 ?Mathematical formulae have been encoded as MathML and are displayed in this HTML version using MathJax in order to improve their display. Uncheck the box to turn MathJax off. This feature requires Javascript. Click on a formula to zoom.
?Mathematical formulae have been encoded as MathML and are displayed in this HTML version using MathJax in order to improve their display. Uncheck the box to turn MathJax off. This feature requires Javascript. Click on a formula to zoom.Abstract
Chikungunya virus (CHIKV) is a medically important human viral pathogen that causes Chikungunya fever accompanied with debilitating and persistent joint pain. Host-elicited or passively-transferred monoclonal antibodies (mAb) are essential mediators of CHIKV clearance. Therefore, this study aimed to generate and characterize a panel of mAbs for their neutralization efficacy against CHIKV infection in a cell-based and murine model.
To evaluate their antigenicity and neutralization profile, indirect enzyme-linked immunosorbent assay (ELISA), an immunofluorescence assay (IFA) and a plaque reduction neutralization test were performed on mAbs of IgM isotype. CHIKV escape mutants against mAb 3E7b neutralization were generated, and reverse genetics techniques were then used to create an infectious CHIKV clone with a single mutation. 3E7b was also administered to neonate mice prior or after CHIKV infection. The survival rate, CHIKV burden in tissues and histopathology of the limb muscles were evaluated. Both IgM 3E7b and 8A2c bind strongly to native CHIKV surface and potently neutralize CHIKV replication. Further analyses of 3E7b binding and neutralization of CHIKV single-mutant clones revealed that N218 of CHIKV E2 protein is a potent neutralizing epitope. In a pre-binding neutralization assay, 3E7b blocks CHIKV attachment to permissive cells, possibly by binding to the surface-accessible E2-N218 residue. Prophylactic administration of 3E7b to neonate mice markedly reduced viremia and protected against CHIKV pathogenesis in various mice tissues. Given therapeutically at 4 h post-infection, 3E7b conferred 100% survival rate and similarly reduced CHIKV load in most mice tissues except the limb muscles. Collectively, these findings highlight the usefulness of 3E7b for future prophylactic or epitope-based vaccine design.
Introduction
Chikungunya virus (CHIKV) is a medically important human alphavirus known to cause distinctive polyarthritis or polyarthralgia.Citation1 Common clinical features include fever, maculopapular rash, myalgia,Citation2 while severe complications such as encephalitis and mortality may occur in immunocompromised individuals,Citation3 the elderlyCitation4 and infants.Citation5,6 As a human arbovirus, CHIKV is transmitted by Aedes aegypti and Aedes albopictus mosquitoes. Since 2004, explosive epidemics in Africa,Citation7 Indian Ocean islandsCitation8 and IndiaCitation9 have propelled CHIKV dissemination to various non-endemic countries in South-East Asia,Citation10 Australia,Citation11 Europe and USA.Citation12,13 At present, millions of CHIKV infection cases have been reported worldwide and virus transmission remains active in various Caribbean countries,Citation14 thus signaling the risk of an imminent global CHIKV epidemic.
CHIKV has a positive-sense RNA genome that encodes 4 non-structural proteins (nsP1, 2, 3, 4), 3 structural proteins (capsid, envelope glycoprotein E1 and E2) and 2 cleavage products (E3 and 6k).Citation15 Structurally, the mature E2 protein adopts 3 immunoglobulin-like folds known as domain A at the N-terminal, domain B at the tip and domain C at the C-terminal, which is closest to the viral membrane. The latter is followed by a stem-like transmembrane helix and cytoplasmic tail.Citation16 The extracellular ectodomain comprising domain A, B and C are interconnected by beta-ribbon. Through extensive array of hydrogen bonds, salt bridges and van der Waals forces, E2 intricately complexed with E1 protein to form heterodimer that arranged as 80 trimeric spikes on the viral lipid envelope.Citation16,17 With such a delicate virion surface architecture, E1 and E2 participate complementarily in CHIKV entry. As a type-I transmembrane protein, E2 first mediates CHIKV attachment to the cellular receptor by interaction with surface-exposed regions on domain A and B.Citation18 E1, being a type-II fusion protein, subsequently promotes viral membrane fusion within acidified endosomal membrane to release CHIKV nucleocapsid into the host cytosol.Citation19
Currently, there are no licensed vaccine or effective antiviral for CHIKV disease. Available treatments based on nonsteroidal anti-inflammatory drugs, analgesics or a combination of corticosteroids are symptomatic,Citation20,21 associated with side effects and ineffective for CHIKV-induced chronic arthritis or neonatal infection from viremic mother.Citation22 A plethora of studies have evaluated chemical compounds and antisense agents as potential CHIKV antivirals, but these therapies may not achieve favorable pharmacosafety and tissue-targeted delivery in vivo.Citation21 In contrast, vaccination strategies have highlighted the importance of humoral immunity in controlling CHIKV infection. Strong long-lasting mAb-mediated protection in infected individuals and animal models was observed after administration of CHIKV-based vaccines.Citation23-27 Passive transfer of anti-CHIKV mAbs purified from the convalescent serum of infected patients or co-administration of pairs of neutralizing mAbs to interferon receptor (IFNR)-deficient mice model was shown to confer significant therapeutic and prophylactic efficacy.Citation28,29 Single dose administration of other mAbs at pre- or post-infection were also effective in enhancing survival, reducing viremia and CHIKV joint swelling. Across various cellular model testing, the neutralizing potency of CHIKV-specific mAbs were also consistently demonstrated.Citation29-34 Some of the neutralizing mAbs identified were also conserved in their efficacy against several CHIKV isolates of different genotypes.Citation29,30,32 Altogether, these studies emphasized mAbs as a promising antiviral strategy for CHIKV infection at both pre- and post-exposure settings.
To our knowledge, all of the reported CHIKV-specific neutralizing mAbs characterized thus far are of the IgG isotype. These IgGs commonly recognize surface-exposed epitopes on E2, prominently in domain A and viral membrane distal-end of domain B.Citation29,34,35 The majority of CHIKV IgG antigenic sequences, when mapped spatially on E2, constituted continuous linearCitation35-37 or discontinuous conformational epitopes.Citation34 Interestingly, patient-derived mAbs were shown to neutralize CHIKV by interacting with alanine 162 located in the acid-sensitive region of E2.Citation33 This highlights that highly exposed epitope(s) on CHIKV E2 ectodomain are antigenically important for mAb binding and neutralization.
To broaden the existing pool of CHIKV-specific mAbs, characterization of more neutralizing epitopes on CHIKV glycoproteins using different mAb isotypes is needed. Herein, a panel of CHIKV-specific mouse IgMs was generated and characterized for their neutralizing efficacy and antigenicity in vitro. Two purified mAbs, 3E7b and 8A2c, bind to native surface of CHIKV and neutralize CHIKV infection potently with IC50 of 4–5 ng/ml. Additionally, 3E7b showed no cross-reactivity to other related alphaviruses, the Ross River virus (RRV) and Sindbis virus (SINV). Sequence analysis of 3E7b escape mutants identified 3 distinct mutations that are solvent-accessible and sequence-conserved on CHIKV E2 protein. However, further studies through reverse genetics identified only E2-N218 as critical for 3E7b binding and neutralization. Prophylactic evaluation of 3E7b in a neonate mouse model revealed a dose-dependent efficacy in prolonging survival, as well as significant protection of CHIKV-induced muscle pathogenesis. When administered 4 h post-CHIKV challenge, 3E7b similarly prolonged survival and reduced CHIKV burden in various tissues except the limb muscles, thus suggesting that it could be less effective in a therapeutic setting. Taken together, our findings can be projected for a rationale epitope vaccine design.
Results
Anti-CHIKV IgM 3E7b and 8A2c bind strongly to surface of native CHIKV virion
In this study, adult BALB/c mice were challenged with whole CHIKV virion to elicit a wide repertoire of antibodies that can recognize the viral envelope glycoprotein, E1 and E2. After the third immunization at 28 day post-infection (p.i.), mice sera were harvested and analyzed in Western blot for antibody detection of CHIKV proteins from CHIKV-infected BHK-21 cell lysates. A 35-38 kD band, suggestive of CHIKV capsid protein was strongly detected by all mice sera, while a weak band of 52–55 kD likely to be E1 or E2 protein was recognized by mice sera 1 to 4 (Fig. S1A). This indicates the presence of CHIKV-specific antibody response following the acute phase of CHIKV infection in mice. The spleen of mouse 1 was subsequently harvested for the production of anti-CHIKV mAbs. Culture supernatant from hybridoma clones were screened by immunofluorescence neutralization assay on BHK cells infected with CHIKV at MOI 10. Among the 30 successful subclones, 4 clones, 3E7b, 8A2c, 8D3d and 10C5b, were shown to neutralize CHIKV infection where at least 90% of cells were stained negatively for CHIKV antigen (Fig. S1B–C). Based on this result, these clones were expanded by production of mice ascites and individual ascites of 3E7b and 8A2c clones were further purified and identified as IgM mAbs.
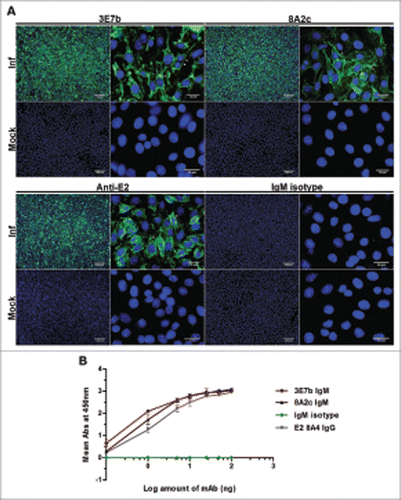
To characterize mAb binding ability to CHIKV, indirect IFA and indirect virion-based ELISA were carried out. Notably, 3E7b and 8A2c IgM strongly detected CHIKV antigen intracellularly, likely due to being in close association with the endoplasmic reticulum (ER) during active virus replication as well as on the plasma membrane of CHIKV-infected cells at day 1 p.i. (; arrowheads). These observations were consistent with the staining by mAb positive control using our laboratory-established rabbit anti-CHIKV E2 polyclonal IgG. Lack of strong staining was seen in all mock-infected cells, thereby suggesting that these mAbs are specific to CHIKV antigen. To further decipher whether 3E7b and 8A2c binds directly to native surface of CHIKV, purified CHIKV was coated onto ELISA plate and a range of mAbs was added. Both 3E7b and 8A2c showed dose-dependent binding to CHIKV, consistent with the mAb positive control, mouse anti-CHIKV E2 IgG. Binding of 3E7b and 8A2c to CHIKV are close to saturation at higher than 25 ng of IgM. Taken together, the results indicate 3E7b and 8A2c IgMs are highly specific to CHIKV antigen, particularly to the native E1 or E2 protein exposed on the surface of CHIKV.
