ABSTRACT
Transforming growth factor (TGF)β levels are elevated in, and drive the progression of, numerous disease states such as advanced metastatic cancer and systemic and ocular fibrosis. There are 3 main isoforms, TGFβ1, 2, and 3. As multiple TGFβ isoforms are involved in disease processes, maximal therapeutic efficacy may require neutralization of 2 or more of the TGFβ isoforms. Fully human antibody phage display libraries were used to discover a number of antibodies that bind and neutralize various combinations of TGFβ1, 2 or 3. The primary panning did not yield any uniformly potent pan-isoform neutralizing antibodies; therefore, an antibody that displayed potent TGFβ 1, 2 inhibition, but more modest affinity versus TGFβ3, was affinity matured by shuffling with a light chain sub-library and further screening. This process yielded a high affinity pan-isoform neutralizing clone. Antibodies were analyzed and compared by binding affinity, as well as receptor and epitope competition by surface plasmon resonance methods. The antibodies were also shown to neutralize TGFβ effects in vitro in 3 assays: 1) interleukin (IL)-4 induced HT-2 cell proliferation; 2) TGFβ-mediated IL-11 release by A549 cells; and 3) decreasing SMAD2 phosphorylation in Detroit 562 cells. The antibodies’ potency in these in vitro assays correlated well with their isoform-specific affinities. Furthermore, the ability of the affinity-matured clone to decrease tumor burden in a Detroit 562 xenograft study was superior to that of the parent clone. This affinity-matured antibody acts as a very potent inhibitor of all 3 main isoforms of TGFβ and may have utility for therapeutic intervention in human disease.
Abbreviations
| TGFβ | = | Transforming growth factor β |
| TβRI | = | Type I TGFβ receptor |
| TβRII | = | Type II TGFβ receptor |
| TβRIII | = | Type III TGFβ receptor |
| SPR | = | Surface plasmon resonance |
| LAP | = | latency associated peptide |
| PPE | = | periplasmic extract |
Introduction
Members of the transforming growth factor (TGF)β family of cytokines are essential for mammalian development and normal tissue homeostasis. Elevated levels of TGFβ have been shown to drive the progression of numerous disease states in model systems and may play a critical role in human diseases such as cancer, as well as systemic and ocular fibrosis. The activity of TGFβ is cell-type specific and context dependent. For example, in normal epithelial cells, TGFβ acts as a mediator of cell cycle arrest and differentiation, but, in many late-stage tumors lacking p53 or with microsatellite instability or an activated RAS oncogene, TGFβ can behave as a potent promoter of tumor growth. It can also impair the ability of host immune cells to check tumor progression, and it can promote metastasis.Citation1-4 Highly conserved in mammals, TGFβ has 3 main isoforms: TGFβ1, TGFβ2, and TGFβ3. These have mechanistically similar receptor activation properties, but varying expression and tissue distribution patterns. Different expression levels of the various isoforms have been shown to translate to differences in prognostic outcome, as well as differential effects on the activation of the immune system.Citation5,6
Each TGFβ isoform is expressed as a pro-protein and processed by a furin-like convertase into a latency-associated peptide (LAP) and mature TGFβ protein. The LAP remains bound to the mature TGFβ and inactivates it by preventing receptor binding. Latent TGFβ binding proteins (LTBPs) in the extracellular matrix bind the inactivated mature TGFβ, forming large latent TGFβ complexes. The mature TGFβ remains bound to the LAP until released by its proteolysis by a matrix metalloprotease or thrombospondin, exposure to low pH, or interaction with a β6 integrin.Citation7-10 TGFβ can only activate its receptors after it is released from the complex and is free of the LAP. Mature TGFβ is a disulfide-linked dimer, composed of 2 112 amino acid monomers.Citation11-14 The TGFβ dimers are generally considered to be homodimeric; heterodimers are not frequently observed, but have been found in TGFβ preparations from porcine platelets and may exist in other contexts at low levels.Citation15
TGFβ signals by binding to a heteromeric kinase receptor complex between Type I and Type II TGFβ receptors: TβRI (also known as ALK5) and TβRII, respectively.Citation7,16-19 This activated complex then phosphorylates SMAD2 and SMAD3, which then bind to SMAD4. The SMAD complex is translocated to the nucleus where it interacts with transcriptional regulators to alter gene expression.Citation3 In addition to the canonical SMAD activation pathway, the activated TGFβ receptor complex can signal via a large number of non-canonical signal transduction pathways, including numerous kinases and cofactors in a cell type and cell context dependent manner.Citation3,20 The TGFβ1 and TGFβ3 isoforms bind with high affinity to the TβRII monomeric receptor, but the TGFβ2 isoform only binds tightly to TβRII when bound and presented by the non-signaling high molecular weight receptor TβRIII (betaglycan).Citation7,21-25 TGFβ signaling requires the formation of a TGFβ/TβRII/TβRI complex, and an antibody or agent that blocks the association of the TGFβ/TβRII interaction should in theory effectively neutralize TGFβ signaling.
It is possible that many disease processes involve elevated activity by more than one TGFβ isoform, and the neutralization of a single isoform may not be sufficient for a therapeutic effect. However, generation of antibodies targeting multiple TGFβ isoforms is challenging for several reasons. There is high species conservation for each of the isoforms with greater than 97% sequence identity among mammals.Citation26 This high interspecies homology and the ubiquitous expression of TGFβ makes breaking tolerance in immunized animals difficult, and low hybridoma yields are typical.Citation27 Moreover, across the 3 isoforms, there is only 68% amino acid sequence identity,Citation28,29 limiting the discovery of antibodies that are cross-reactive with, and able to neutralize, multiple TGFβ isoforms.
