ABSTRACT
The coupling of growth to nutritional status is an important adaptive response of living organisms to their environment. For this ability, animals have evolved various strategies, including endocrine systems that respond to changing nutritional conditions. In animals, nutritional information is mostly perceived by peripheral organs, such as the digestive tract and adipose tissues, and is subsequently transmitted to other peripheral organs or the brain, which integrates the incoming signals and orchestrates physiological and behavioral responses. In Drosophila melanogaster, adipose tissue, known as the fat body, functions as an endocrine organ that communicates with the brain. This fat body-brain axis coordinates growth with nutritional status by regulating the secretion of Drosophila insulin-like peptides (Dilps) from the brain. However, the molecular nature of the fat body-brain axis remains to be elucidated. We recently demonstrated that a small peptide, CCHamide-2 (CCHa2), expressed in the fat body and gut, directly stimulates its receptor (CCHa2-R) in the brain, leading to Dilp production. Notably, the expression of CCHa2 is sensitive to the presence of nutrients, particularly sugars. Our results, together with the results of previous studies, show that signaling between peripheral organs and the brain is a conserved strategy that couples nutritional availability to organismal physiology.
Introduction
Organismal growth is controlled in response to intrinsic and environmental cues. Among the environmental factors, nutritional availability has the most significant impact on an animal's growth. An animal's ability to adjust its growth rate in response to changing nutritional conditions allows it to survive in a volatile nutritional environment. For such flexible growth control, animals have evolved a variety of nutrient-sensing signals. Target of rapamysin (TOR), an evolutionarily conserved protein kinase required for protein biosynthesis, is at the core of nutrient-sensing signaling.Citation1 TOR is under the control of cellular amino acid and energy levels, thereby coordinating cell growth with nutritional status. In addition to such cellular mechanisms, multicellular organisms require a system for sharing the detected nutritional information among different organs. Nutritional information is primarily sensed by peripheral organs, such as the digestive tract and adipose tissues, before being relayed to other organs and the brain, which modulates the physiological responses. Endocrine systems, in which hormones are secreted from specialized endocrine cells, have evolved to facilitate this type of organ-to-organ communication.
In Drosophila, adipose tissue known as the fat body acts as a nutrient sensor and coordinates tissue growth by endocrine mechanisms. The fat-body-specific down-regulation of the slimfast gene, which encodes an amino acid transporter, or downregulation of the TOR pathway, reduces systemic growth in larvae.Citation2 These findings suggest that the fat body secretes an amino-acid-dependent signal(s) that remotely controls tissue growth. The Unpaired-2 (Upd2) ligand, which activates the JAK/STAT pathway, is another growth-promoting signal derived from the fat body. Although the nutrient sensitivity of upd2 in larvae remains unknown, its expression is sugar- and lipid-sensitive in adult flies. Therefore, upd2 is apparently independent of the amino acid-dependent signal.Citation3
These fat body-derived signals ultimately control Drosophila insulin-like peptide (Dilp) secretion from the brain.Citation3,4 Ex vivo co-culture experiments demonstrated that humoral factor(s) secreted by the fat body induces Dilp secretion from insulin-producing cells (IPCs) in the brain; however, the molecules involved in this fat body to brain communication mechanism were not identified.Citation4 We recently showed that a small peptide, CCHamide-2 (CCHa2), and its receptor (CCHa2-R), form a direct signaling system between peripheral organs and the brain. In this article, I discuss our recent findings suggesting that CCHa2/CCHa2-R signaling promotes the coupling of nutritional availability to growth in Drosophila.