Figure 1. Characterization of mouse IgM mAbs in (A) IFA on CHIKV-infected BHK-21 cells and (B) indirect virion-based ELISA. (A) Purified mAbs produced from immunization of CHIKV was tested at 1:100 or 1:300 dilution in PBS. Rabbit anti-CHIKV E2 polyclonal IgG and mouse IgM isotype antibodies serve as positive and negative control, respectively. Cell nuclei were stained with DAPI while CHIKV antigens (white arrowhead) were secondarily stained with goat anti-mouse or anti-rabbit IgG FITC. Images were captured under 10x and 100x magnification and representative images from 2 independent experiments are shown. (B) Virion-based indirect ELISA. Sucrose-purified CHIKV were coated onto 96-well plate and incubated with anti-CHIKV IgMs, IgM isotype antibody or mouse anti-CHIKV E2 8A4 IgG mAb positive control at a range of 0.1 to 100 ng. 8A4 mAb was previously validated to be strongly positive for CHIKV binding in indirect ELISA.Citation67 The negative control consists of wells not coated with CHIKV. Absorbance value (Abs) was measured at 450 nm. Mean abs is derived from 3 independent experiments performed in duplicates and ± SD values are shown as error bars.
Anti-CHIKV IgM specifically and strongly neutralizes CHIKV
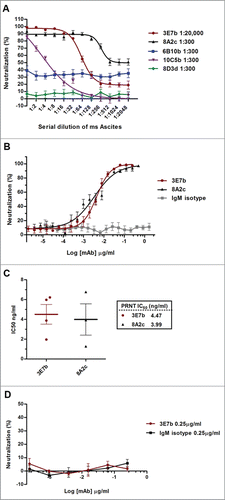
To evaluate if there is a dose-dependent neutralizing efficacy of mice ascites and purified IgMs against CHIKV, an in vitro Plaque Reduction Neutralization Test (PRNT) was optimized for this purpose. In brief, mice ascites or purified IgMs were serially-diluted in sterile phosphate-buffered saline (PBS) before each dilution was incubated with 100 plaque-forming units (PFU) of CHIKV for 1 h at 37°C.The mixture was then added to confluent BHK-21 cells. Consistent with results from the earlier immunofluorescence neutralization assay screening, mice ascites of 3E7b, 8A2c and 10C5b clones strongly inhibited CHIKV replication (). In particular, 3E7b ascites remained effective at a high dilution of 1:20,000. In contrast, 8D3d ascites failed to be neutralizing even though it showed strong efficacy in previous neutralization screening. As the exact IC50 could not be determined from mice ascites test, PRNT was further performed on purified IgM. Both the purified 3E7b and 8A2c are strongly neutralizing against 100 PFU of CHIKV across the wide range of mAb concentration tested (). Based on the dose-dependent neutralization curves obtained, 3E7b has an IC50 of 4.47 ng/ml while 8A2c has IC50 of 3.99 ng/ml, suggesting that 8A2c is slightly more neutralizing than 3E7b (). Taken together, both 3E7b and 8A2c were remarkably potent against CHIKV replication.
Figure 2. PRNT analysis of (A) complement-inactivated mice ascites and (B) purified IgM against CHIKV infection. Both ascites and purified IgM were 2-fold serially diluted in PBS before incubating with 100 PFU of CHIKV. Non-linear regression analysis of 3E7b and 8A2c are performed and best-fitted to dose-dependent inhibition curves (GraphPad Prism 6). Due to non-convergence of IgM isotype mAb data points, dose-inhibition curve is not applicable. (C) Grouped scatter plot of IC50 values from 3E7b and 8A2c non-linear regression describes quantitative evaluation of the mAb neutralizing potency. The thick line represents the mean value. (D) Post-CHIKV binding neutralization assay was performed by having CHIKV attached to BHK cells prior to 3E7b binding. Error bars are representative of ± SEM where at least 3 independent sets in duplicates were performed.
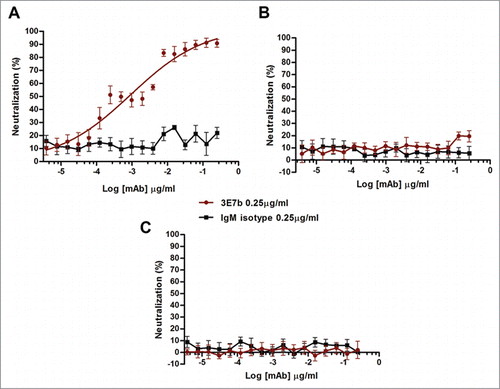
To elucidate the plausible mechanism of neutralization, a post-PRNT was performed to determine if the neutralization efficacy is maintained when 3E7b was added after CHIKV has bounded to the cell surface. CHIKV of 100 PFU was added to BHK cells at 4°C to allow virus binding for 1 h, followed by incubation with 3E7b for 30 min at 4°C. Subsequently, PRNT was performed as described earlier. Here, it is notable that 3E7b failed to block CHIKV replication after virus-host cell attachment has occurred (). This suggests that 3E7b-mediated neutralization of CHIKV requires a pre-incubation of the mAb with CHIKV, probably to allow its binding to the viral E2 epitopes to form 3E7b-CHIKV complex. Therefore, this leads to ineffective CHIKV attachment when the mixture is added to BHK cells.
Next, we investigated whether 3E7b could have a broad neutralizing efficacy against other CHIKV strain or alphaviruses. Here, the standard PRNT was performed by incubating 3E7b with 100 PFU of CHIKV Ross strain, Sindbis virus (SINV) or Ross River virus (RRV), respectively. SINV is a representative member of the Sindbis serocomplex while RRV shares the same Semliki Forest Virus serocomplex with CHIKV. Relative to the IgM isotype control, 3E7b neutralized CHIKV Ross dose-dependently, but no evident neutralization was observed against SINV and RRV replication (). This suggests that 3E7b is specific in CHIKV neutralization. However, compared to CH122508 strain (), 3E7b was less effective in neutralizing CHIKV Ross replication. Though both CH122508 and Ross strains belong to the same ECSA lineage, differences in the 3E7b neutralizing epitopes may contribute to the lower potency against CHIKV Ross replication.
Figure 3. PRNT analysis of 3E7b IgM against (A) CHIKV Ross strain, (B) Sindbis virus (SINV) or (C) Ross River Virus (RRV). Non-linear regression analysis of 3E7b against CHIKV Ross strain is performed and best-fitted to dose-dependent inhibition curve. However due to wide variability, the derived IC50 is not applicable. Non-convergence analysis are obtained in (B) standard PRNT of SINV and (C) RRV at 100 PFU. Error bars are representative of ± SEM where at least 3 independent sets in duplicates were performed.
CHIKV escape mutants against 3E7b neutralization contained 3 mutations in viral E2 protein
To identify critical amino acid residues required for CHIKV neutralization, escape mutant virus against 3E7b was generated. CH122508 strain was sequentially passaged in BHK cells for 10 rounds under increasing selective pressure from 3E7b, followed by 2 additional rounds of virus plaque-purification to isolate individual escape clones. In parallel to 3E7b, CHIKV was also passaged in PBS (the absence of mAb as mock selection) and in the presence of IgM isotype (as IgM specificity control). A total of 69 escape clones, namely 57 CHIKV/3E7b clones, 8 CHIKV/IgM clones and 4 CHIKV/PBS clones were obtained. When representative escape clones were further evaluated in IFA, it was shown that CHIKV selected from 3E7b neutralization abolished binding to 3E7b while CHIKV selected from IgM isotype and PBS treatment retained binding ability to 3E7b (data not shown). This implies a likely escape mutation on CHIKV protein that conferred resistance to 3E7b binding.
To further assess if this mutation was associated with a loss of neutralization, a representative escape clone CHIKV/3E7b 1a was analyzed in PRNT. Both CHIKV/PBS 1a and CHIKV/IgM 1a clones were susceptible to 3E7b neutralization with IC50 of 0.12 μg/ml and 0.17 μg/ml, respectively (). However, CHIKV/3E7b 1a clone was not neutralized by 3E7b at 2.5 μg/ml, the highest concentration used earlier for the generation of escape mutation. Viral RNA from CHIKV/3E7b escape clones were reverse-transcribed and sequenced for the entire length of viral E1 and E2 gene. Notably, all 7 representative CHIKV/3E7b clones showed 3 single point mutation in E2 gene that results in E→D at residue 24 (E24D), N→D at residue 218 (N218D) and D→G at residue 223 (D223G) in the E2 protein (). No mutation was identified in CHIKV/PBS 1a, CHIKV/IgM 1a, CHIKV/IgM 4a and the wildtype CH122508 strain E2 sequence, thereby suggesting that escape mutations are specific only to CHIKV that has been selected under the neutralizing pressure of 3E7b. In addition, none of the escape clones, represented by CHIKV/3E7b 3a and 7a clones, as well as CHIKV/IgM 4a and CHIKV/PBS 1a control clones have any mutation in E1 gene (data not shown). All E2 mutations are single amino acid substitutions that did not cause translational frameshift and CHIKV escape mutants remained detectable by a positive mAb control, the anti-CHIKV E2 IgG in IFA (data not shown). Taken together, the escape mutations did not cause any global effect on E2 conformation.
Figure 4. Identification of Escape Mutations in CHIKV E2. (A) PRNT was performed to investigate the escape ability of plaque-purified CHIKV/3E7b clone to mAb 3E7b. 2.5 μg/ml of 3E7b, 5 μg/ml of IgM isotype control or PBS (no mAb) was evaluated against a representative clone CHIKV/3E7b 1a diluted to 1000 PFU. One independent experiment was performed in duplicates as it is not possible to reproduce variability from the same mutant clone stock. CH stands for CHIKV while ~ indicates neutralization. (B) Sequencing chromatogram of CHIKV/3E7b 1a clone shows 3 single base A substitution in CHIKV E2 gene, which translates to E→D, N→D and D→G amino acid mutation at position 24, 218 and 233 of E2 protein, respectively. All CHIKV/3E7ba 1a-7a clones showed the same escape mutations at these residues when compared to sequences of wildtype CH122508 strain. No mutation was identified in CHIKV/PBS 1a, CHIKV/IgM 1a and 4a clones. Sequence analysis was performed using SeqTrace software.Citation63 8585, 9166 and 9186 refers to the nucleotide position. (C) Structural localization of 3E7b escape epitopes on CHIKV E1/E2 heterodimer modeled on CH122508 strain (GenBank Accession number: FJ445502). A top-down view is shown with E1 (orange), and E2- domain A (yellow), domain B (violet), C (cyan), beta-ribbon connectors (green) and transmembrane helix (gray). Arrow denotes the direction of E1/E2 protein complex relative to the viral membrane. Side-chain of escape residues, E24, N218 and D223, are presented as stick structures that show carbon and hydrogen atoms (white), oxygen atom (red) and nitrogen (blue). Amino acid distance is measured in angstroms (1Å = 0.1 nm) and figure is prepared using Pymol software.Citation65
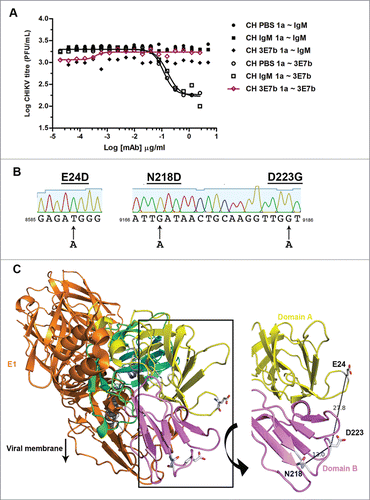
Various studies have demonstrated that an escape mutation on viral envelope protein represents a critical mAb epitope for virus neutralization. Using the crystal structure of mature E1/E2 glycoprotein complex (PDB: 3N43) of CHIKV, we mapped the 3 identified escape mutations onto this model. E24 is found to be located on the N-terminal hairpin of domain A while N218 and D223 are both located on the outermost tip of domain B (). Collectively, these residues are positioned on random linker coils between the beta sheets of domain A and B. Relative to E24, both N218 and D223 are spatially closer to each other, with a proximity of 13.0 Å. Hence, N218 and D223 could constitute the epitope for 3E7b binding to the native conformation of CHIKV E1/E2 heterodimer. Notably, all 3 residues are also highly surface accessible, with estimated values of 89.9%, 69.2%, 77.6%, respectively.Citation38 This suggests that 3E7b can freely bind to these epitopes on the native virion without much steric hindrance. Given its high surface accessibility, it is also favorable for these mutations, E24D, N218D and D233G, to occur for escape neutralization from 3E7b.