Phage display is a powerful antibody discovery method that allows access to a large and diverse antibody repertoire, and can circumvent much of the self-censoring of the immune system response that would prevent the development of most auto-antibodies. This self-censoring is a significant obstacle to generating neutralizing, high-affinity anti-TGFβ antibodies by conventional techniques such as hybridoma technology. Given TGFβ's ubiquitous expression, there is a low abundance of high affinity TGFβ antibodies in the human and mouse antibody repertoires.Citation30-33 In addition, active TGFβ has physical properties that make it a challenging target for use in therapeutic monoclonal antibody development, in particular, its poor stability at neutral pH in purified form. XOMA's ADAPTTM antibody platform, utilizing some of the largest available antibody libraries,Citation34 is well-suited for targeting challenging antigens, and was used in an effort to generate a number of TGFβ neutralizing antibodies with varying specificities, including pan-specific antibodies that neutralize all 3 human TGFβ isoforms.
XOMA has constructed several very large fully human antibody phage display libraries with diversities of roughly 1×1011 unique antibody sequences. These libraries were constructed from multiple naïve human donor sources as described in Schwimmer et al.Citation34 The interrogation of these libraries with TGFβ generated several hundred unique TGFβ binding antibodies with a variety of properties. Several antibodies targeting multiple TGFβ isoforms were screened and ranked biochemically and in functional cell-based assays for activity and utility in neutralizing TGFβ function. The discovery and characterization of antibodies that bind and neutralize TGFβ1, TGFβ2, and TGFβ3 and the evolution of a pan TGFβ1/2/3 neutralizer (XPA.42.681) are highlighted here. These antibodies were assessed for effectiveness in blocking the receptor-ligand interaction, affinity, and neutralization of TGFβ signaling in cell-based functional assays. Furthermore, certain clones were evaluated in a xenograft tumor model using the human pharyngeal carcinoma cell line Detroit 562 for their ability to inhibit tumor growth in vivo.
Results
Primary panning and screening
Phage selection utilizing TGFβ adsorbed to ELISA plates did not yield any useful binders (data not shown). Another strategy utilized biotinylated TGFβ, which was captured out of solution in the presence of carrier protein by streptavidin-coated magnetic beads after incubation with the phage library. Unbound phage were washed away while remaining phage, displaying a fragment antibody that bound the antigen, were amplified to use for the next round. Three selection rounds were performed using decreasing amounts of biotinylated TGFβ and increasing numbers of washes to increase stringency and bias selection for specific binders to antigen. Each isoform and sub-library was panned independently. Of the 3 isoforms, only the panning campaign that used TGFβ2 was successful in yielding a large diversity of antibodies. This difference in isoform panning results is likely due to the greater observed stability of the TGFβ2 isoform at neutral pH.
DNA from clones that passed the primary surface plasmon resonance (SPR)-based binding screen was sequenced to determine antibody diversity. Clones were defined as unique if they had one or more amino acids in their complementarity-determining regions (CDRs) that were different from other clones. shows the number and binding properties of clones identified, predominantly from the panning campaigns against TGFβ2.
Table 1. Soluble Panning Results. Number of binders identified from soluble panning using biotinylated TGFβ2. Comparative data is provided for both XOMA phage display library formats: the human Fab library (XFab1) and the human single chain antibody, scFv, (XscFv2).
DNA from clones identified as pan binders or dual binders was sequenced to determine their diversity. The results are shown in .
Table 2. Sequence Diversity of Binders. Number of unique sequences from binders to more than one TGFβ isoform.
Light chain shuffling
Despite the generation of hundreds of unique TGFβ-binding monoclonal antibodies, no antibodies that bound to all 3 isoforms with equally high affinity and potency were found from the primary screens, regardless of the panning strategy employed. Therefore, a clone (XPA.42.068) that bound to and neutralized TGFβ1 and TGFβ2 with high affinity, but had a lower affinity and relatively weak neutralization of TGFβ3, was affinity matured using a light chain shuffling approach. Two sub-libraries were constructed, each consisting of the single parent heavy chain sequence combined with a diverse kappa or lambda light chain library, respectively. Panning and screening of these sub-libraries resulted in the identification of an antibody (XPA.42.681) with a new light chain sequence that allowed high affinity binding to, and neutralization of, all 3 TGFβ isoforms. XPA.42.068 parent antibody had a lambda light chain, and TGFβ binding clones were discovered from the lambda XPA.0168 light chain library, but no TGFβ binders were identified from the kappa library. These data demonstrate the importance of the light chain to the binding characteristics of the clones derived from this heavy chain.
Following two rounds of panning, 372 clones were screened by SPR for TGFβ binding. Most of the clones were TGFβ binders with kinetics similar to the parent clone. Sequencing results showed that the majority of the 372 clones had the parental antibody light chain sequence, which was not excluded from the library. Only three of the clones showed an improved (slower) off-rate from TGFβ3. The light chain sequences of these 3 clones had changes in both the framework and CDR regions compared with the parental light chain sequence. compares the light chain sequence of the parent XPA.42.068 and the most improved affinity matured variant XPA.42.681. The L-CDR2 of XPA.42.681 had one substitution from a tyrosine to a phenylalanine (YDR→FDR), whereas the matured L-CDR3 sequence showed several differences (QVWDNTSEHV → QVWDSDSDDL). The three antibodies were cloned into a full-length human IgG2 vector for expression, purification, and further testing.
Figure 1. Sequence Comparison of Parent XPA.42.068, Affinity Matured Variant XPA.42.681, and XPA.42.089. The heavy and light chain sequence of the parent XPA.42.068 is compared to the affinity improved light chain variant, XPA.42.681. Amino acid positions that are different between these clones are highlighted in bold black type. For XPA.42.089, amino acids are highlighted which are different than the XPA.42.068 clone.