CCHa2 is a bioactive peptide conserved in insects
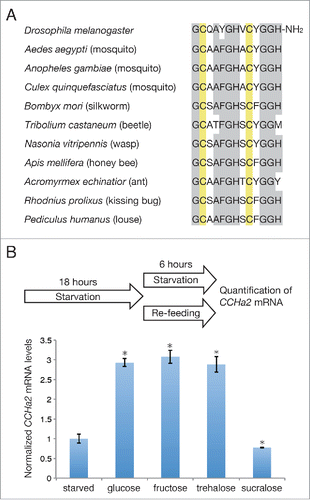
CCHa2 is a 13 amino acid peptide containing 2 disulfide-bonded cysteines and an amidated C-terminus (). The CCHa2 gene was first identified in the silkworm Bombyx mori.Citation5 The CCHa2 gene is also present in the Drosophila genome, and the synthetic CCHa2 peptide was found to activate a G protein-coupled receptor (GPCR) encoded by CG14593.Citation6 Subsequently, endogenous CCHa2 peptide was detected in the larval gut by mass spectrometric analysis and was biochemically purified from adult flies.Citation7,8 There are several CCHa2 isoforms; one matches the predicted amino acid sequence, and the others have 2 or 8 additional amino acids in the N-terminus. No functional differences in CCHa2-R activation in vitro have been detected among these isoforms.Citation7 CCHa2 is highly conserved in insects, including flies, mosquitos, silkworms, beetles, wasps, ants, kissing bugs, and liceCitation6 (); however, it is currently unclear whether its functions are similarly conserved.
Figure 1. (A) Alignment of CCHa2 sequences in the indicated insect species. Conserved amino acids and bisulfide-bonded cysteine residues are highlighted in gray and yellow, respectively. (B) Effects of sugars on CCHa2 expression. Larvae were starved for 18 h, and then either further starved or re-fed with the indicated sugars for 6 h. Glucose, fructose, and trehalose, but not sucralose induced CCHa2 expression. *P < 0.05.
CCHa2 and CCHa2-R mediate signaling between peripheral organs and the brain
To determine the function(s) of CCHa2, we first examined the CCHa2 mRNA expression pattern in Drosophila larval tissues.Citation9 The results indicated that CCHa2 is primarily expressed in the fat body. Low levels of CCHa2 expression were also detected in the gut and central nervous system (CNS). In contrast, CCHa2-R mRNA was found to be highly enriched in the CNS. Histochemical analysis revealed that CCHa2-R is expressed in the IPCs and in neuropeptide F (NPF)- and SIFamide (SIFa)-secreting cells in the brain. Thus, CCHa2-R is expressed in several different types of neuroendocrine cells.
These observations raised the possibility that CCHa2 and CCHa2-R function to transmit signals between peripheral organs and the brain. To investigate this possibility, we generated Drosophila strains harboring null mutations in CCHa2-R, and analyzed calcium signaling in brain explants from the wild-type and mutant larvae by using the fluorescent calcium sensor, GCaMP6s, which was expressed in the IPCs. The dissected brains were immersed in phosphate-buffered saline and then treated with medium containing synthetic CCHa2 peptide. Notably, the CCHa2 peptide treatment resulted in dramatically increased calcium signaling in the brain IPCs of the wild-type larvae, but not in those of the CCHa2-R mutant larvae. Taken together, these results indicated that CCHa2 and CCHa2-R function to relay information between peripheral organs and the IPCs of the brain.
CCHa2 and CCHa2-R control insulin-like peptide production in the brain IPCs
The CCHa2-CCHa2-R-mediated activation of IPCs in the brain explants suggested that CCHa2 produced by peripheral organs controls the functioning of IPCs in the brain. Since IPCs produce Dilp2, −3, and −5,Citation10,11 we examined the transcription and secretion of Dilp2 and Dilp5 in the CCHa2-R mutants. The results showed that CCHa2-R is required for the transcription of dilp5 and the secretion of both Dilp2 and Dilp5 in the brain of Drosophila larvae.
In mammalian pancreatic β-cells, the insulin secretion pathway can be activated by a group of Gαs- and Gαq/11-coupled GPCRs.Citation12 Since CCHa2-R is a GPCR, these Gα subunits could be involved in transducing intracellular signals downstream of this receptor. Thus, it will be interesting to determine which Gα subunit is involved in the CCHa2/CCHa2-R-mediated Dilp release.
A conserved transcription factor complex consisting of Dachshund (Dac) and Eyeless (Ey) has been shown to activate dilp5 transcription in IPCs,Citation13 and a recent study identified a signaling cascade that activates dilp5 transcription in a nutrient-dependent manner.Citation14 Circulating Dilps and amino acids promote Dilp6 production in glial cells at the surface of the brain. Dilp6 secreted inside the brain activates the insulin receptor (InR) expressed on cholinergic neurons, leading to Jelly belly (Jeb) function, which in turn activates its receptor, Anaplastic lymphoma kinase (Alk), expressed on IPCs. The activation of Alk signaling releases the Dac-Ey complex from Forkhead box O (FoxO)-dependent inhibition, allowing dilp5 expression.