E2-N218D mutation confers CHIKV resistance to 3E7b binding and neutralization
In the interest of investigating the functional role of individual E2 mutation in mediating escape from 3E7b neutralization, reverse genetics technique was employed to generate full-length infectious CHIKV clones that contained a single mutation of E24D, N218D or D223G. These single mutant clones are named as CHIKV-E24D, CHIKV-N218D, CHIKV-D223G. Together with CHIKV-IVT (CHIKV generated from in vitro transcribed-infectious clone cDNA template) and CHIKV-WT (wildtype CH122508 strain), they were evaluated for their replication competency in BHK cells. Viral supernatant were collected at specific timepoint p.i. and quantitated by plaque assay. All CHIKV mutant clones demonstrated similar growth kinetic profile to CHIKV-IVT and CHIKV-WT virus controls (Fig. S2A). Collectively, the virus titer rapidly increased within the first 36 h p.i and infectious cDNA clone-generated CHIKV displayed higher replication efficacy relative to CHIKV-WT control. Among these clones, minor differences in the peak of virus production were also observed. However, there were no evident differences in the plaque size and morphology of the mutant clones relative to CHIKV-IVT and wildtype controls at 24 h p.i (Fig. S2B). Taken together, these data suggest that the engineered mutation in viral E2 protein did not greatly affect mutant virus replication or extracellular spreading of progeny virus in BHK cells.
Next, we investigated if the single, surface-exposed mutation on CHIKV E2 protein could affect virus binding to 3E7b. A dual-color IFA assay was performed on BHK cells-infected with CHIKV-E24D, CHIKV-N218D and CHIKV-D223G at MOI 10, respectively. CHIKV E2 binding to 3E7b was tested by co-staining of viral-infected cells with 3E7b IgM and anti-CHIKV E2 IgG positive control followed by quantification of the respective signals. Interestingly, the binding of CHIKV-N218D to 3E7b was significantly reduced where 3E7b-FITC signal was absent on the cell membrane and only some diminished signal was seen in the cytosol of the infected cells at day 1 p.i. (; white arrowheads). On the contrary, CHIKV-E24D and CHIKV-D223G retained strong binding efficacy to 3E7b similar to CHIKV-WT and CHIKV-IVT controls. Comparatively, only 2.1% of the cells infected with CHIKV-N218D were positively stained with 3E7b relative to 95.3% and 99.5% binding in CHIKV-WT and CHIKV-IVT, respectively (). In the latter CHIKV control-infected cells, colocalized signal arising from bound 3E7b-FITC and anti-E2 594 was consistently observed in the ER and plasma membrane. This indicates intracellular CHIKV E2 synthesis at the ER and localization of CHIKV E2 at the plasma membrane for progeny virus budding. From these observations, only N218D mutation, but not E24D and D223G, is critical for 3E7b binding.
Figure 5. Evaluation of mutant CHIKV binding and neutralization by mAb 3E7b. (A) Confluent BHK-21 cells were infected with CHIKV-WT, CHIKV-IVT, CHIKV-E24D, CHIKV-N218D or CHIKV-D223G clones at MOI 10. At day 1 p.i., cells were fixed and co-stained with 3E7b IgM and rabbit anti-E2 polyclonal antibody as a positive control. The difference in binding was visualized with goat anti-mouse IgM FITC and anti-rabbit IgG 594 secondary antibodies. Cell nuclei were stained DAPI and images were captured under 10x and 100x magnification. White arrowhead indicates reduced 3E7b binding. Representative images from 3 independent experiments are shown. (B) FITC signal of 3E7b binding is expressed as a percentage over 594 signal that represents the number of infected cells, n, quantitated. Error bars represent ± SD. One-way ANOVA followed by a post-Dunnett test, with statistical significance defined as ***p < 0.001. (C) PRNT of single mutant CHIKV clones was performed where 100 PFU of the respective CHIKV clone was neutralized with serially diluted 3E7b from 3.81 pg/ml to 0.25 μg/ml. Non-linear regression analysis was performed and data was best-fitted to dose-dependent inhibition curves for CHIKV-WT, CHIKV-IVT, CHIKV-E24D and CHIKV-D223G. Due to non-convergence of CHIKV-N218D data points, dose-inhibition curve and IC50 are not applicable. Mean value from at least 3 independent experiments performed in duplicates are presented. Error bars represent ± SEM.CH; CHIKV. a; ambiguous.
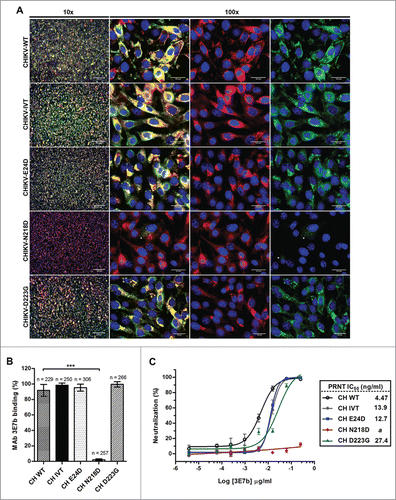
Poor binding of 3E7b to CHIKV due to E2-N218D mutation suggests that N218 could be a critical neutralizing epitope. To validate this hypothesis, PRNT was performed using 0.25 μg/ml of 3E7b, the neat concentration optimized previously. Respective IC50 values of mutant clones were calculated by non-linear regression analysis and compared to CHIKV-WT and CHIKV-IVT controls. Evidently, CHIKV-N218D was able to confer resistance to 3E7b neutralization, whereas CHIKV-E24D and CHIKV-D223G mutants were neutralized effectively over the tested range of 3E7b concentration from 3.81 pg/ml to 0.25 μg/ml (). Relative to CHIKV-WT, the IC50 values of 3E7b from neutralization of CHIKV-IVT, CHIKV-E24D and CHIKV-D223G are at least 2.6-fold higher. Decreased neutralization efficiency could be due to inherent differences in infectious-clone generated CHIKV clones. Nonetheless, the overall neutralization trend was similar among CHIKV-WT, CHIKV-IVT and mutant clones except for CHIKV-N218D.
In order to determine if N218 vary among different lineages of CHIKV, sequences flanking the mutation were aligned with all existing CHIKV structural polyprotein and E2 sequences in GenBank and PDB. These escape mutations were found to be 100% identical with >178 published sequences. In this alignment, high sequence homology is also shared between different CHIKV genotypes, namely East/Central/South African (ECSA), Asian and West Africa lineage (data not shown). Therefore, this implies that the conserved escape residues on E2 could be critical for CHIKV replication across different genotypic strains. Taken together, our findings validate that N218 residue is highly conserved on CHIKV E2, and it is an important epitope for potent neutralization by mAb 3E7b.
Prophylactic administration of 3E7b protects mice from CHIKV disease
Having shown that 3E7b was potently neutralizing in vitro, it is necessary to investigate if the mAb can similarly exert antiviral efficacy in vivo. Herein, 3E7b was tested in a neonate mouse model of CHIKV infection that was previously well-established in our laboratory. In this model, 6-day old BALB/c mouse was shown to be highly susceptible to CHIKV infection, specifically to CH6708 strain (LK(EH)CH6708, GenBank Accession number: FJ513654). As they have yet to develop a mature innate immunity, this model displayed clinical features similar to those reported in CHIKV-infected patients.Citation39,40 This suggests that neonate mouse is an ideal in vivo model for recapitulating CHIKV pathogenesis. Based on our previous laboratory findings, CHIKV infection was strain-specific in our neonate mouse model where mice succumb only to CH6708 infection with CHIKV disease manifestations. Indeed, we observed that during the acute phase of CHIKV infection at day 2–4 p.i., neonate mice developed high viremia and viral loads in various tissues including the hind limb muscles, liver, spleen and brain (data yet to be published). High CHIKV load is accompanied with an onset of hind limb paralysis at day 3–4 p.i., which is characteristic of CHIKV disease, followed by lethality within day 5–9 p.i. Because CH6708 strain shares a high 99.7% genomic similarity with CH122508 strain, it is considered identical to CH122508 strain for testing of 3E7b efficacy in vivo. Using this 6-day old BALB/c mouse model, the prophylactic efficacy of 3E7b was assessed over a mAb dose range of 0.2, 2, 20 or 200 μg per mouse that was given intraperitoneally at 24 h and 8 h prior to infection with 4 × 105 PFU of CH6708 strain. IgM isotype antibody and PBS (mock-mAb) were included as irrelevant mAb treatment and diluent control, respectively.
All control mice that received PBS succumbed to lethal CHIKV infection by day 9 p.i., while IgM isotype-treated mice showed poor survival (40%) at day 14 p.i. (). However, mice pre-treated with 3E7b were able to overcome CHIKV-induced lethality, with a good baseline level of 80% survival across the given doses. Dose-dependent protection is highly significant by log-rank test (p < 0.001), with 80% survival in 0.2 μg and 2 μg 3E7b treatment groups and 100% survival in 20 μg and 200 μg treatment. To further assess the minimum prophylactic dose required during the acute phase of CHIKV infection, mice were given the same treatment regimen, sacrificed on day 2 p.i. and virus titer in the serum and various tissues were quantitated. Relative to PBS-infected control, pre-treatment with 2 μg of 3E7b significantly cleared CHIKV burden in serum () and other virus-targeted tissues, namely, limb muscles () (p < 0.05), brain () (p < 0.001), liver () and spleen () (p < 0.01). Dose-dependency is also consistently observed where 0.2 μg of 3E7b pre-treatment failed to confer much protection against CHIKV infection in these mice tissues.
Figure 6. Prophylactic evaluation of mAb 3E7b against CHIKV-induced lethality in a neonate mouse model. Five-day oldBALB/c mice were intraperitoneally (i.p.) injected with PBS, IgM isotype or mAb 3E7b at 24 h and 8 h prior to infection with CHIKV of 4 × 10Citation5 PFU via i.p. injection route. Infected mice were then monitored daily for their (A) survival and other clinical symtoms of CHIKV disease. Kaplan-Meier survival curves of the respective mAb dose are plotted and statistically analyzed by log-rank test (Graphpad prism 6). (B–F) Neonate mice were subjected to the same treatment and infection regimen as described. Serum and various tissues of infected mice were harvested at day 2 p.i and quantitated by CHIKV titer in standard plaque assay. Thick line represents the mean virus titer and dashed line indicates the limit of the detection in which data points on the x-axis refer to zero virus titer from zero plaque. Kruskal-Wallis test was performed across all treatment followed by a Dunn's post test to PBS infected control. Statistical significance is defined as *p < 0.05 and **p < 0.01 (GraphPad Prism 6).