Neutralization of TGFβ-mediated Inhibition of HT-2 cell proliferation
TGFβ inhibits interleukin (IL)-4-dependent growth of mouse T-cell line HT-2 through transactivation of genes promoting cell cycle arrest. IL-4 transactivates a mitogenic gene expression program by activating targets such as c-myc and GM-CSF, whereas TGFβ signaling transactivates genes that suppress c-myc and GM-CSF expression.Citation35 Inhibition of TGFβ signaling by a neutralizing antibody results in HT-2 cell proliferation.
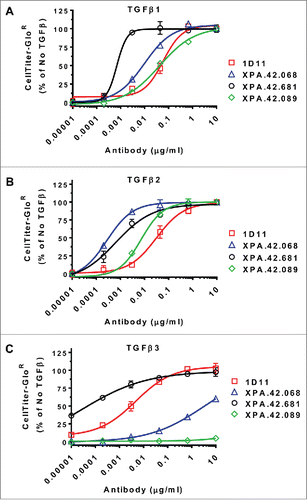
Antibody clones were first screened in a HT-2 assay for neutralization of TGFβ2 activity. Active clones were then advanced into an additional HT-2 proliferation assay to measure the antibody dose response for the neutralization of TGFβ1 and TGFβ3. Representative data demonstrating dose-dependent inhibition of TGFβ1, TGFβ2 by the most potent TGFβ1 and TGFβ 2 inhibiting antibodies, XPA.42.089 and XPA.42.681 is shown in A-B. The affinity-matured clone XPA.42.681 showed markedly better potency against TGFβ3 than the lower affinity XPA.42.068 parental antibody, whereas the XPA.42.089 antibody had little TGFβ3 neutralization (C). XPA.42.681 was the most potent inhibitor of TGFβ in all HT-2 assays. The clones’ potency of neutralization correlates well with their relative affinities for the various TGFβ isoforms.
Figure 2. Antibody Neutralization of TGFβ Mediated HT-2 Growth Arrest. HT-2 cells were treated with a fixed dose of TGFβ1, 2, or 3 (EC80 level) in the presence of neutralizing antibody titrations. Cell growth was measured after 48 hours of incubation at 37°C using a luminescent cell-viability reagent (CellTiter-Glo®). Values are presented as percent of a no TGFβ control.
Neutralization of TGFβ-mediated IL-11 release by A549 cells
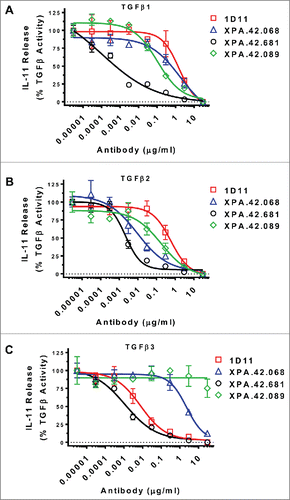
Another screening assay assessed TGFβ-mediated secretion of IL-11 by A549 lung carcinoma cells, which is part of a pro-fibrotic response in lung fibroblasts and epithelial cells and may be implicated in metastatic disease. Representative data are shown in , and indicate dose-dependent inhibition of TGFβ-mediated IL-11 release by the neutralizing anti-TGFβ antibodies. Similar to the HT-2 assay, the results show that the affinity matured clone XPA.42.681 was more potent against all isoforms of TGFβ than any of the other clones, and showed an improvement in potency of 3 logs EC50 value to TGFβ3 over the parent XPA.42.068 (XPA.42.681 EC50 of 0.002 μg/mL vs. an EC50 of 2 μg/mL for XPA.42.068). XPA.42.089 had weak activity against TGFβ3, but modestly better potency than XPA.42.068 against TGFβ1 and TGFβ2.
Figure 3. Antibody Neutralization of TGFβ Stimulated IL-11 release from A549 Cells. A549 cells were treated with a fixed dose of TGFβ1, 2, or 3 (EC80 level) in the presence of neutralizing antibody titrations. After 24 hours at 37°C, IL-11 levels in cell culture supernatants were measured by ELISA. Values are reported as percent of a no antibody control.
Characterization of antibody affinities
Two SPR-based kinetic methods were used to compare the binding affinities and rate constants of the antibodies XPA.42.089, XPA.42.068, and XPA.42.681. The first SPR method utilized immobilized antibodies and injected TGFβ, and the second method used the inverse orientation with immobilized TGFβ and injected antibodies. In both assays, the ranking and relative binding affinities to the different isoforms was maintained despite a difference in absolute KD (affinity or equilibrium dissociation constant in units M) and ka (on-rate in units M−1s−1) values between the 2 methods. For all antibodies, the first method ( and .) yielded lower KD values (higher affinity) than the immobilized TGFβ assays ( and Fig. S1). For example, XPA.42.681 binding to TGFβ2 had estimated KDs of ≤10 pM and 31 pM using the immobilized antibody method and the immobilized antigen method, respectively. We observed no direct evidence of any non-specific binding of TGFβ to the reference surface, which is nearly identical chemically to the low-density antibody surface.
Table 3. Antibody Affinity Using IgG Capture Method. Antibodies were immobilized on a CM5 sensor chip of a Biacore 2000 via amine chemistry. TGFβ was injected at 10 nM, 2 nM, 0.4 nM, and 0.08 nM. Data were analyzed for binding rate parameters using the Scrubber software after double referencing.
Table 4. Antibody Affinity Using Immobilized TGFβ. TGFβs were immobilized on a CM5 chip via amine chemistry. Antibodies were then injected at 33.33 nM, 6.67 nM, 1.33 nM, 267 pM, and 53 pM. Data were analyzed for binding rate parameters using the Scrubber software after double referencing.