The relationship between Jeb/Alk and CCHa2/CCHa2-R signaling in the regulation of dilp5 is unclear. One possibility is that the abnormal Dilp secretion observed under conditions of impaired CCHa2/CCHa2-R signaling affects Dilp6 production in the glial cells, which in turn inhibits the full activation of Jeb/Alk, resulting in dilp5 down-regulation in IPCs. Alternatively, the Jeb/Alk and CCHa2/CCHa2-R signaling pathways may operate independently at different stages of larval development. It has been shown that Dac-Ey mediates dilp5 transcription only until the early-third instar larval stage, after which it is mediated by an unknown mechanism.Citation13 In our studies, the effects of CCHa2/CCHa2-R signaling on dilp5 expression were examined in mid- to late-third instar larvae; thus, it is unclear whether CCHa2/CCHa2-R signaling is also required for dilp5 expression in earlier stages. These are interesting issues that can be addressed in future studies.
Intriguingly, we found that CCHa2 acts directly on IPCs to induce Dilp expression. The IPC-specific knockdown of CCHa2-R down-regulated dilp5 expression, which mimicked the phenotype of the global CCHa2-R mutant. This result, together with the results from the calcium imaging experiments, supported the idea that CCHa2 crosses the blood-brain barrier (BBB) to regulate IPCs in the brain. The Drosophila BBB consists of 2 different glial cell layers. The inner layer, adjacent to the neurons of the brain, forms septate junctions, which separate the humoral space and the brain.Citation15, 16 Although BBB development has been extensively studied, little is known about certain functional aspects of the BBB. For instance, it is unclear whether peptide or protein signals can pass through the BBB, and if so, what mechanisms may be involved. Thus, CCHa2-CCHa2-R-mediated IPC signaling could serve as a useful model for studying BBB function as well as peptide or protein drug delivery across the BBB.
Dilp2 and Dilp5 play major roles in regulating larval growth.Citation17 The body weight of CCHa2-R mutant larvae is markedly lower than that of controls; the weight of CCHa2-R mutant larvae is approximately half of that of wild-type larvae from 72 to 108 h after egg laying (AEL). After this period, the larval growth is recovered in response to premature dilp6 up-regulation, which is a consequence of the impaired Dilp2 and 5 production. These findings indicate that CCHa2/CCHa2-R-mediated signaling plays a role in physiological growth control. Notably, we found that CCHa2 expression is responsive to altered nutritional conditions. CCHa2 transcription is significantly downregulated following an 18-h starvation period and is recovered by re-feeding with yeast. These results suggest that CCHa2 is synthesized in response to the presence of nutrients and couples organismal growth to nutritional availability by regulating Dilp production.
Nutrient responsiveness of CCHa2 expression
Since yeast contains various nutrients, including amino acids and carbohydrates, we examined which nutrient was responsible for the CCHa2 upregulation. Our analyses indicated that CCHa2 expression is responsive to glucose, as well as to fructose and trehalose (), although the mechanism by which these sugars activate CCHa2 expression is currently unknown. In addition, it was demonstrated that circulating fructose levels can be used as an indicator of nutritional status in adult flies.Citation18 Blood fructose levels undergo rapid changes following sugar meals, while the glucose and trehalose levels remain unaltered. This sensitivity of fructose to sugar intake is probably a consequence of the very low concentration of fructose in the hemolymph, which is approximately 1% of the glucose and trehalose levels. Circulating fructose is also elevated after glucose feeding, a response that is probably mediated by the polyol pathway, in which glucose is reduced to sorbitol, which is then converted to fructose;Citation19 thus, sugar availability is reflected in the blood fructose level. It is therefore possible that the presence of fructose triggers CCHa2 expression in the larval fat body and gut. Consistent with this possibility, non-nutritious sucralose affects neither the fructoseCitation18 nor the CCHa2 mRNA level ().