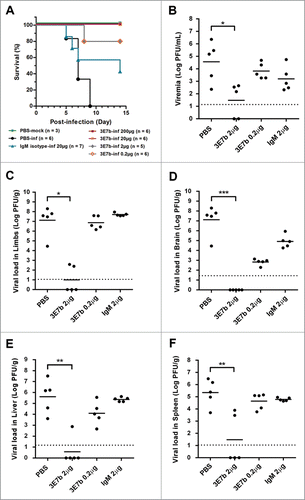
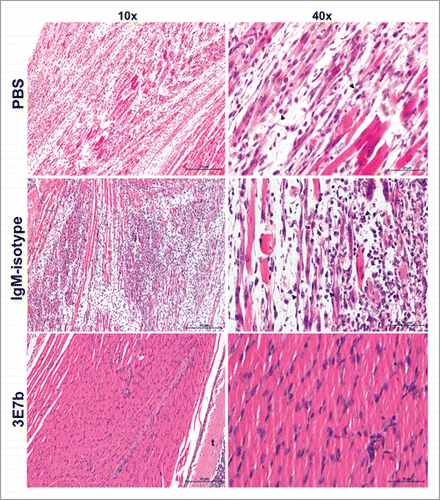
Figure 7. Histological analysis of limb muscles of mice pre-treated with 3E7b. Neonate mice that were mock-infected or pre-treated with PBS, 20 μg of IgM isotype or 3E7b were sacrificed on day 7 p.i. and harvested for their limbs. Limb muscles were paraformaldehyde-fixed, decalcified and processed according to the standard procedures for hematoxylin & eosin staining. Images are viewed and captured under 10× and 40× magnification of BX43 Olympus microscope. Representative images are shown with scale bar of 20 μm. b, bone; t, tendon.
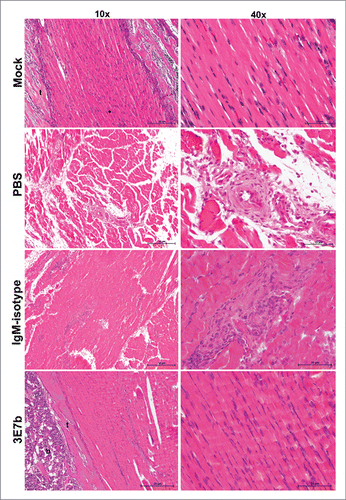
Having shown that 3E7b effectively reduced CHIKV load in limb muscles at day 2 p.i.,we further examined if the protective efficacy of 3E7b can be sustained in the skeletal muscles during the course of CHIKV infection. Mice hind limbs were harvested at day 7 p.i, paraformaldehyde-fixed and procesed for Hematoxylin and Eosin (H&E) staining. The skeletal muscles of PBS-treated mice showed extensive necrosis with loss of striations and infiltration of inflammatory cells (). IgM isotype-treated mice showed less severe muscle necrosis and inflammation relative to PBS control. In contrast, hind limb muscles of 3E7b-treated mice retained healthy striated morphology as seen by absence of abnormality or necrosis. Therefore, pre-treatment of 3E7b could have prevented CHIKV dissemination to muscle tissues and thereby protects against CHIKV-induced muscle pathogenesis.
Therapeutic administration of 3E7b protects mice from CHIKV disease
Based on the findings from prophylactic treatment, consecutive doses of 20 μg 3E7b were effective in preventing CHIKV-induced lethality and limb muscle pathology. Therefore, it is interesting to assess if the same dose could also protect mice from CHIKV infection when given at post-CHIKV exposure. Using the same neonate mouse model, 6-day old mice were infected with 4 × 105 PFU of CHIKV prior to receiving a single 20 μg dose of 3E7b at 4 h or 8 h p.i. Mice given PBS diluent or IgM isotype showed poor survival (less than 20%; ). For mice post-treated with 3E7b at 8 h p.i., survival was improved to 50% (p < 0.05). However, when given at 4 h p.i., 3E7b was significantly effective (p < 0.01), protecting all mice against CHIKV-induced lethality. This suggests a time-dependent neutralizing efficacy of 3E7b during acute infection where there is high CHIKV burden.
Figure 8. Therapeutic evaluation of mAb 3E7b against CHIKV-induced lethality in a neonate mouse model. Six-day old BALB/c mice were i.p injected with CHIKV of 4 × 105 PFU prior to administration of PBS, 20 μg of IgM isotype control or 3E7b at 8 h or 4 h p.i. (A) Survival (%) till 14 day p.i. is presented as Kaplan-Meier survival curves and statistically analyzed by log-rank test (GraphPad Prism 6). (B–F) Neonate mice were CHIKV-infected as described and post-treated with 20 μg of 3E7b at 4 h p.i. At day 2 p.i., serum and various tissues were harvested and quantitated for CHIKV titer. Thick line represents the mean virus titer and dashed line indicates the limit of the detection. Kruskal-Wallis test was performed across all treatment followed by a Dunn's post test to PBS infected control. Statistical significance is defined as *p < 0.05 and **p < 0.01 (GraphPad Prism 6).
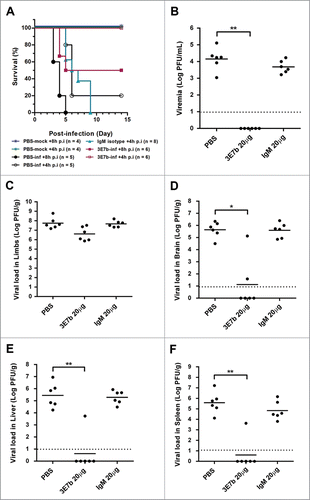
With a defined 3E7b therapeutic window at 4 h p.i., we further evaluated the level of CHIKV load in the serum and various mice tissues at day 2 p.i. Relative to PBS-infected control, 3E7b was significantly effective (p < 0.01) in reducing viral loads in the serum (), liver () and spleen (), while a lower significance is observed in the brain tissue () (p < 0.05). There was at least a 4-log decrease in CHIKV titer consistently in these tissues. However, a high level of CHIKV load was consistently detected in the hind limb muscles (). Histological analysis of mice limb muscles at day 7 p.i. revealed partial protection, where some areas of the muscles retained healthy morphology () while some areas were evidently necrotized with infiltration of inflammatory cells. Despite such muscle pathology, these mice did not display limb paralysis and were able to survive against CHIKV infection over 14 days p.i. Taken together, this suggests that the protective effect of 3E7b against CHIKV infection in other tissues could have helped to overcome CHIKV-induced lethality. Therefore, these data demonstrate that therapeutic administration of 20 μg 3E7b was protective in most CHIKV-targeted tissues and could improve the overall CHIKV disease outcome.
Figure 9. Histological analysis of limb muscles of mice post-treated with 3E7b after CHIKV infection. As described previously, limb muscles of infected mice at day 7 p.i. were harvested, fixed and processed for hematoxylin & eosin staining as described. Images are viewed and captured under 10× and 40× magnification of BX43 Olympus microscope. Representative images are shown with scale bar of 20 μm. Black arrowhead points to inflammatory cell. t, tendon.
Discussion
At present, neutralizing CHIKV-specific mAbs that were characterized and reported belong to IgG isotype, and they were either produced in mice by a combination of passive immunization and infectious CHIKV challengeCitation29,31 or derived from the sera of patients.Citation30,33,34 In this study, we inoculated live CHIKV in mice to generate envelope protein-specific mAbs that could confer strong neutralization potency. From PRNT analysis, 2 of our anti-CHIKV IgMs, 3E7b and 8A2c, demonstrated strong neutralization potency of 4-5 ng/ml in CHIKV-infected BHK cells. The most potent anti-CHIKV mAb previously described, CHK-152 IgG, has an in vitro IC50 of 1–3 ng/ml in Vero and mouse fibroblast cells.Citation29 Because IgM is a pentamer with 10 antigen-binding sites, the antibody has high avidity and could more effectively neutralize CHIKV antigen compared with bivalent IgG. Further neutralization studies of 3E7b and 8A2c on other cell lines can be performed to determine if there are cell type-specific factors involved in IgM-mediated CHIKV neutralization.
The efficacy of antibody neutralization is pre-determined by factors such as epitope affinity, accessibility and occupancy.Citation41 The effective binding of 3E7b to CHIKV observed in IFA and indirect ELISA suggests that the mAb epitope(s) are situated conformationally on the surface of native CHIKV E1 or E2 protein. Because not all binding epitopes are neutralizing, we further identified a few amino acids that constitute or associate with the neutralizing epitope of 3E7b. Escape mutant CHIKV, namely, E24D, N218D and D233G were generated from 10 neutralization rounds under 3E7b selective pressure and they collectively localized to surface-exposed regions on CHIKV E2 ectodomain with high solvent accessibility. E24 is situated on the N-terminal beta-hairpin known as the N-flap, N218 is at the tip of domain B on strand E and D223 is part of a 310 helix secondary structure.Citation16 Of these 3 residues, N218 and D223 are spatially closest to each other in 13.0 Å, indicating that they could be within the footprint of an antibody paratope. Further analysis using infectious CHIKV mutant clone showed that only N218D mutation significantly reduced CHIKV binding (p < 0.0001) and resisted neutralization by 3E7b (), thereby highlighting that E2-N218 is a critical epitope for 3E7b binding and neutralization.
Based on these findings, we postulate a plausible mechanism of 3E7b-mediated CHIKV neutralization. The IgM binds to a surface-exposed residue, N218, on viral E2 to form 3E7b-CHIKV aggregates that prevent virus attachment to the cell surface receptor. E2-N218 interaction with 3E7b might be associated with neighboring E24 and D223 residues within the antibody paratope where these residues indirectly stabilize 3E7b-CHIKV aggregates. Further Fab-docking studies are required to elucidate a detailed 3E7b binding mechanism to CHIKV E2 protein. As IgM binds with high flexibility to poliovirus in stellate and staple-like conformations,Citation42 it may also be useful to unveil the binding conformation of 3E7b to CHIKV for better understanding on the mechanistic role of IgM in virus neutralization. Functionally, the role of E2-N218 in CHIKV neutralization has yet to be reported in other CHIKV-specific IgG studies. However, the residue might be an important antigenic determinant involved in alphavirus neutralization. Consistent with analysis on SINV escape variants that charged residues are important for antibody interaction,Citation43 N218D mutation has resulted in substitution of a non-charged residue, Asn, to a charged Asp residue. In addition, a nearby residue, E2-N216 of RRV, SINV and Venezuelan equine encephalitis virus, was selected to confer virus escape from neutralizing antibodies.Citation43-45 Mutation on E2-218 was also found to adapt RRV binding to heparan sulfate, the cell surface glycosaminoglycan involved in alphavirus receptor binding.Citation46 Taken together, these studies suggest that N218 might be a conserved epitope associated in neutralization of other alphaviruses.
