01, ***p < .001, compared with si-NC.
Discussion
The PI3K family, comprising lipid kinase proteins, plays a crucial role in translating signals from growth factors, cytokines, and other environmental cues into intracellular messages [Citation16]. Overactivity of PI3K signaling, commonly observed in human tumors, is associated with altered tumor cell functions and genomic instability [Citation17]. Besides, abnormally activated PI3K contributes to the formation of the tumor microenvironment, tumor angiogenesis, and the recruitment of inflammatory factors [Citation18,Citation19]. In the present study, we focused on the regulatory subunit of PI3Kγ, PIK3R6 (also known as PI3K-P84), in CCRCC. Through bioinformatics analysis, we found that PIK3R6 was upregulated in CCRCC, and higher levels of PIK3R6 were linked to poorer survival outcomes. This correlation was further validated in patient samples, where PIK3R6 expression was significantly elevated compared to adjacent non-cancerous tissues. Our findings suggest that aberrant accumulation of PIK3R6 in CCRCC could serve as a marker for adverse disease progression in patients.
Targeting Class-I PI3Ks, including the PI3Kγ isoform, is crucial in cancer therapy, but their complete inhibition can lead to liver damage, hyperglycemia, and insulinemia, which paradoxically activates PI3K in tumors and reduces the effectiveness of pan-PI3K inhibitors [Citation20,Citation21]. Our research found that specifically suppressing PIK3R6 via si-PIK3R6 transfection significantly reduced the proliferation, migration, and invasion of CCRCC cells. Furthermore, our in vivo experiments corroborated these findings, showing that silencing PIK3R6 impairs tumor growth in a CCRCC xenograft mouse model. Our findings align with previous research showing that PIK3R6 is a target gene of RBBP6, which positively regulates it and contributes to RBBP6’s oncogenic role in ovarian cancer cells [Citation13]. Additionally, high PIK3R6 expression in tumor tissues has been found to have a negative correlation with patient prognosis and to enhance angiogenesis in HCC cells [Citation14]. These insights suggest that PIK3R6 may act as a tumor-promoting gene in CCRCC. However, further research is required to elucidate the specific mechanisms by which PIK3R6 influences the malignancy of CCRCC cells. The cell cycle is a tightly regulated process that controls cell growth and division. In cancer, this regulation is disrupted, leading to uncontrolled cell proliferation. Oncogenes and tumor suppressor genes are crucial in cell cycle regulation [Citation22]. Given that alterations in the G1/S phase cell-cycle modulators occur in a wide range of tumors, targeting this pathway is a promising therapeutic approach [Citation23]. Eukaryotic cell cycles are regulated by cyclins, cyclin-dependent kinases (CDKs), and cyclin-dependent kinase inhibitors (CDKIs) [Citation24], with the transition from the G1 phase to the S phase for mitosis being regulated by the CDK4/Cyclin D1 complex [Citation25]. Apoptosis, or programmed cell death, is a mechanism by which cells can self-destruct when they are damaged or no longer needed. Cancer cells often gain the ability to evade apoptosis, allowing them to survive despite genetic abnormalities or damage that would normally trigger cell death. This evasion of apoptosis is a hallmark of cancer, contributing to tumor growth and the accumulation of further genetic alterations [Citation26]. Caspase-cascade is a central part of cell apoptosis and regulated by various kinds of molecules, such as Bcl-2 family proteins. Bcl-2 family plays a pivotal role in either inhibiting (Bcl-2) or promoting (Bax) cell death [Citation27]. Our data indicated that knockdown of PIK3R6 led to G0/G1 phase arrest and apoptosis in cells, accompanied by decreased CDK4/Cyclin D1 and increased Bax expression. This is consistent with findings that umbelliferone causes G0/G1 phase arrest and apoptosis in human