Although sugars can induce CCHa2 expression, they are insufficient for inducing Dilp2 secretion.Citation4, 20 The transcription of dilp5 is also unaffected by sugars.Citation14 One possibility is that multiple signals are required for Dilp production, and that CCHa2 alone cannot promote Dilp2 secretion and dilp5 transcription. Supporting this hypothesis, CCHa2 overexpression in the brain, fat body, or gut did not restore the dilp5 expression in starved larvae (unpublished observation). Alternatively, although CCHa2 transcription is activated by sugars, CCHa2 secretion may require the presence of other nutrients. Amino acids are a good candidate for the factor inducing CCHa2 secretion, given that amino acids alone can induce Dilp2 secretion from the Drosophila brain.Citation4
Future perspectives
Signaling between peripheral organs involved in nutritional uptake and the brain has been well described in mammals. A variety of hormones are secreted from peripheral organs and target the brain, in particular the hypothalamus, which regulates energy metabolism and controls behavior.Citation21 Our studies showed that in Drosophila, peripheral signals from nutrient-sensing organs are transmitted to the brain to facilitate the coordination of nutritional status and growth. These findings indicate that vertebrates and invertebrates use common strategies in regulating physiological responses to nutritional intake.
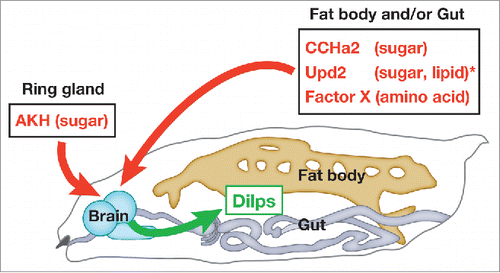
Our data indicate that the activation of CCHa2/CCHa2-R signaling promotes the sensation of good nutritional status in the larval brain, leading to Dilp-dependent tissue growth (). CCHa2/CCHa2-R signaling is not the sole nutrient-dependent Dilp regulator. Recent findings indicate that Dilps are regulated by multiple signals that respond to the presence of different nutrients during larval developmentCitation3, 4, 20 (). Further dissection of the roles of Dilp regulators and their interactions with specific nutrients will lead to a better understating of the mechanisms by which animals couple growth and physiology to nutritional availability.
Figure 2. Periphery-to-brain signaling involved in Dilp regulation during larval development. Dilps are regulated by multiple signals that respond to the presence of different nutrients. CCHa2, Upd2, and an as-yet unidentified factor(s) expressed in the fat body and/or gut, and Adipokinetic hormone (AKH) produced in the ring gland positively regulate the synthesis and/or secretion of Dilps. *The nutrient sensitivity of Upd2 has only been determined in adults.
In mammals, several nutrient-responsive hormones regulate not only physiological, but also behavioral responses. For example, orexin, which is produced in the hypothalamus in response to reduced blood glucose levels, regulates feeding behavior and sleep.Citation22, 23, 24 Leptin, produced in adipose tissue in response to the amount of lipid stored in adipocytes, acts on receptors in the hypothalamus to control feeding behavior and energy expenditure.Citation25 These observations suggest that dual regulation by a single hormone may be an important mechanism for orchestrating biological reactions to nutritional conditions and maintaining internal homeostasis. Thus, it will be interesting to explore the functions of CCHa2/CCHa2-R signaling beyond growth regulation.
The small size of Drosophila has hindered the molecular analysis of its endocrine system. However, recent advances in mass spectrometry have enabled the detection of peptides in whole flies and even the detection of minute amounts of peptides in dissected tissues.Citation8, 26, 27, 28 In addition, advanced genetic tools, such as the GAL4 and LexA-based binary transcriptional systems, have facilitated the investigation of signaling systems that promote communication between different organs. Thus, future studies using Drosophila are expected to elucidate molecular mechanisms underlying endocrine regulation and the biological significance of various endocrine systems.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Acknowledgments
I thank Akira Nakamura and Naoki Okamoto for their helpful discussions and comments on the manuscript.
Funding
The work discussed here was supported by Grants-in-Aid for Scientific Research (24770203, 15K14530, 25115007, 22126003) from MEXT and JSPS (to Hiroko Sano, Akira Nakamura, Azusa Kamikouchi, and Masayasu Kojima), the Tomizawa Jun-ichi and Keiko Fund of the Molecular Biology Society of Japan for Young Scientists, a research grant from the Ishibashi Foundation, the Program of the Joint Usage/Research Center for Developmental Medicine, at the Institute of Molecular Embryology and Genetics, Kumamoto University (to H.S.), and the Howard Hughes Medical Institute (to James W. Truman).