Understanding that CHIKV E1-A226V mutation and epistatic mutations on E2 protein have enabled CHIKV to adapt to a new Aedes mosquito vector,Citation47,48 future mutation may occur on less conserved regions of the viral structural proteins. Hence, there is a need to generate neutralizing mAbs recognizing conserved epitope to help mitigate disease severity caused by CHIKV strains from a wide geographical range.Citation49 In our study, we further evaluated whether mAb 3E7b could neutralize: 1) Ross, another CHIKV strain of ECSA lineage; 2) RRV, which shares the same SFV antigenic complex as CHIKV; and 3) SINV, a related alphavirus that has different antigenic complexity to CHIKV. 3E7b was found to cross-react with CHIKV Ross strain at a lower neutralization potency. Although E2-N218 is highly conserved in the ECSA lineage of CH122508 and Ross strains, there could be differences in other neutralization epitopes on CHIKV Ross. Similarly, the lack of neutralization efficacy against RRV and SINV could be due to sequence differences in E2 antigenicity that involved other neutralizing epitopes, or other virus-specific factors. Nevertheless, sequence analysis of N218 residue with more than 178 published CHIKV strains in GenBank and PDB database revealed 100% conservation of this residue. Thus, our present findings can be expanded to evaluate 3E7b on a broader range of CHIKV genotypic isolates, as demonstrated by anti-CHIKV mAb 5F10 and 8B10.Citation30 With the identification of cross-reactive IgM epitopes, 3E7b can be useful for broad-spectrum epitope vaccine design against CHIKV and other related alphaviruses.
Upon CHIKV infection, naturally occurring antibodies are predominantly IgMs, which provide the first line of defense by neutralizing CHIKV in the serum. Due to its multivalency, IgM can efficiently mediate virus aggregation, thereby facilitating their recognition and destruction by cytotoxic T cells.Citation42,50 However, the role of IgM in pre- and actual clinical setting is not well-understood. CHIKV-specific IgMs were found to persist in patients with chronic arthritis,Citation4,51,52 thus they could serve as biomarkers for CHIKV disease. In a broader context, analysis of IgM response in humans with alphaviruses infection has revealed the potential usefulness of IgM as a diagnostic tool especially during endemics.Citation53 Indeed, IgM-based ELISA has been developed for CHIKV serodiagnosis of adult patients infected with La Reunion strain.Citation54 At present, there are no prior studies done on understanding the use of CHIKV-specific IgM as an immunotherapy in patients and animal models.
To evaluate in vivo efficacy of anti-CHIKV IgM, passive transfer of 3E7b was performed in our laboratory-established mouse model. Neonate mice are susceptible to CHIKV infection due to the lack of a fully-developed immune system.Citation39 Hence, they can recapitulate hallmarks of CHIKV pathogenesis similar to clinical observations in patients. In this study, prophylactic dosage of 3E7b at 2 μg greatly improved disease outcome, with a survival of 80% and efficient virus clearance in the serum and CHIKV-targeted tissues, namely the limb, liver, spleen and brain. Relative to prior studies that achieved 80-100% survival with 4 μg or 250 μg of prophylactic anti-CHIKV mAbs,Citation33,55 2 μg of 3E7b is a potent dose against CHIKV infection. Recent studies showed that passive mAb transfer prior to CHIKV challenge protected mice against CHIKV-induced arthritis and swelling in the foot and joint.Citation29,31,33 However, there are no studies evaluating mAb efficacy against CHIKV-induced myalgia. Moreover, because IgM has a short serum half-life and elicited response during the acute phase,Citation36 we investigated if the protective efficacy of 3E7b could be sustained in the muscle tissues. We thus examined at day 7 p.i. the limb histology of neonate mice pre-treated with 20 μg of 3E7b. These mice showed healthy limb muscle morphology, in contrast to severe necrosis and inflammation in the control groups and other CHIKV mice models that revealed necrotizing myositis and mixed inflammatory infiltrates at day 7 p.i.Citation56-59 This sustained protection against CHIKV-induced musclo-pathological changes implies that 20 μg of 3E7b is an optimal prophylactic dose.
To investigate whether 3E7b can be additionally protective in a post-infection setting, a single dose of 20 μg at 3E7b was administered to the mice at 4 h or 8 h after CHIKV challenge. Notably, 4 h post-treatment conferred 100% protection against CHIKV-induced lethality whereas protection was reduced to 50% when 3E7b was administered at 8 h p.i. As CHIKV caused high virus burden during the acute phase of infection, therapeutic dose of mAb is critical to improve the disease outcome. High virus load was detected in the limb muscles at day 2 p.i () and evident CHIKV-induced inflammation and necrosis was observed in some parts of the right limb muscle while other regions of the muscles retained healthy morphology at day 7 p.i (). This suggests that 3E7b could have partially protected against the onset of CHIKV-induced myalgia, possibly by supporting the host immunity and reducing CHIKV dissemination to muscle tissues. Mice sera, liver and spleen showed that CHIKV was significantly cleared (p < 0.01) to levels below the limit of the detection assay (< 10 PFU/ml), while CHIKV clearance was less significant in the brain tissue (p < 0.05). Because liver, spleen and brain are more vascularized than the muscles, we postulate that 3E7b was delivered efficiently to neutralize or prevent CHIKV infection in these tissues.
In conclusion, we successfully generated and characterized 3E7b, a novel IgM mAb that potently neutralized CHIKV infection in cell-based and neonate murine models. As 3E7b protected mice from CHIKV disease, including CHIKV-induced myalgia, when given 24 h and 8 h prior to CHIKV infection, its potential role as a prophylaxis can be further explored. Fab fragments of 3E7b can be derived for use in antibody docking on cryo-EM map of CHIKV to better elucidate the neutralization mechanism. This knowledge can be useful in future design of an epitope-based vaccine against CHIKV infection.
Materials and Methods
Cells and virus culture
BHK-21 (ATCC No. CCL-10) cells were maintained in RPMI-1640 medium supplemented with 10% FCS and incubated at 37ºC in 5% CO2. C6/36 mosquito cells (ATCC No. CRL-1660) were maintained in L-15 medium with 10% heat-inactivated FCS and incubated at 28ºC. Chikungunya virus SGEHICHD122508 (GenBank Accession number: FJ445502) or named as CH122508 strain was used in all cellular experiments in this study. CHIKV LK(EH)CH6708 (GenBank Accession number: FJ513654), named as CH6708, was used in mice experiments because this virus strain produced better clinical manifestations of CHIKV infection compared to CH122508 strain. These virus isolates were obtained from the serum of locally-infected patients and kindly provided by Environmental Health Institute of the National Environmental Agency (Singapore). Sequence analyses performed in our laboratory showed that CH122508 and CH6708 belong to the East/Central/South African (ECSA) lineage, and they represent currently circulating strains of CHIKV. CHIKV Ross (GenBank Accession number: AF490259), kindly provided by Dr. Ooi Eng Eong from DUKE-NUS, Sindbis virus (SINV) (GenBank Accession number: NC_001547) and Ross River Virus (RRV) (ATCC) were also used in the PRNT. To establish an infectious virus pool, confluent C6/36 cells were infected with local CHIKV strains, CHIKV Ross, RRV or SINV and maintained for 72 h to 96 h post-infection (p.i.). Viral supernatant was collected and clarified by centrifugation at 290 g for 10 min before quantitating the virus titer by standard plaque assays. For use in indirect virion-based ELISA, a large pool of CH122508 strain was grown up from 10 T75 flasks of C6/36 cell culture. Clarified viral supernatants were filter-sterilized through 0.22 μM filter and concentrated by 100-fold by centrifugation at 3000 g for 20 min in an Amicon Ultra 100K filter unit (Millipore, UFC910024). Concentrated virus was ultracentrifuged in 25% sucrose cushion at 25,000 rpm for 2.5 h at 4°C (Beckman L-90 and rotor SW41 Ti). Following which, virus pellet was resuspended in PBS and subjected to another round of ultracentrifugation. Purified CHIKV pellet obtained was then resuspended in PBS and plaque-assayed for virus titer.
Mouse mAb production and purification
BALB/c mice used in this study were housed in a pathogen-free BSL2-facility in Vivarium, CeLS building in National University of Singapore. All animal experiments were reviewed and approved by Institutional Animal Care and Use Committee (IACUC) of YLLSoM faculty, NUS (Protocol No. R14/78), while ascites production was performed according to the protocol approved by IACUC of the Biological Resource Centre, A*Star, Singapore (Protocol No. 110694). All procedures were carried out in strict accordance with the recommendations of the National Advisory Committee for Laboratory Animal Research guidelines in Singapore. All efforts were made to minimize animal suffering and euthanasia was performed using an overdose of carbon dioxide.
Three-week old female BALB/c mouse (n = 5) were intraperitoneally injected with 800 μl of CH122508 strain (5 × 107 PFU) for 3 times consecutively at 1-week interval. Mice serum was harvested after the second and third CHIKV challenge and analyzed via Western blotting for presence of immune response to CHIKV. After the fourth CHIKV boost, mouse spleen was harvested and splenocytes were fused with myeloma cells according to the instructions in ClonaCell-HY Hybridoma Cloning kit (StemCell Technologies, 03800). The resultant successful hybridoma cells were cultured according to a previously described protocol.Citation60 Hybridoma clones were screened via IFA for the secretion of neutralizing anti-CHIKV mAbs in the crude culture supernatant. Positive hydridoma clones were further cloned by limiting dilution and expanded. Resultant subclones secreting neutralizing anti-CHIKV mAbs were further screened by immunofluorescence neutralization assay. Hybridoma subclones were then introduced into the peritoneal cavities of BALB/c mice. Following the production of ascites, mAbs were purified as described according to an optimized protocolCitation61 and with the use of an IgM purification kit (Pierce, 44897). Final mAb purity and yield were analyzed by SDS-PAGE gel electrophoresis and BCA assay (Pierce, 23225), respectively. Purified mAb was isotyped with isostrip mouse monoclonal antibody isotyping kits (Roche, 11493027001).
Characterization of mAb antigenicity
(i) Immunofluorescence Assay
Confluent BHK cells on coverslips were infected with CHIKV CH122508 strain or single mutant CHIKV clones at MOI 10. At 24 h p.i, cells were fixed with methanol at −20°C, washed extensively with PBS and stained with mouse anti-CHIKV IgM, 3E7b or 8A2c at 1:100 dilution in PBS. Double-staining of mutant CHIKV-infected cells were performed using 3E7b and rabbit anti-E2 13893 B3 (in-house produced) at 1:100 or 1:300. Secondary antibody staining was performed with goat anti-mouse IgM FITC (Pierce, 31992) or anti-rabbit DyLight 594 conjugate (Pierce, 35560) at 1:300 or 1:500 dilution. Coverslips were washed with PBS after every staining step and finally mounted onto glass slide using Duolink in situ mounting media with DAPI (Sigma-Aldrich, DUO82040). Images were captured at 10x and 100x under fluorescence microscopy of DAPI, FITC or TRITC channel (Olympus IX81 inverted microscope).