Figure 4. Antibody Affinity Using Immobilized IgG Method. Antibodies were immobilized on a CM5 sensor chip of a Biacore 2000 via amine chemistry. TGFβ was injected at 10 nM, 2 nM, 0.4 nM, and 0.08 nM. Data were analyzed for binding rate parameters using the Scrubber software after double referencing.
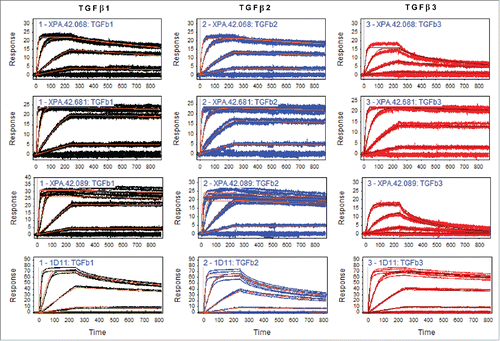
The affinities of the XPA.42.089 antibody in the immobilized TGFβ assay were in the picomolar range for TGFβ1 and TGFβ2, whereas binding to TGFβ3 was too weak to effectively estimate the KD at the antibody concentrations (≤33 nM) that were used. In the immobilized antibody format, the XPA.42.089 antibody bound to TGFβ1 and TGFβ2 with an affinity of 4 pM and 25 pM, respectively, but it bound to TGFβ3 with at least 50-fold weaker affinity (∼1.4 nM). These data demonstrated that XPA.42.089 was the highest affinity antibody identified from the primary panning and screening to TGFβ1 and TGFβ2, but the much weaker affinity to TGFβ3 would most likely make it unable to effectively neutralize TGFβ3 activity, as seen in and .
XPA.42.681, which was affinity enhanced by light chain shuffling from the parental XPA.42.068, had higher binding affinity (lower KD) to all of the TGFβ isoforms than any other antibody we have tested, including the commercially available mouse antibody 1D11.
Competition assays
Competition-based SPR assays were performed to determine the ability of the isolated antibodies to compete with each other, 2 of the TGFβ receptors (TβRII, TβRIII), and the TGFβ1 LAP. Detailed results of the various assays are described below. In general, the antibody competition results indicate that when the TGFβ is saturated with antibody, the antibodies are almost universally competitive with the TGFβ receptors and the TGFβ1 LAP. Only one clone was found that was not competitive with the TGFβ receptors, and it appeared to be a negative allosteric ligand modifying antibody (ALMA) of TGFβ binding and signaling (data not shown).Citation36 This clone was not highly potent in in vitro studies and was therefore not tested further. All of the antibodies were also competitive for TGFβ binding, the other antibodies, and with 1D11.
Receptor competition assays
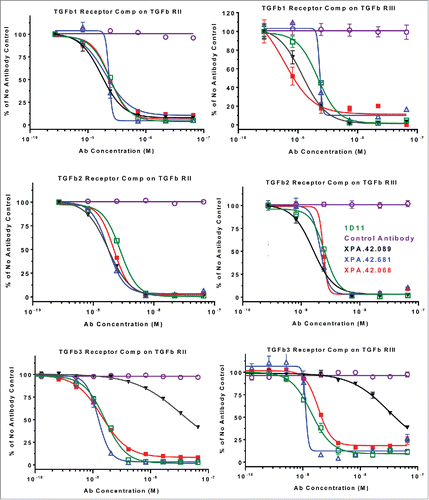
The ability of the Antibodies to inhibit or block the binding of the TGFβ ligands to the TGFβ receptors was analyzed by SPR competition assays. TGFβ signals through TβRII, which is a serine/threonine kinase transmembrane protein and requires the cytoplasmic association of TβRI for activation. The ligand-binding role of TβRI is complex and dependent on TβRII.Citation16 Recombinant TβRI was tested in the SPR assays, but the soluble recombinant form did not bind TGFβ at concentrations up to 100 nM, or to TGFβ bound to TβRII, and could not be used in receptor competition experiments. Therefore, the receptor competition experiments were only performed against TβRII and TβRIII. The TβRII only binds tightly to TGFβ1 and TGFβ3, whereas the TβRIII binds with similar affinity to all 3 TGFβ isoforms and its expression can confer sensitivity of cells to TGFβ2.Citation26 The 3 antibodies all blocked TGFβ activity in vitro, so the receptor competition assays were performed to assess whether this action was a result of blocking the ligand binding to the receptor or if the activity was due to an allosteric effect on receptor signaling. Results from competition assays utilizing the type II, and III receptors are shown in . These results suggest that all antibodies blocked the association of TGFβ with TβRII and TβRIII. The XPA.42.089 clone showed much weaker inhibition TGFβ3 receptor binding (∼15 fold) than the other TGFβs, and this is consistent with its lower affinity to the TGFβ3 isoform.
Figure 5. TβRII and TβRIII competition by antibodies utilizing TGFβ1, TGFβ2, and TGFβ3. TGFβRII or TGFβRIII protein was immobilized on a SPR surface. Antibodies were incubated at various concentrations with 4 nM TGFβ2 or 1.6 nM TGFβ3 prior to injection over the receptor-coated surfaces. Binding levels at the end of a 2 minute injection were recorded and values were normalized to a no antibody control samples. The data were fit using a 4 parameter fit in GraphPad Prism.