RCC cells, along with a reduction in Bcl-2, CDK4, and Cyclin D1, by lowering the p110γ catalytic subunit of PI3K [Citation12]. The epithelial-mesenchymal transition (EMT) is a process through which epithelial cells lose their cell polarity and cell-cell adhesion and gain migratory and invasive properties to become mesenchymal stem cells. This transition is critical in embryonic development, wound healing, and tissue regeneration [Citation28]. Our results revealed that PIK3R6 knockdown led to increased E-cadherin expression and decreased N-cadherin and Vimentin levels in CCRCC cells. E-cadherin, encoded by CDH1, is known as a metastatic suppressor during EMT. Conversely, N-cadherin and Vimentin, encoded by CDH2 and VIM respectively, are recognized as indicators of EMT [Citation29]. To our knowledge, the role of PI3Kγ in the progression of EMT has been highlighted by Yuan et al. [Citation30], who showed that mJPYZ impedes EMT in gastric cancer cells through a PI3Kγ-dependent mechanism involving TAM reprogramming, ultimately leading to the suppression of gastric cancer growth and metastasis. Increasing evidence suggests that the activation of the PI3K/AKT signaling pathway is implicated in CCRCC initiation and progression [Citation31]. Our data showed that knockdown of PIK3R6 downregulating PI3K/AKT signaling pathway in CCRCC cells. As our best knowledge, the pathways governing cell cycle, apoptosis, and EMT are not isolated; they cross-regulate each other. For instance, certain signals that promote EMT can also inhibit apoptosis, enabling cancer cells to migrate and invade other tissues while resisting cell death. Similarly, the dysregulation of the cell cycle can lead to abnormal cell survival and proliferation, facilitating the EMT process and metastatic spread. Here, specific PI3K/AKT signaling can induce EMT in cancer cells while also having the potential to inhibit the cell cycle or promote apoptosis under different contexts [Citation32]. Drawing on this evidence, we hypothesize that PIK3R6 may contribute to the development of CCRCC by influencing both cell cycle progression and EMT processes via regulating PI3K/AKT signaling. Certainly, the limitations of our study have been documented, notably the limited number of animals used in the in vivo experiments and the omission of PIK3R6 overexpression analysis.
In conclusion, our study has identified an oncogenic role for PIK3R6 in the development of CCRCC, establishing it as an independent prognostic biomarker for this disease. Our findings also suggest that PIK3R6 could be a viable therapeutic target for CCRCC treatment.
Authors’ contributions
Peilin Zhang: Data curation, Formal analysis, Funding acquisition, Investigation, Writing-original draft. Jia Yang: Data curation, Formal analysis, Methodology, Writing-original draft. Xiaoni Zhong: Validation, Methodology. Xiaoling Gao: Software, Formal analysis. Wenyi Xie: Supervision, Formal analysis, Validation. Yaokai Chen and Yuanjiang Liao: Formal analysis, Investigation. Peilin Zhang and Jia Yang: Project administration, Conceptualization, Supervision, Writing – review & editing. The author(s) read and approved the final manuscript
Ethics approval and consent to participate
All patients provided their written informed consent for participation in the study. This study was performed in line with the principles of the Declaration of Helsinki. The research adhered to strict medical ethical standards and received approval from the Institutional Ethics Committee of Chongqing Ninth People’s Hospital (Approval No. CNP20210356A). All animal studies were performed in accordance with the Chongqing Medical University’s Guidelines for the Care and Use of Laboratory Animals. Studies involving laboratory animals followed the ARRIVE guidelines (https://arriveguidelines.org/).
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
Data will be made available by the corresponding author upon reasonable request.