References
- Hietakangas V, Cohen SM. Regulation of tissue growth through nutrient sensing. Annu Rev Genet 2009; 43:389-410; PMID:19694515; http://dx.doi.org/10.1146/annurev-genet-102108-134815
- Colombani J, Raisin S, Pantalacci S, Radimerski T, Montagne J, Leopold P. A nutrient sensor mechanism controls Drosophila growth. Cell 2003; 114:739-49; PMID:14505573; http://dx.doi.org/10.1016/S0092-8674(03)00713-X
- Rajan A, Perrimon N. Drosophila cytokine unpaired 2 regulates physiological homeostasis by remotely controlling insulin secretion. Cell 2012; 151:123-37; PMID:23021220; http://dx.doi.org/10.1016/j.cell.2012.08.019
- Geminard C, Rulifson EJ, Leopold P. Remote control of insulin secretion by fat cells in Drosophila. Cell metabolism 2009; 10:199-207; PMID:19723496; http://dx.doi.org/10.1016/j.cmet.2009.08.002
- Roller L, Yamanaka N, Watanabe K, Daubnerova I, Zitnan D, Kataoka H, Tanaka Y. The unique evolution of neuropeptide genes in the silkworm Bombyx mori. Insect Biochem Mol Biol 2008; 38:1147-57; PMID:19280707; http://dx.doi.org/10.1016/j.ibmb.2008.04.009
- Hansen KK, Hauser F, Williamson M, Weber SB, Grimmelikhuijzen CJ. The Drosophila genes CG14593 and CG30106 code for G-protein-coupled receptors specifically activated by the neuropeptides CCHamide-1 and CCHamide-2. Biochem Biophys Res Commun 2011; 404:184-9; PMID:21110953; http://dx.doi.org/10.1016/j.bbrc.2010.11.089
- Ida T, Takahashi T, Tominaga H, Sato T, Sano H, Kume K, Ozaki M, Hiraguchi T, Shiotani H, Terajima S, et al. Isolation of the bioactive peptides CCHamide-1 and CCHamide-2 from Drosophila and their putative role in appetite regulation as ligands for G protein-coupled receptors. Front Endocrinol 2012; 3:177; PMID:23293632; http://dx.doi.org/10.3389/fendo.2012.00177
- Reiher W, Shirras C, Kahnt J, Baumeister S, Isaac RE, Wegener C. Peptidomics and peptide hormone processing in the Drosophila midgut. J Proteome Res 2011; 10:1881-92; PMID:21214272; http://dx.doi.org/10.1021/pr101116g
- Sano H, Nakamura A, Texada MJ, Truman JW, Ishimoto H, Kamikouchi A, Nibu Y, Kume K, Ida T, Kojima M. The Nutrient-Responsive Hormone CCHamide-2 Controls Growth by Regulating Insulin-like Peptides in the Brain of Drosophila melanogaster. PLoS genetics 2015; 11:e1005209; PMID:26020940; http://dx.doi.org/10.1371/journal.pgen.1005209
- Brogiolo W, Stocker H, Ikeya T, Rintelen F, Fernandez R, Hafen E. An evolutionarily conserved function of the Drosophila insulin receptor and insulin-like peptides in growth control. Curr Biol 2001; 11:213-21; PMID:11250149; http://dx.doi.org/10.1016/S0960-9822(01)00068-9
- Rulifson EJ, Kim SK, Nusse R. Ablation of insulin-producing neurons in flies: growth and diabetic phenotypes. Science 2002; 296:1118-20; PMID:12004130; http://dx.doi.org/10.1126/science.1070058
- Blad CC, Tang C, Offermanns S. G protein-coupled receptors for energy metabolites as new therapeutic targets. Nat Rev Drug Discovery 2012; 11:603-19; PMID:22790105; http://dx.doi.org/10.1038/nrd3777
- Okamoto N, Nishimori Y, Nishimura T. Conserved role for the Dachshund protein with Drosophila Pax6 homolog Eyeless in insulin expression. Proc Natl Acad Sci U S A 2012; 109:2406-11; PMID:22308399; http://dx.doi.org/10.1073/pnas.1116050109
- Okamoto N, Nishimura T. Signaling from Glia and Cholinergic Neurons Controls Nutrient-Dependent Production of an Insulin-like Peptide for Drosophila Body Growth. Dev Cell 2015; 35:295-310; PMID:26555050; http://dx.doi.org/10.1016/j.devcel.2015.10.003