Recombinant human TGFβ1 LAP (rhLAP) competition assay
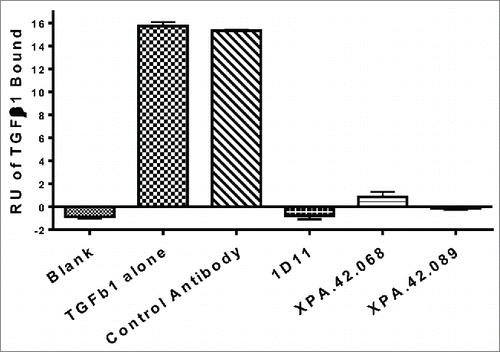
The vast majority of TGFβ in the body is present as the inactive latent form and can be readily activated by enzymes, proteases, and other factors.Citation8,37 To determine whether or not the antibodies interact with only the active form of TGFβ or also bind to the latent form, a LAP competition assay was performed. The results indicated that antibodies XPA.42.068, XPA.42.089, and the 1D11 control antibody were all competitive with rhLAP and only bind to active TGFβ (). The affinity enhanced light chain mutant XPA.42.681 antibody was not tested in this assay, but the parental XPA.42.068 showed strong competition and we would predict that the affinity matured version would share this property.
Figure 6. Antibody competition with TGFβ1 binding to rhLAP. rhLAP was immobilized on a CM5 sensor chip of a Biacore 2000. TGFβ1 was injected at 10 nM in the presence of 66.6 nM IgG. Level of TGFβ1 bound to rhLAP at the end of the injection is shown. A lack of signal shows competition with TGFβ1 binding by the antibodies.
Stoichiometry evaluation
The binding of a divalent IgG to a TGFβ homodimer creates the possibility of generating antibodies with different binding stoichiometries. For example, a likely scenario, and one that seems to hold true for all of the antibodies evaluated in these studies, is that each of the 2 Fab arms of an IgG is capable of binding to a TGFβ molecule and that each TGFβ homodimer has 2 antibody-binding epitopes as well.Citation14 A portion of the antibody molecules appear to bind bivalently to a single TGFβ homodimer creating a 1:1 TGFβ homodimer to IgG ratio, but this arrangement is not the dominant situation. Instead, it appears that, especially under conditions where the antibodies are in excess, each TGFβ molecule is bound by 2 antibodies. All antibodies tested were capable of a considerable amount of self-pairing to the TGFβ homodimer, supporting the idea that each TGFβ homodimer has 2 antibody binding sites.
To evaluate the binding stoichiometry of XPA.42.681, XPA.42.089 and 1D11, the antibodies were immobilized onto a planar carboxy surface in a Biacore 2000. TGFβ was injected at saturating concentrations and then another antibody was injected. This evaluation showed that TGFβ captured by each of the 3 antibodies was still available for binding by a subsequently injected antibody, and that there was no obvious difference in stoichiometry across the clones by this method (Fig. S2). To verify that the direct immobilization of the antibodies was not introducing error, the XPA.42.068, XPA.42.681, and XPA.42.089 antibodies were also captured on an anti-human-Fc surface at similar densities and then evaluated for maximal binding levels (Rmax) to TGFβ2 (). The mass ratio of antibody to TGFβ was estimated to be 6:1. Assuming the antibody is 150 kDa and TGFβ2 is 25 kDa, an estimate of the number of TGFβ molecules bound per captured antibody can be calculated as follows: [TGFβ RU/(Ab RU/6)]. This calculation yielded roughly 1.5-1.7 TGFβ/antibody, suggesting that most of the immobilized antibodies bind to 2 TGFβ molecules. It is likely that the TGFβ/antibody ratio is somewhat less than 2 because a portion of the antibodies are binding divalently to a single TGFβ molecule. This is consistent with the values from the previously discussed assay format (Fig. S2) where mass calculated binding levels suggested that roughly 0.6 Abs were bound for each captured TGFβ molecule. This apparent ability of the antibodies to bind both bivalently and monovalently to the TGFβ homodimer could account for the modest amount of binding heterogeneity seen in the kinetics of the TGFβ binding. The dissociation kinetics would be slowed for the population of bivalently bound TGFβ, and would be faster for the monovalently bound forms. Interestingly, the TβRII protein did not allow for the same stoichiometry as the IgGs and bound to the TGFβ dimer bivalently as has been previously reported,Citation38 and could not self-pair (data not shown).
Table 5. Stoichiometry evaluation: Data summary from antibody capture kinetics RUmax test using TGFβ2. Antibodies were captured at 107-117 RU onto an anti-human-Fc capture surface and TGFβ2 was injected in duplicate at 20 nM, 4 nM, and 0.8nM. Data was then double referenced and fit kinetically to establish the RUmax using the formula TGFβ/antibody = [TGFβ RU/(Ab RU/6)].
Inhibition of TGFβ-stimulated pSMAD2 response in Detroit 562 cells
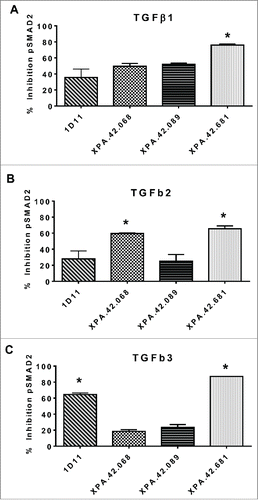
The three antibody clones were evaluated in a SMAD2 phosphorylation assay using the Detroit 562 pharyngeal carcinoma cell line that was to be used in a xenograft tumor model. This assay was developed to evaluate neutralization of TGFβ signaling through the TGFβRII/TGFβRI receptor complex.Citation7 shows the percent inhibition of the TGFβ-induced pSMAD2 response (normalized to total SMAD2) for each clone relative to the anti-keyhole limpet hemocyanin (KLH) control antibody. A two-tailed T-test revealed that the parent XPA.42.068 was significantly more potent than 1D11 against TGFβ2 in neutralizing pSMAD2 signaling. XPA.42.681 was significantly more potent than 1D11 against all 3 isoforms of TGFβ (p < 0.05), and significantly better than the parent XPA.42.068 against TGFβ1 and TGFβ3. Against TGFβ3, the affinity matured variant XPA.42.681 showed a dramatic improvement over the parent, with neutralization up to 80%.