(ii) Indirect virion-based ELISA
Purified CHIKV was diluted in 50 μl of coating buffer (BD OptEIA, 550534) to a titer of 6 × 108 PFU and coated onto 96-well plate at 4°C overnight with shaking. Wells with coating buffer served as negative control. Following incubation, wells were washed with PBST buffer (PBS with 0.01% Tween-20) and BSA blocking buffer was added (2% BSA in PBST). After blocking at 37°C for 2h, wells were washed twice with PBST and anti-CHIKV IgM diluted to 0.1, 5, 10, 25, 50 or 100 ng, mouse mAb anti-CHIKV E2 8A4 IgG (a kind gift from Dr. Philippe Desprès, Institute Pasteur, France) or mouse IgM isotype clone GC323 (Millipore, MABC008) was added to each well. Concentration of CHIKV IgM was pre-quantitated by BCA assay. After the addition of primary mAbs, the ELISA plate was further incubated at 37°C for 1.5 h with shaking, washed thrice with PBST, and incubated with secondary anti-mouse IgM HRP at dilution of 1:5000 for another 1.5 h at 37°C with shaking. Wells were washed with a final round in PBST for 3 times and incubated with 100 μl of TMB substrate per well (BD OptEIA, 550534) for 10 min at room temperature. Reaction was stopped with 100 μl stop solution (BD OptEIA, 550534) and OD absorbance values were read at 450 nM using Tecan plate reader (Tecan i-control infinite 200). Negative controls consisting of “IgM + no CHIKV antigen coated” and blank wells consisting of “no IgM + no antigen coating” were included in the experiment. Mean OD values were expressed by subtracting negative control reading from positive “IgM + CHIKV coated wells” readings.
Neutralization Assay
Immunofluorescence neutralization assay screening
BHK-21 cells were seeded on 96-well plate to achieve 100% confluent monolayer. Equal volumes of CHIKV (CH122508 strain) of MOI 0.1 and culture supernatant of the hybridoma clone were mixed and incubated at 37°C for 1 h. Following that, the mixture was added to the confluent BHK cells and CHIKV infection was carried out at 37°C for 1.5 h. Cells were washed with PBS twice and maintained in RPMI with 2% FCS. At 24 h p.i., cells were methanol-fixed and standard IFA was carried out as described above. Primary antibody staining was performed using mouse monoclonal anti-CHIKV E2 IgG (a kind gift from Dr. Philippe Desprès, Pasteur Institute of France) at 1:300 dilution for 1 h at 37°C, followed by goat anti-mouse IgG FITC (Pierce, 31992) at 1:500 dilution for the next 1 h. Cells were finally stained with DAPI and viewed under DAPI and FITC channel at 10x magnification (Olympus IX81 microscope).
Plaque Reduction Neutralization Test
PRNT assay was adapted and modified from reported studies.Citation29,30,62 Mice ascites were complement-inactivated by heating at 55°C for 30 min prior to the assay. Anti-CHIKV IgM mAb at starting concentration of 0.25 μg/ml, 0.5 μg/ml or 1 μg/ml or mouse IgM isotype clone GC323 (Millipore, MABC008) at 1.7 μg/ml were 2-fold serially diluted in PBS. CHIKV (CH122508 strain) of 100 PFU was mixed with equal volume (1:1) of mAb for 1 h at 37°C. Following which, CHIKV-mAb mixture was added to confluent BHK-21 cell monolayer for 1.5 h at 37°C. Mock antibody control consists of CHIKV-PBS mixture. Following infection and PBS wash, cells were overlaid with 1% (w/v) carboxymethylcellulose (CMC/Aquacide II, Calbiochem, 9004-32-4) in RPMI supplemented with 2% FCS. After 72 h p.i., cells were stained with crystal violet dye containing 4% paraformaldehyde (PFA, Sigma-Aldrich, 158127). Relative percent of neutralization was calculated by expressing the total number of plaques per mAb dilution as a percentage of the number of plaques formed in PBS diluent (mock mAb control).
Values were fitted onto a dose-inhibition curve, from which non-linear regression was performed and IC50 was generated (Graphpad Prism 6). To validate cross-reactivity of 3E7b IgM with other strains of CHIKV or alphavirus, the above-described PRNT was also performed with CHIKV Ross strain as a representative of CHIKV East/Central/South African (ECSA) genotype, Ross River virus and Sindbis virus as representative of Semliki forest and Sindbis serocomplex, respectively. CHIKV Ross- and RRV-infected cells were crystal violet dye-fixed at 72 h p.i., while SINV-infected cells were fixed after 48 h p.i.
Post-binding neutralization assay
BHK-21 cells of 100% confluency were prechilled to 4°C before the cells were incubated with CHIKV 100 PFU for 1 h at 4°C. Cells were then gently washed twice with cold PBS, followed by incubation with mAb 3E7b or IgM isotype at 0.25 μg/ml diluted in PBS and RPMI with 2% FCS media for additional 30 min at 4°C. After which, cells were warmed to 37°C for 1 h to allow virus entry. Washes (3 times) with PBS were performed and infected cells were maintained in CMC overlay in RPMI media supplemented with 2% FCS. At 72 h p.i., cells were fixed with crystal violet dye and scored for plaques.
Neutralization escape mutant assay
CHIKV (CH122508 strain) was passaged consecutively under selective pressure from neutralizing 3E7b and under non-neutralizing condition using an equivalent mouse IgM isotype or PBS diluent. In brief, CHIKV of 105 PFU/ml was 10-fold serially diluted and each virus dilution was incubated at 1:1 volume with 3E7b 0.25 μg/ml, IgM isotype 0.5 μg/ml or PBS for 1 h at 37°C. Similar to PRNT, the mixture was used to infect confluent BHK-21 cells that were later maintained in RPMI media with 3E7b or IgM isotype at respective concentration used for neutralization. Cells were monitored for a cytopathic effect (CPE) for the next 4 days and virus supernatant was then harvested from the wells with the highest dilution of virus that showed extensive CPE of estimated 80-100%. The supernatant was subjected to additional 9 rounds of passage in BHK cells as described above. At every 2 to 3 consecutive passage, viral supernatant was incubated with increasing concentration of 3E7b at 0.5 μg/ml, 1 μg/ml or 2.5 μg/ml, respectively. After the 10th passage, the harvested viral supernatant was plaque-purified in BHK cells. In brief, cells infected with the viral supernatant were overlaid with 0.5% agarose in RPMI media containing 3E7b 2.5 μg/ml, IgM isotype 5 μg/ml or PBS. Following 48 h to 72 h p.i, visible plaques were isolated individually and further propagated in BHK cells for one passage. Viral supernatant was harvested at 48 h pi, when extensive CPE occur, and this was subjected to another round of plaque purification as described above.
Viral RNA extraction, reverse-transcription and sequencing
CHIKV genomic RNA was extracted from the viral supernatant of escape clones using QIAmp viral RNA mini kit (Qiagen, 52906) according to the manufacturer's protocol. Eluted CHIKV RNA was reversed transcribed by MMLV Reverse transcriptase (Promega) to obtain viral E1 or E2 cDNA. Following which, viral cDNA were amplified by PCR using GoTaq Green Master Mix (Promega, M7122) and primers designed (Sigma-Aldrich) to flank the complete CHIKV E1 or E2 gene. PCR products were electrophoresed, gel extracted and spin-column purified according to the manufacturer's protocol (Qiagen, 28106). Purified PCR products were sequenced at CHIKV E1 or E2 gene and resulting sequences were analyzed using SeqTrace software.Citation63
Computational analyses of neutralization epitopes
CHIKV E1/E2 protein structure of CH122508 strain was predicted based on the published CHIKV mature glycoprotein complex (PDB code: 3N43) by molecular superimposition using Phyre 2 software (http://www.sbg.bio.ic.ac.uk/phyre2/html/page.cgi?id=index).Citation64 Visualization and distance quantification of single epitopes were performed on Pymol software (www.pymol.org).Citation65 Single-point mutation was highlighted in the respective colors. Calculation of surface accessibility was additionally performed using POPS algorithm (http://mathbio.nimr.mrc.ac.uk/wiki/POPS).Citation38
CHIKV mutant clone generation
Site-directed mutagenesis inverse PCR
Site-directed mutagenesis of CHIKV E2 gene was carried out by inverse PCR cycling using CH122508 full-length genomic DNA pSMART-LCKan vector (Lucigen, 40821-2; GenBank Accession number: AF532106) that was established previously in our laboratory. Three sets of inverse PCR primers (Sigma-Aldrich) were constructed where each pair of forward and reverse primer contains the desired single-base escape mutation in E2 (E24D, N218D and D223G). Inverse PCR reaction was carried out using Long PCR enzyme mix (ThermoScientific, K0181) following manufacturer's conditions. PCR products were digested by Dpn-I (NEB, R0176S) at 37°C for 6 h and transformed into competent Escherichia coli strain XL10 gold in 1:5 ratio. Transformed culture was spread onto LB agar supplemented with 30 μg/ml kanamycin (MP Biomedicals, 0219453105) and incubated overnight at 30°C. Plasmid from colonies were isolated by Pureyield miniprep kit (Promega, A1222) and subjected to digestion by BglII (NEB, R0144S) and SmaI (NEB, R0141S) at 37°C for 1 h. After visualization on 0.7% ethidium bromide gel, positive clones were DNA sequenced using CHIKV E2 forward primer to confirm the presence of desired mutation (1st base sequencing, Singapore). Successful bacterial clones were further expanded, plasmid-extracted and column-purified.
In vitro transcription (IVT) of full-length virus mutant clone
CHIKV mutant DNA plasmid clones of 4 μg were linearized with NotI enzyme (NEB, R0189S) at 37°C overnight. Complete linearization was verified by gel electrophoresis and 1 μg of the linearized plasmid was subjected to IVT reaction using mMESSAGE mMACHINE T7 transcription kit (Life Technologies, AM1344). After 2 h incubation at 37°C, the transcribed RNA was recovered using RNeasy Mini kit (Qiagen, 74106) and quantitated at 260 nm by NanoDrop 2000 spectrophotometer (ThermoScientific).
Viral RNA Transfection
Confluent BHK-21 cells in 24-well plates were transfected with 1 μg of CHIKV RNA-Lipofectamine 2000 complex (Life Technologies, 11668-019) and maintained in RPMI supplemented with 2% FCS. Cells were observed for presence of CPE in the 48 h to 72 h p.i. Harvested supernatant was clarified and further passaged once in C6/36 cells to establish an infectious CHIKV pool. DNA sequencing of entire length of CHIKV mutant cDNA clone was performed using a set of optimized CHIKV-specific primers established previously.
MAb immunotherapy in mice model
For prophylaxis treatment, 5-day old neonate mice (n = 5 or 6 per group) were pre-treated with 0.2 μg, 2 μg, 20 μg or 200 μg of anti-CHIKV 3E7b or a purified mouse IgM isotype (Bethyl laboratories, MI10-100) diluted in sterile PBS or mock-treated with PBS by intraperitoneal (i.p) injection at 24 h and 8 h prior to CHIKV infection. Following this, mice were i.p injected with 50 μl of CHIKV at 4 x 105 PFU (strain LK(EH)CH6708, GenBank Accession number: FJ513654). For therapeutic study, 6-day old neonate mice were given CHIKV by i.p injection, followed by administration of 3E7b, IgM isotype or PBS at 8 h or 4 h p.i. To investigate virus clearance in the tissues and serum, mice were sacrificed at 48 h p.i. by CO2 overdose and whole blood, brain, spleen, liver and muscle of the hind limbs were harvested for quantification of CHIKV titer by virus plaque assay. Tissues were suspended in 1 ml PBS and homogenized using Precellys CK28 beads (Bertin Technologies, KT03961-1-002.2). Blood and homogenized tissues were then further clarified at 10,000 g before quantification by viral plaque assay. For survival analysis, neonate mice (n = 5 or 6) were treated as described above. Following CHIKV infection, mice were monitored and scored daily for sign and symptom of CHIKV disease. A clinical score of 6 or death was determined as endpoint of the study. At the end of 2 weeks, the number of survivals were noted and all mice were euthanized by CO2 overdose and sacrificed.