- DeSalvo MK, Mayer N, Mayer F, Bainton RJ. Physiologic and anatomic characterization of the brain surface glia barrier of Drosophila. Glia 2011; 59:1322-40; PMID:21351158; http://dx.doi.org/10.1002/glia.21147
- Stork T, Engelen D, Krudewig A, Silies M, Bainton RJ, Klambt C. Organization and function of the blood-brain barrier in Drosophila. J Neurosci 2008; 28:587-97; PMID:18199760; http://dx.doi.org/10.1523/JNEUROSCI.4367-07.2008
- Gronke S, Clarke DF, Broughton S, Andrews TD, Partridge L. Molecular evolution and functional characterization of Drosophila insulin-like peptides. PLoS genetics 2010; 6:e1000857; PMID:20195512; http://dx.doi.org/10.1371/journal.pgen.1000857
- Miyamoto T, Slone J, Song X, Amrein H. A fructose receptor functions as a nutrient sensor in the Drosophila brain. Cell 2012; 151:1113-25; PMID:23178127; http://dx.doi.org/10.1016/j.cell.2012.10.024
- Hers HG. [The mechanism of the transformation of glucose in fructose in the seminal vesicles]. Biochimica et biophysica acta 1956; 22:202-3; PMID:13373872; http://dx.doi.org/10.1016/0006-3002(56)90247-5
- Kim J, Neufeld TP. Dietary sugar promotes systemic TOR activation in Drosophila through AKH-dependent selective secretion of Dilp3. Nat Commun 2015; 6:6846; PMID:25882208; http://dx.doi.org/10.1038/ncomms7846
- Cummings DE, Overduin J. Gastrointestinal regulation of food intake. J Clin Investig 2007; 117:13-23; PMID:17200702; http://dx.doi.org/10.1172/JCI30227
- Cai XJ, Widdowson PS, Harrold J, Wilson S, Buckingham RE, Arch JR, Tadayyon M, Clapham JC, Wilding J, Williams G. Hypothalamic orexin expression: modulation by blood glucose and feeding. Diabetes 1999; 48:2132-7; PMID:10535445; http://dx.doi.org/10.2337/diabetes.48.11.2132
- Chemelli RM, Willie JT, Sinton CM, Elmquist JK, Scammell T, Lee C, Richardson JA, Williams SC, Xiong Y, Kisanuki Y, et al. Narcolepsy in orexin knockout mice: molecular genetics of sleep regulation. Cell 1999; 98:437-51; PMID:10481909; http://dx.doi.org/10.1016/S0092-8674(00)81973-X
- Sakurai T, Amemiya A, Ishii M, Matsuzaki I, Chemelli RM, Tanaka H, Williams SC, Richardson JA, Kozlowski GP, Wilson S, et al. Orexins and orexin receptors: a family of hypothalamic neuropeptides and G protein-coupled receptors that regulate feeding behavior. Cell 1998; 92:573-85; PMID:9491897; http://dx.doi.org/10.1016/S0092-8674(00)80949-6
- Ahima RS, Flier JS. Leptin. Annu Rev Physiol 2000; 62:413-37; PMID:10845097; http://dx.doi.org/10.1146/annurev.physiol.62.1.413
- Baggerman G, Cerstiaens A, De Loof A, Schoofs L. Peptidomics of the larval Drosophila melanogaster central nervous system. The Journal of biological chemistry 2002; 277:40368-74; PMID:12171930; http://dx.doi.org/10.1074/jbc.M206257200
- Baggerman G, Boonen K, Verleyen P, De Loof A, Schoofs L. Peptidomic analysis of the larval Drosophila melanogaster central nervous system by two-dimensional capillary liquid chromatography quadrupole time-of-flight mass spectrometry. J Mass Spectrom 2005; 40:250-60; PMID:15706625; http://dx.doi.org/10.1002/jms.744
- Yew JY, Wang Y, Barteneva N, Dikler S, Kutz-Naber KK, Li L, Kravitz EA. Analysis of neuropeptide expression and localization in adult drosophila melanogaster central nervous system by affinity cell-capture mass spectrometry. J Proteome Res 2009; 8:1271-84; PMID:19199706; http://dx.doi.org/10.1021/pr800601x