Figure 7. Inhibition of SMAD2 phosphorylation. Detroit 562 cells were pre-incubated with neutralizing or control antibodies (50 ng/mL), then stimulated with 5 ng/ML TGFβ1, -2, or -3 for 30 minutes. Cell lysate were prepared and analyzed by ELISA for total and phosphorylated SMAD2. Percent inhibition of pSMAD2 was normalized to total SMAD2 for each clone relative to the anti-KLH control antibody.
Tumor growth inhibition in a Detroit 562 xenograft study
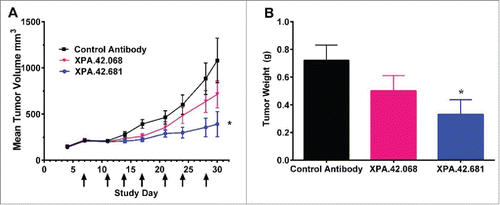
The Detroit 562 pharyngeal carcinoma cell line has been shown to express TGFβ1, 2, and 3 proteins as measured by ELISA (Fig. S3A) and is responsive to all 3 isoforms of TGFβ in the pSMAD2 assay (Fig. S3B). Inhibition of growth in Detroit 562 xenografts by rTGFβrII-HuFc (a putative pan inhibitor of TGFβ function) has previously been reported in the literature.Citation39 In the current study, we used the Detroit 562 xenograft model to determine the effects of TGFβ neutralizing antibody XPA.42.068 in vivo and compare its potency with that of the higher affinity light chain variant XPA.42.681. Female nude mice were implanted subcutaneously with Detroit 562 tumor cells on Day 0 and showed significant tumor grow by Day 4 reaching 145.8 ± 31.7 mm3. The mice were then randomized into treatment groups (n=12/group) and received twice weekly intraperitoneal (IP) injections of either XPA.42.068, XPA.42.681, or the anti-KLH HuIgG2 isotype control antibody at 3 mg/kg starting at Day 7 when the tumors had reached an average size of 214.5 ± 45.4 mm3. The results of this study are shown in . Mice treated with the control antibody had a mean tumor volume of 1080.9 ± 835.7 by Day 30. The data showed a 33.5% reduction in mean tumor volume (p=0.1) by Day 30 following treatment with XPA.42.068 at 3 mg/kg. In contrast, mice treated with the engineered XPA.42.681 showed a statistically significant (p < 0.05) 63.4% reduction in tumor size versus the control antibody. At Day 30, tumor weights were recorded immediately after harvest and showed a significant decrease in the group treated with XPA.42.681 vs. the control antibody (0.33 ± 0.11g versus 0.72 ± 0.11g). These data indicate that, although XPA.42.068 had a modest effect on mean tumor volumes and tumor weight, XPA.42.681 had a dramatic and statistically significant effect on tumor growth. End of study serum antibody levels were measured for the XPA.42.068 and XPA.42.681 treated groups and antibody levels were comparable (data not shown) suggesting that the observed potency differences were not due to differences in pharmacokinetics.
Figure 8. Detroit 562 xenograft tumor volumes following twice weekly IP injections of XPA.42.068, XPA.42.681 or control antibody.Animals were implanted at Day 0 and randomized at Day 4 following establishment of tumors. Each group with 12 mice per group was treated twice weekly with 3mg/kg XPA.42.068, XPA.42.681 or isotype control antibody. Tumor volumes were determined prior to dosing. A) Change in mean tumor volume over time for each dose group. Arrows indicate days of antibody injection. Control Antibody (squares), XPA.42.068 (inverted triangles), and XPA.42.681 (Circles) B). Mean tumor weight for each group taken at Day 30 post-implantation. (*=p < 0.05).
Discussion
The generation of specific high affinity antibodies to TGFβ, in particular a fully human, pan-TGFβ neutralizing antibody that is capable of inhibiting all 3 isoforms equally, has proven to be challenging. A few such anti-TGFβ antibodies have been described in the literature. In 1989 Dasch et al.,Citation27 despite only screening for binding to TGFβ1 and TGFβ2 at the time, found a hybridoma-derived mouse pan-TGFβ neutralizing antibody 1D11.16 (1D11). While attempting to generate a TGFβ1-specific antibody, Lucas et al. identified another pan-TGFβ neutralizing mouse hybridoma antibody (2G7).Citation31 Hoefer et al reported a pan-isoform reactive antibody that was raised against a 12 AA peptide comprising amino acids 48 to 60 of TGFβ1.Citation36 There have been no reports of other unrelated pan-neutralizing TGFβ monoclonal antibodies in the literature.Citation40
This work demonstrates that panning of large naïve recombinant phage display libraries of human antibody fragments can be used to generate high-affinity, neutralizing antibodies to TGFβ. In the current study, antibodies that bound and neutralized individual TGFβ isoforms, as well as dual-specific antibodies that neutralized 2 isoforms were observed and characterized. However, from primary panning efforts, we were not able to isolate a pan- high affinity clone that uniformly neutralized all 3 TGFβ isoforms.