Histological examination
Whole limbs of infected mice were harvested at day 7 p.i. and fixed in 4% PFA for 3 days at room temperature. Hind limbs were decalcified in Surgipath decalcifying solution II (Leica, DC3800460II) for an additional 2 h. Following which, tissues were stepwise dehydrated in 50% and 70% ethanol, paraffin-embedded and sectioned to 4 μm before staining with Hematoxylin and Eosin (H&E). Images were captured using BX43 Olympus microscope at ×10 and ×40 magnification.
Statistical analysis
In accordance with our approved IACUC protocol guidelines, a minimum number of mice were used in this study without compromising the statistical power of our findings. For survival analysis, sample size (n) required in a time to event occurrence is calculated using the following equation:Citation66
Here, pc is defined as the estimated proportion of mice in the control group that will succumb to CHIKV infection, pe is the desired proportion of mice in the treatment group that will succumb to CHIKV infection, d is given as pc–qc and C is 10.51 given a significance level of 0.05 and confidence level of 90%. As for CHIKV load quantification, sample size is calculated using the following equation:Citation66
Here, s is defined as the group standard deviation and d as the desired treatment effect. C is 10.51 given a significance level of 0.05 and confidence level of 90%. Using these equations, n = 5 or 6 mice used in our study is determined to have sufficient statistical power to detect at a significant level of 0.05, if there is any different effect between PBS (mock mAb) and mAb treatment.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Supplemental Material
Supplemental data for this article can be accessed on the publisher's website.
Supplemental_Material.docx
Download MS Word (1.2 MB)Acknowledgments
The authors wish to thank Dr. Ng Lee Ching from Environmental Health Institute, NEA, Singapore, for her kind provision of CHIKV strain SGEHICHD122508 (GenBank Accession number: FJ445502) and Dr. Ooi Eng Eong, Duke-NUS, for kindly providing CHIKV Ross strain.
Funding
This study was supported by BMRC (A*STAR R182-000-158-305) and MINDEF DIRP Grant (R182-000-210-232).
References
- Fourie ED, Morrison JG. Rheumatoid arthritic syndrome after chikungunya fever. S Afr Med J 1979; 56:130-2; PMID:494034
- Brighton SW, Prozesky OW, de la Harpe AL. Chikungunya virus infection. A retrospective study of 107 cases. S Afr Med J 1983; 63:313-5; PMID:6298956
- Kee AC, Yang S, Tambyah P. Atypical chikungunya virus infections in immunocompromised patients. Emerg Infect Dis 2010; 16:1038-40; PMID:20507772; http://dx.doi.org/10.3201/eid1606.091115
- Hoarau JJ, Jaffar Bandjee MC, Krejbich Trotot P, Das T, Li-Pat-Yuen G, Dassa B, Denizot M, Guichard E, Ribera A, Henni T, et al. Persistent chronic inflammation and infection by Chikungunya arthritogenic alphavirus in spite of a robust host immune response. J Immunol 2010; 184:5914-27; PMID:20404278; http://dx.doi.org/10.4049/jimmunol.0900255
- Pellot AS, Alessandri JL, Robin S, Samperiz S, Attali T, Brayer C, Pasquet M, Jaffar-Bandjee MC, Benhamou LS, Tiran-Rajaofera I, et al. Severe forms of chikungunya virus infection in a pediatric intensive care unit on Reunion Island. Med Trop (Mars) 2012; 72 Spec No:88-93; PMID:22693937
- Robin S, Ramful D, Le Seach F, Jaffar-Bandjee MC, Rigou G, Alessandri JL. Neurologic manifestations of pediatric chikungunya infection. J Child Neurol 2008; 23:1028-35; PMID:18287573; http://dx.doi.org/10.1177/0883073808314151
- Schuffenecker I, Iteman I, Michault A, Murri S, Frangeul L, Vaney MC, Lavenir R, Pardigon N, Reynes JM, Pettinelli F, et al. Genome microevolution of chikungunya viruses causing the Indian Ocean outbreak. PLoS Med 2006; 3:e263; PMID:16700631; http://dx.doi.org/10.1371/journal.pmed.0030263
- Njenga MK, Nderitu L, Ledermann JP, Ndirangu A, Logue CH, Kelly CHL, Sang R, Sergon K, Breiman R, Powers AM. Tracking epidemic Chikungunya virus into the Indian Ocean from East Africa. J Gen Virol 2008; 89:2754-60; PMID:18931072; http://dx.doi.org/10.1099/vir.0.2008/005413-0
- Ravi V. Re-emergence of chikungunya virus in India. Indian J Med Microbiol 2006; 24:83-4; PMID:16687855; http://dx.doi.org/10.4103/0255-0857.25175
- Pulmanausahakul R, Roytrakul S, Auewarakul P, Smith DR. Chikungunya in Southeast Asia: understanding the emergence and finding solutions. Int J Infect Dis 2011; 15:e671-6; PMID:21775183; http://dx.doi.org/10.1016/j.ijid.2011.06.002
- Viennet E, Knope K, Faddy HM, Williams CR, Harley D. Assessing the threat of chikungunya virus emergence in Australia. Commun Dis Intell Q Rep 2013; 37:E136-43; PMID:24168087
- Dogan AD, Bunes K, Skarphédinsson S. The tropical disease Chikungunya fever has come to Europe. Ugeskr Laeger 2013; 175:1716-9; PMID:23763933
- Powers AM. Risks to the Americas associated with the continued expansion of chikungunya virus. J Gen Virol 2015; 96:1-5; PMID:25239764; http://dx.doi.org/10.1099/vir.0.070136-0
- Van Bortel W, Dorleans F, Rosine J, Blateau A, Rousset D, Matheus S, Leparc-Goffart I, Flusin O, Prat C, Cesaire R, et al. Chikungunya outbreak in the Caribbean region, December 2013 to March 2014, and the significance for Europe. Euro Surveill 2014; 19:pii=20759
- Strauss JH, Strauss EG. The alphaviruses: gene expression, replication, and evolution. Microbiol Rev 1994; 58:491-562; PMID:7968923
- Voss JE, Vaney MC, Duquerroy S, Vonrhein C, Girard-Blanc C, Crublet E, Thompson A, Bricogne G, Rey FA. Glycoprotein organization of Chikungunya virus particles revealed by X-ray crystallography. Nature 2010; 468:709-12; PMID:21124458; http://dx.doi.org/10.1038/nature09555
- Mukhopadhyay S, Zhang W, Gabler S, Chipman PR, Strauss EG, Strauss JH, Baker TS, Kuhn RJ, Rossmann MG. Mapping the structure and function of the E1 and E2 glycoproteins in alphaviruses. Structure 2006; 14:63-73; PMID:16407066; http://dx.doi.org/10.1016/j.str.2005.07.025
- Smith TJ, Cheng RH, Olson NH, Peterson P, Chase E, Kuhn RJ, Baker TS. Putative receptor binding sites on alphaviruses as visualized by cryoelectron microscopy. PNAS 1995; 92:10648-52; PMID:7479858; http://dx.doi.org/10.1073/pnas.92.23.10648
- Li L, Jose J, Xiang Y, Kuhn RJ, Rossmann MG. Structural changes of envelope proteins during alphavirus fusion. Nature 2010; 468:705-8; PMID:21124457; http://dx.doi.org/10.1038/nature09546
- Mohan A. Chikungunya fever: clinical manifestations & management. Indian J Med Res 2006; 124:471-4; PMID:17213512
- Kaur P, Chu JJ. Chikungunya virus: an update on antiviral development and challenges. Drug Discov Today 2013; 18:969-83; PMID:23684571; http://dx.doi.org/10.1016/j.drudis.2013.05.002
- Gerardin P, Barau G, Michault A, Bintner M, Randrianaivo H, Choker G, Lenglet Y, Touret Y, Bouveret A, Grivard P, et al. Multidisciplinary prospective study of mother-to-child chikungunya virus infections on the island of La Reunion. PLoS Med 2008; 5:e60; PMID:18351797; http://dx.doi.org/10.1371/journal.pmed.0050060
- Edelman R, Tacket CO, Wasserman SS, Bodison SA, Perry JG, Mangiafico JA. Phase II safety and immunogenicity study of live chikungunya virus vaccine TSI-GSD-218. Am J Trop Med Hyg 2000; 62:681-5; PMID:11304054
- Hallengard D, Kakoulidou M, Lulla A, Kummerer BM, Johansson DX, Mutso M, Lulla V, Fazakerley JK, Roques P, Le Grand R, et al. Novel attenuated Chikungunya vaccine candidates elicit protective immunity in C57BL/6 mice. J Virol 2014; 88:2858-66; PMID:24371047; http://dx.doi.org/10.1128/JVI.03453-13
- Tretyakova I, Hearn J, Wang E, Weaver S, Pushko P. DNA vaccine initiates replication of live attenuated chikungunya virus in vitro and elicits protective immune response in mice. J Infect Dis 2014; 209:1882-90; PMID:24585894; http://dx.doi.org/10.1093/infdis/jiu114
- Khan M, Dhanwani R, Rao PV, Parida M. Subunit vaccine formulations based on recombinant envelope proteins of Chikungunya virus elicit balanced Th1/Th2 response and virus-neutralizing antibodies in mice. Virus Res 2012; 167:236-46; PMID:22610133; http://dx.doi.org/10.1016/j.virusres.2012.05.004
- Akahata W, Yang ZY, Andersen H, Sun S, Holdaway HA, Kong WP, Lewis MG, Higgs S, Rossmann MG, Rao S, et al. A virus-like particle vaccine for epidemic Chikungunya virus protects nonhuman primates against infection. Nat Med 2010; 16:334-8; PMID:20111039; http://dx.doi.org/10.1038/nm.2105
- Couderc T, Khandoudi N, Grandadam M, Visse C, Gangneux N, Bagot S, Prost JF, Lecuit M. Prophylaxis and therapy for Chikungunya virus infection. J Infect Dis 2009; 200:516-23; PMID:19572805; http://dx.doi.org/10.1086/600381
- Pal P, Dowd KA, Brien JD, Edeling MA, Gorlatov S, Johnson S, Lee I. Development of a Highly Protective Combination Monoclonal Antibody Therapy against Chikungunya Virus. PLoS Pathog 2013; 9:e1003312; PMID:23637602; http://dx.doi.org/10.1371/journal.ppat.1003312
- Warter L, Lee CY, Thiagarajan R, Grandadam M, Lebecque S, Lin RT, Bertin-Maghit S, Ng LF, Abastado JP, Despres P, et al. Chikungunya virus envelope-specific human monoclonal antibodies with broad neutralization potency. J Immunol 2011; 186:3258-64; PMID:21278338; http://dx.doi.org/10.4049/jimmunol.1003139