Additional information
Funding
References
- Bukavina L, Bensalah K, Bray F, et al. Epidemiology of renal cell carcinoma: 2022 update. Eur Urol. 2022;82(5):529–542. doi: 10.1016/j.eururo.2022.08.019
- Moch H, Cubilla AL, Humphrey PA, et al. The 2016 WHO classification of tumours of the urinary system and male genital organs—part A: renal, penile, and Testicular tumours. Eur Urol. 2016;70(1):93–105. doi: 10.1016/j.eururo.2016.02.029
- Jiang Z, Chu PG, Woda BA, et al. Combination of quantitative IMP3 and tumor stage: a new system to predict metastasis for patients with localized renal cell carcinomas. Clin Cancer Res. 2008;14(17):5579–5584. doi: 10.1158/1078-0432.CCR-08-0504
- Eggener SE, Yossepowitch O, Pettus JA, et al. Renal cell carcinoma recurrence after nephrectomy for localized disease: predicting survival from time of recurrence. J Clin Oncol. 2006;24(19):3101–3106. doi: 10.1200/JCO.2005.04.8280
- Sciarra A, Gentile V, Salciccia S, et al. New anti-angiogenic targeted therapy in advanced renal cell carcinoma (RCC): current status and future prospects. Rev Recent Clin Trials. 2008;3(2):97–103. doi: 10.2174/157488708784223808
- Zhang H, Ma M. Circ_0101692 knockdown retards the development of clear cell renal cell carcinoma through miR-384/FN1 pathway. Transl Oncol. 2023;28:101612. doi: 10.1016/j.tranon.2022.101612
- Thorpe LM, Yuzugullu H, Zhao JJ. PI3K in cancer: divergent roles of isoforms, modes of activation and therapeutic targeting. Nat Rev Cancer. 2015;15(1):7–24. doi: 10.1038/nrc3860
- Vanhaesebroeck B, Guillermet-Guibert J, Graupera M, et al. The emerging mechanisms of isoform-specific PI3K signalling. Nat Rev Mol Cell Biol. 2010;11(5):329–341. doi: 10.1038/nrm2882
- Li J, Kaneda MM, Ma J, et al. PI3Kγ inhibition suppresses microglia/TAM accumulation in glioblastoma microenvironment to promote exceptional temozolomide response. Proc Natl Acad Sci USA. 2021;118(16):118. doi: 10.1073/pnas.2009290118
- Xu Y, Afify SM, Du J, et al. The efficacy of PI3Kγ and EGFR inhibitors on the suppression of the characteristics of cancer stem cells. Sci Rep. 2022;12(1):347. doi: 10.1038/s41598-021-04265-w
- Becattini B, Breasson L, Sardi C, et al. PI3Kγ promotes obesity-associated hepatocellular carcinoma by regulating metabolism and inflammation. JHEP Rep. 2021;3(6):100359. doi: 10.1016/j.jhepr.2021.100359
- Wang X, Huang S, Xin X, et al. The antitumor activity of umbelliferone in human renal cell carcinoma via regulation of the p110γ catalytic subunit of PI3Kγ. Acta Pharm. 2019;69(1):111–119. doi: 10.2478/acph-2019-0004
- Liu YJ, Cui LL, Liu ZS, et al. RBBP6 aggravates the progression of ovarian cancer by targeting PIK3R6. Eur Rev Med Pharmacol Sci. 2020;24(20):10366–10374. doi: 10.26355/eurrev_202010_23386
- Du Y, Zhao W, Han H, et al. PIK3R6 promotes angiogenesis in hepatocellular carcinoma by activating STAT3 signaling pathway. Altern Ther Health Med. 2023;29(8):704–709.