TGF-β ligand isoforms are expressed in and are important drivers of many diseases.Citation3,41 Although all 3 TGFβ ligands have high amino acid homology, each mature protein has unique heterologous motifs that have been maintained throughout evolution. Conservation of these heterologous motifs suggests that each homolog could have some isoform-specific properties.Citation29,42,43 Indeed, differentiated roles for each of the TGFβ isoforms in disease have been suggested by the isoform-specific properties of the individual TGFβ1, 2, and 3 isoforms in in vivo studies.Citation44,45 and by the unique phenotypes for each individual TGFβ knock-out mouse. In general, TGFβ1 expression is induced by immediate early genes in response to signals such as injury and stress, whereas TGFβ2 and TGFβ3 appear to be more hormonally and developmentally regulated.Citation45 As specific examples, all TGFβ isoforms are expressed at variable levels in HaCaT keratinocytes, suggesting separate roles in skin carcinogenesis and therefore the use of multi-ligand inhibitors for increased therapeutic benefit. In another example,Citation46 TGFβ1 is generally considered to be the major isoform involved in fibrosis, however a combination of TGFβ isoforms demonstrated additive effects. In addition, the ability of TGFβ isoforms to stimulate production of other TGFβ ligands suggests that inhibition of multiple TGFβ ligands may yield the best therapeutic effect in reducing renal fibrosis.Citation47 Consistent with these data Cambridge Antibody Technologies and Genzyme developed monoclonal antibodies specific to individual ligands, CAT‑152.Citation48,49 and CT‑192,Citation50 targeting TGFβ2 or TGFβ1 respectively, or the pan-isoform specific GC‑1008.Citation40,51-53 All antibodies were advanced through preclinical and clinical development, but GC1008 has progressed furthest in the clinic for both cancer and fibrotic indications. It was found to be well tolerated and safe in Phase I trials for metastatic melanoma and renal cell carcinoma,Citation52 and for the fibrotic disorder focal segmental glomerulosclerosis,Citation51 and showed some clinical benefit,Citation54 and is reported to be continuing development. However, both mono-specific anti-TGFβ antibodies, CAT‑152 and CT‑192, despite being safe and well tolerated, failed to show efficacy in corneal scarring and systemic sclerosis, respectively, and were discontinued.Citation14 These data suggest that inhibition of at least 2 TGFβ isoforms is critical and that inhibition of individual isoforms is not sufficient for therapeutic benefit.
From a practical standpoint, active TGFβ is a very challenging target for therapeutic monoclonal antibody development because it is not stable at neutral pHs in purified form, and given its ubiquitous expression, there is a low abundance of high affinity TGFβ antibodies in the human and mouse antibody repertoires.Citation30-33 Despite generating hundreds of unique TGFβ binding monoclonal antibodies using phage display, the low homology of the TGFβ isoforms meant we were unable to discover high affinity pan-neutralizing antibodies regardless of the panning strategy employed. The panning produced a fairly large number of “unique” clones that had reactivity to more than one TGFβ isoform. However, surprisingly, diversity was primarily derived from the light chains rather than the heavy chains. Antibodies with shared heavy chains had different potencies according to variations in their light chain sequences. Based on our recognition of the important contribution of the light chain to TGFβ binding in the identified clones, we used a light chain shuffling affinity maturation strategy. The XPA.42.068 clone, which had high affinity and potent neutralization to TGFβ1 and TGFβ2, but weaker binding to TGFβ3 by at least an order of magnitude, was selected for affinity maturation and 2 sub-libraries were constructed consisting of the single parental heavy chain sequence combined with a diverse lambda or kappa light chain library. This light chain library utilized the same library of light chains initially used to construct the Fab primary library and did not require de novo creation, allowing rapid library generation. Panning and screening of this light chain library resulted in the identification of an affinity maturated clone, XPA.42.681 that had higher binding affinity and neutralization against all 3 TGFβ isoforms in vitro, as well as increased anti-tumor potency in vivo.
Latent TGFβ is relatively abundant and widely distributed in the body, and could potentially act as a large target sink for an anti-TGFβ therapeutic antibody that was not specific to only the active form of TGFβ. In competition assays with the TGFβ1 LAP, all of the potent neutralizing antibodies we identified bound only to free mature TGFβ protein that was not associated with the LAP, indicating their specificity for the active form of TGFβ.
The binding properties of the antibodies generated in this study were compared using 2 SPR-based kinetic methods: one using immobilized antibody and the other using immobilized TGFβ. The affinity constants (KD) for these antibodies ranged from 1.7 pM to 1400 pM using the first method and 31 pM to 2700 pM using the second method, with the immobilized antibody method (injected TGFβ) yielding higher affinity estimates due mainly to large increases in on-rate (ka). There are a number of technical as well as biological issues that might explain the differences between the 2 assay methods. Most likely, the immobilized TGFβ may be conformationally altered or partially obscured by coupling to the surface, which inhibits the on-rate of the antibody to the surface bound TGFβ.Citation55 This conformational alteration would be much less of an issue for the antibody immobilization because the antibodies are large (150 kDa), and it is unlikely that both of the antibodies’ independent binding domains would be hindered by immobilization. It is also possible that there is some form of charge attraction between the soluble TGFβ and the chip surface that either: 1) enhances the on-rate by essentially pre-concentrating the TGFβ near the chip surface; or 2) causes an electrostatic steering effect that accelerates the association of the smaller TGFβ molecule, which has a smaller radius and increased diffusivity over the antibody. When a protein is immobilized, its diffusion coefficient drops to zero, and given that the TGFβ is much smaller and has a higher diffusivity than an IgG, immobilization of the smaller TGFβ may have a larger effect on the observed rate kinetics.Citation56-58
There is no strong evidence to support which set of affinity constants more accurately reflects the in vivo situation, and therefore these data are presented as dual data sets with 2 values, both of which are meaningful in the context of their own methodologies. We are not the first to report orientation-dependent affinity differences using TGFβ. In a SPR-based study of TGFβ binding to recombinant TBRII extracellular domains by De Crescenzo,Citation59 a 4-order of magnitude orientation-dependent shift in binding affinity was seen, and it was found that the higher affinity values from the receptor immobilized assay orientation were more consistent with the cell-based radio-ligand binding assays previously performed by others. This result is consistent with the idea that the immobilization of TGFβ negatively affects the TGFβ structure or binding epitopes. When interpreting the kinetic results, it is also important to note that neither assay orientation completely eliminates the potential for avidity effects in the analysis because both the TGFβ proteins and the antibodies are bivalent dimers. To minimize the avidity effects, these analyses were performed at very low surface densities,Citation60 but it is unlikely that those effects were completely eliminated.