- Goh LY, Hobson-Peters J, Prow NA, Gardner J, Bielefeldt-Ohmann H, Pyke AT, Suhrbier A, Hall RA. Neutralizing monoclonal antibodies to the E2 protein of chikungunya virus protects against disease in a mouse model. Clin Immunol 2013; 149:487-97; PMID:24239837; http://dx.doi.org/10.1016/j.clim.2013.10.004
- Masrinoul P, Puiprom O, Tanaka A, Kuwahara M, Chaichana P, Ikuta K, Ramasoota P, Okabayashi T. Monoclonal antibody targeting chikungunya virus envelope 1 protein inhibits virus release. Virol J 2014; 464-465:111-7; PMID:24927852
- Selvarajah S, Sexton NR, Kahle KM, Fong RH, Mattia KA, Gardner J, Lu K, Liss NM, Salvador B, Tucker DF, et al. A neutralizing monoclonal antibody targeting the acid-sensitive region in chikungunya virus E2 protects from disease. PLoS Negl Trop Dis 2013; 7:e2423; PMID:24069479; http://dx.doi.org/10.1371/journal.pntd.0002423
- Fong RH, Banik SS, Mattia K, Barnes T, Tucker D, Liss N, Lu K, Selvarajah S, Srinivasan S, Mabila M, et al. Exposure of epitope residues on the outer face of the chikungunya virus envelope trimer determines antibody neutralizing efficacy. J Virol 2014; 88:14364-79; PMID:25275138; http://dx.doi.org/10.1128/JVI.01943-14
- Kam YW, Lum FM, Teo TH, Lee WW, Simarmata D, Harjanto S, Chua CL, Chan YF, Wee JK, Chow A, et al. Early neutralizing IgG response to Chikungunya virus in infected patients targets a dominant linear epitope on the E2 glycoprotein. EMBO Mol Med 2012; 4:330-43; PMID:22389221; http://dx.doi.org/10.1002/emmm.201200213
- Lum FM, Teo TH, Lee WW, Kam YW, Renia L, Ng LF. An essential role of antibodies in the control of Chikungunya virus infection. J Immunol 2013; 190:6295-302; PMID:23670192; http://dx.doi.org/10.4049/jimmunol.1300304
- Kam YW, Lee WW, Simarmata D, Harjanto S, Teng TS, Tolou H, Chow A, Lin RT, Leo YS, Renia L, et al. Longitudinal analysis of the human antibody response to Chikungunya virus infection: implications for serodiagnosis and vaccine development. J Virol 2012; 86:13005-15; PMID:23015702; http://dx.doi.org/10.1128/JVI.01780-12
- Cavallo L, Kleinjung J, Fraternali F. POPS: A fast algorithm for solvent accessible surface areas at atomic and residue level. Nucleic Acids Res 2003; 31:3364-6; PMID:12824328; http://dx.doi.org/10.1093/nar/gkg601
- Couderc T, Chretien F, Schilte C, Disson O, Brigitte M, Guivel-Benhassine F, Touret Y, Barau G, Cayet N, Schuffenecker I, et al. A mouse model for chikungunya: Young age and inefficient type-I interferon signaling are risk factors for severe disease. PLoS Pathog 2008; 4:e29; PMID:18282093; http://dx.doi.org/10.1371/journal.ppat.0040029
- Dhanwani R, Khan M, Lomash V, Rao P, Ly H, Parida M. Characterization of Chikungunya Virus Induced Host Response in a Mouse Model of Viral Myositis. PLoS One 2014; 9:e92813; PMID:24667237; http://dx.doi.org/10.1371/journal.pone.0092813
- Klasse PJ, Sattentau QJ. Occupancy and mechanism in antibody-mediated neutralization of animal viruses. J Gen Virol 2002; 83:2091-108; PMID:12185262
- Feinstein A, Munn EA, Richardson NE. The three-dimensional conformation of M and A globulin molecules. Ann N Y Acad Sci 1971; 190:104-21; PMID:5290007; http://dx.doi.org/10.1111/j.1749-6632.1971.tb13526.x
- Strauss EG, Stec DS, Schmaljohn AL, Strauss JH. Identification of antigenically important domains in the glycoproteins of Sindbis virus by analysis of antibody escape variants. J Virol 1991; 65:4654-64; PMID:1714515
- Vrati S, Fernon CA, Dalgarno L, Weir RC. Location of a major antigenic site involved in Ross River virus neutralization. Virol J 1988; 162:346-53; http://dx.doi.org/10.1016/0042-6822(88)90474-6
- Agapov EV, Razumov IA, Frolov IV, Kolykhalov AA, Netesov SV, Loktev VB. Localization of four antigenic sites involved in Venezuelan equine encephalomyelitis virus protection. Arch Virol 1994; 139:173-81; PMID:7529989; http://dx.doi.org/10.1007/BF01309462
- Heil M, Albee A, Strauss J, Kuhn R. An amino acid substitution in the coding region of the E2 glycoprotein adapts Ross River virus to utilize heparan sulfate as an attachment moiety. J Virol 2001; 75:6303-9; PMID:11413296; http://dx.doi.org/10.1128/JVI.75.14.6303-6309.2001
- Tsetsarkin K, McGee C, Volk S, Vanlandingham D, Weaver S, Higgs S. Epistatic Roles of E2 Glycoprotein Mutations in Adaption of Chikungunya Virus to Aedes Albopictus and Ae. Aegypti Mosquitoes. PLoS One 2009; 4:e6835; PMID:19718263; http://dx.doi.org/10.1371/journal.pone.0006835
- Tsetsarkin K, Vanlandingham D, McGee C, Higgs S. A Single Mutation in Chikungunya Virus Affects Vector Specificity and Epidemic Potential. PLoS Pathog 2007; 3:e201; PMID:18069894; http://dx.doi.org/10.1371/journal.ppat.0030201
- Powers AM, Logue CH. Changing patterns of chikungunya virus: re-emergence of a zoonotic arbovirus. J Gen Virol 2007; 88:2363-77; PMID:17698645; http://dx.doi.org/10.1099/vir.0.82858-0
- Ochsenbein AF, Zinkernagel RM. Natural antibodies and complement link innate and acquired immunity. Immunol Today 2000; 21:624-30; PMID:11114423; http://dx.doi.org/10.1016/S0167-5699(00)01754-0
- Chopra A, Anuradha V, Lagoo-Joshi V, Kunjir V, Salvi S, Saluja M. Chikungunya virus aches and pains: an emerging challenge. Arthritis Rheum 2008; 58:2921-2; PMID:18759351; http://dx.doi.org/10.1002/art.23753
- Malvy D, Ezzedine K, Mamani-Matsuda M, Autran B, Tolou H, Receveur MC, Pistone T, Rambert J, Moynet D, Mossalayi D. Destructive arthritis in a patient with chikungunya virus infection with persistent specific IgM antibodies. BMC Infect Dis 2009; 9:200; PMID:20003320; http://dx.doi.org/10.1186/1471-2334-9-200
- Calisher CH, el-Kafrawi AO, Al-Deen Mahmud MI, Travassos da Rosa AP, Bartz CR, Brummer-Korvenkontio M, Haksohusodo S, Suharyono W. Complex-specific immunoglobulin M antibody patterns in humans infected with alphaviruses. J Clin Microbiol 1986; 23:155-9; PMID:3009526
- Borgherini G, Poubeau P, Staikowsky F, Lory M, Le Moullec N, Becquart JP, Wengling C, Michault A, Paganin F. Outbreak of chikungunya on Reunion Island: early clinical and laboratory features in 157 adult patients. Clin Infect Dis 2007; 44:1401-7; PMID:17479933; http://dx.doi.org/10.1086/517537
- Fric J, Bertin-Maghit S, Wang C-I, Nardin A, Warter L. Use of Human Monoclonal Antibodies to Treat Chikungunya Virus Infection. J Infect Dis 2013; 207:319-22; PMID:23125446; http://dx.doi.org/10.1093/infdis/jis674
- Morrison TE, Oko L, Montgomery SA, Whitmore AC, Lotstein AR, Gunn BM, Elmore SA, Heise MT. A mouse model of chikungunya virus-induced musculoskeletal inflammatory disease: evidence of arthritis, tenosynovitis, myositis, and persistence. Am J Pathol 2011; 178:32-40; PMID:21224040; http://dx.doi.org/10.1016/j.ajpath.2010.11.018
- Gardner J, Anraku I, Le TT, Larcher T, Major L, Roques P, Schroder WA, Higgs S, Suhrbier A. Chikungunya virus arthritis in adult wild-type mice. J Virol 2010; 84:8021-32; PMID:20519386; http://dx.doi.org/10.1128/JVI.02603-09
- Ziegler SA, Lu L, da Rosa AP, Xiao SY, Tesh RB. An animal model for studying the pathogenesis of chikungunya virus infection. Am J Trop Med Hyg 2008; 79:133-9; PMID:18606777
- Teo TH, Lum FK, Lee WWL, Ng LFP. Mouse models for Chikungunya virus: deciphering immune mechanisms responsible for disease and pathology. Immunol Res 2012; 53:136-47; PMID:22418724; http://dx.doi.org/10.1007/s12026-012-8266-x
- Oh HL, Akerstrom S, Shen S, Bereczky S, Karlberg H, Klingstrom J, Lal SK, Mirazimi A, Tan YJ. An antibody against a novel and conserved epitope in the hemagglutinin 1 subunit neutralizes numerous H5N1 influenza viruses. J Virol 2010; 84:8275-86; PMID:20519402; http://dx.doi.org/10.1128/JVI.02593-09
- Ng OW, Keng CT, Leung CS, Peiris JS, Poon LL, Tan YJ. Substitution at aspartic acid 1128 in the SARS coronavirus spike glycoprotein mediates escape from a S2 domain-targeting neutralizing monoclonal antibody. PLoS One 2014; 9:e102415; PMID:25019613; http://dx.doi.org/10.1371/journal.pone.0102415
- Lok SM, Kostyuchenko V, Nybakken GE, Holdaway HA, Battisti AJ, Sukupolvi-Petty S, Sedlak D, Fremont DH, Chipman PR, Roehrig JT, et al. Binding of a neutralizing antibody to dengue virus alters the arrangement of surface glycoproteins. Nat Struct Mol Biol 2008; 15:312-7; PMID:18264114; http://dx.doi.org/10.1038/nsmb.1382
- Stucky B. SeqTrace: a graphical tool for rapidly processing DNA sequencing chromatograms. J Biomol Tech 2012; 23:90-3; PMID:22942788; http://dx.doi.org/10.7171/jbt.12-2303-004
- Kelley L, Sternberg M. Protein structure prediction on the web: a case study using the Phyre server. Nat Protoc 2009; 4:363-71; PMID:19247286; http://dx.doi.org/10.1038/nprot.2009.2
- Schrodinger L. The PyMOL molecular graphics system, version 1.3 r1. New York: Oxford University Press; 2010.
- Dell RB, Holleran S, Ramakrishnan R. Sample Size Determination. ILAR J 2002; 43:207-13; PMID:12391396; http://dx.doi.org/10.1093/ilar.43.4.207
- Bréhin AC, Rubrechtb L, Navarro-Sancheza ME, Maréchala V, Frenkiela MP, Lapaludb P, Launeb D, Sallc AA, Desprès P. Production and characterization of mouse monoclonal antibodies reactive to Chikungunya envelope E2 glycoprotein. Virol J 2008; 371:185-95; http://dx.doi.org/10.1016/j.virol.2007.09.028