- Liu M, Liao L, Gao Y, et al. BCAM deficiency May contribute to preeclampsia by suppressing the PIK3R6/p-STAT3 signaling. Hypertension. 2022;79(12):2830–2842. doi: 10.1161/HYPERTENSIONAHA.122.20085
- Lee MJ, Jin N, Grandis JR, et al. Alterations and molecular targeting of the GSK-3 regulator, PI3K, in head and neck cancer. Biochim Biophys Acta, Mol Cell Res. 2020;1867(6):118679. doi: 10.1016/j.bbamcr.2020.118679
- Okkenhaug K, Graupera M, Vanhaesebroeck B. Targeting PI3K in cancer: impact on tumor cells, their protective stroma, angiogenesis, and immunotherapy. Cancer Discov. 2016;6(10):1090–1105. doi: 10.1158/2159-8290.CD-16-0716
- Liu X, Xu Y, Zhou Q, et al. PI3K in cancer: its structure, activation modes and role in shaping tumor microenvironment. Future Oncol. 2018;14(7):665–674. doi: 10.2217/fon-2017-0588
- Kobialka P, Sabata H, Vilalta O, et al. The onset of PI3K-related vascular malformations occurs during angiogenesis and is prevented by the AKT inhibitor miransertib. EMBO Mol Med. 2022;14(7):e15619. doi: 10.15252/emmm.202115619
- Hopkins BD, Pauli C, Du X, et al. Suppression of insulin feedback enhances the efficacy of PI3K inhibitors. Nature. 2018;560(7719):499–503. doi: 10.1038/s41586-018-0343-4
- Janku F, Yap TA, Meric-Bernstam F. Targeting the PI3K pathway in cancer: are we making headway? Nat Rev Clin Oncol. 2018;15(5):273–291. doi: 10.1038/nrclinonc.2018.28
- Tang SW, Chang WH, Su YC, et al. MYC pathway is activated in clear cell renal cell carcinoma and essential for proliferation of clear cell renal cell carcinoma cells. Cancer Lett. 2009;273(1):35–43. doi: 10.1016/j.canlet.2008.07.038
- Helsten T, Kato S, Schwaederle M, et al. Cell-cycle gene alterations in 4,864 tumors analyzed by next-generation sequencing: implications for targeted therapeutics. Mol Cancer Ther. 2016;15(7):1682–1690. doi: 10.1158/1535-7163.MCT-16-0071
- Kõivomägi M, Swaffer MP, Turner JJ, et al. G1cyclin–cdk promotes cell cycle entry through localized phosphorylation of RNA polymerase II. Science. 2021;374(6565):347–351. doi: 10.1126/science.aba5186
- Sun F, Li N, Tong X, et al. Ara-c induces cell cycle G1/S arrest by inducing upregulation of the INK4 family gene or directly inhibiting the formation of the cell cycle-dependent complex CDK4/cyclin D1. Cell Cycle. 2019;18(18):2293–2306. doi: 10.1080/15384101.2019.1644913
- Xu H, He L, Feng X, et al. Specific reduction of fas-associated protein with death domain (FADD) in clear cell renal cell carcinoma. Cancer Invest. 2009;27(8):836–843. doi: 10.1080/07357900902849681
- Brunelle JK, Letai A. Control of mitochondrial apoptosis by the Bcl-2 family. J Cell Sci. 2009;122(4):437–441. doi: 10.1242/jcs.031682
- Hanahan D. Hallmarks of cancer: new dimensions. Cancer Discov. 2022;12(1):31–46. doi: 10.1158/2159-8290.CD-21-1059
- Zhao J, Dong D, Sun L, et al. Prognostic significance of the epithelial-to-mesenchymal transition markers e-cadherin, vimentin and twist in bladder cancer. Int Braz J Urol. 2014;40(2):179–189. doi: 10.1590/S1677-5538.IBJU.2014.02.07
- Yuan M, Zou X, Liu S, et al. Modified jian-pi-yang-zheng decoction inhibits gastric cancer progression via the macrophage immune checkpoint PI3Kgamma. Biomed Pharmacother. 2020;129:110440. doi: 10.1016/j.biopha.2020.110440
- Fan D, Liu Q, Wu F, et al. Prognostic significance of PI3K/AKT/mTOR signaling pathway members in clear cell renal cell carcinoma. PeerJ. 2020;8:e9261. doi: 10.7717/peerj.9261
- Chen L, Qing J, Xiao Y, et al. TIM-1 promotes proliferation and metastasis, and inhibits apoptosis, in cervical cancer through the PI3K/AKT/p53 pathway. BMC Cancer. 2022;22(1):370. doi: 10.1186/s12885-022-09386-7