The potency of the antibodies in receptor competition assays correlated directly with their affinities to the various isoforms, despite the fact that the EC50 values for the high potency clones cluster around 2 nM. This clustering of result is a demonstration of the stoichiometric limitations of the competition assay, where a 4nM TGFβ concentration was used. This 4 nM TGFβ concentration is well above the KD value of the high affinity antibodies, and therefore the binding curve, and EC50 values, represent a stoichiometric titration of the antibody, where the majority of the antibody is bound, even in the low concentrations, and 50% inhibition is achieved when antibody and TGFβ concentrations are equivalent. This makes it difficult to distinguish between antibodies that may have different binding affinities if all of their KD values are well below the concentration of the TGFβ used in the assay, and relative potency was more accurately estimated by in vitro potency assays and affinity measurements. Lower levels of TGFβ could not be used given the modest affinity (on-rate) of the TGFβ to the monomeric recombinant receptors. Also evaluated was the binding stoichiometry of the antibody, TGFβ interactions. There did not appear to be any major differences among clones in binding stoichiometry. Each TGFβ homodimer has 2 independent binding sites that can be bound by either 2 separate antibodies or simultaneously by both arms of the same IgG molecule, and both binding species are evident.
In vitro TGFβ neutralization assays support the biophysical characterization profiles of isoform specificity and competition described above. Two primary cell-based assays were applied as screening and ranking tools for the candidate antibodies, the HT-2 proliferation assay and the A549 IL-11 release assay. The ability to suppress TGFβ-induced SMAD2 phosphorylation was also evaluated for select clones.
TGFβ isoforms inhibit IL-4 dependent growth of mouse T-cell line HT-2 through trans-activation of genes promoting cell cycle arrest. IL-4 transactivates a mitogenic gene expression program by activating targets such as c-myc and GM-CSF, whereas TGFβ signaling trans-activates genes that suppress c-myc and GM-CSF expression and inhibit cell growth.Citation35 Addition of any of the 3 main isoforms of TGFβ to these cells results in a similar level of growth inhibition, and, when the TGFβ is neutralized by an antibody, the HT-2 cells proliferate. This assay was highly responsive to TGFβ inhibition, and the most potent antibody XPA.42.681 showed a partial effect even at sub ng/mL concentrations. The XPA.42.089 clone showed potent inhibition of TGFβ1 and TGFβ2, but almost no activity on TGFβ3.
Another in vitro assay utilized TGFβ-mediated secretion of IL-11 from A549 lung carcinoma cells, which is part of a pro-fibrotic response in lung fibroblasts and epithelial cells. This assay models some of the TGFβ-mediated biological responses that contribute to fibrosis and metastatic disease.Citation61 The antibody clones showed expected patterns of neutralization to the various TGFβ isoforms. The XPA.42.681 clone showed the most potent neutralization of all 3 TGFβ isoforms and XPA.42.089 showed strong neutralization to TGFβ1 and 2, but almost no neutralization of TGFβ3.
The Detroit 562 pharyngeal carcinoma cell line was also used to evaluate the XPA.42.068, XPA.42.681, and XPA.42.089 antibodies’ ability to neutralize the proximal signal transduction event of SMAD2 2 phosphorylation (pSMAD2). In this assay, the XPA.42.681 clone was a potent neutralizer of SMAD2 phosphorylation for all TGFβ isoforms, whereas XPA.42.068 and XPA.42.089 were unable to inhibit SMAD2 phosphorylation induced by TGFβ3.
The in vitro cell-based results demonstrate dose-dependent neutralization of TGFβ-mediated suppression of cell proliferation, TGFβ-mediated IL-11 release, and phosphorylation of Smad2 by the neutralizing anti-TGFβ antibodies. The HT-2 proliferation and A549 IL-11 release assays were consistent with the affinity data, with the potency of the anti-TGFβ antibodies in vitro correlating with their affinity for the particular TGFβ isoform used to elicit the cellular response.
In vivo efficacy of the neutralizing antibody XPA.42.068 in inhibiting tumor growth was confirmed in a Detroit 562 xenograft model. The affinity-matured and pan-neutralizing XPA.42.681 was more efficacious at reducing tumor volumes than the parental antibody. With the selection of the high affinity pan-TGFβ neutralizing antibody XPA.42.681, the data suggest that we have developed the highest affinity and most potent TGFβ neutralizing antibody yet described. Fresolimumab (GC1008) is a fully human IgG4 monoclonal antibody that was developed by Genzyme by cloning the 1D11.16 antibody CDR regions into libraries of human framework sequences and performing various rounds of CDR swapping, mutagenesis, library construction, and screening to identify a humanized and optimized form of the 1D11.16 parent antibody.Citation62 Fresolimumab was evaluated in several Phase 1 clinical studies for focal segmental glomerulosclerosis, systemic sclerosis, myelofibrosis, idiopathic pulmonary fibrosis, renal cell carcinoma, malignant melanoma, metastatic breast cancer, and mesothelioma.Citation3 The treatment has been well tolerated in patients, supporting the idea that antibodies such as XPA.42.681 that neutralize all 3 main isoforms of TGFβ have potential uses in the treatment of a broad range of human disease. Importantly, these data suggest that although both 1D11 and XPA.42.681 are both pan-TGFβ isoform reactive antibodies, they are remarkably different in 2 ways that have direct implications for clinical application. First, the binding affinity of 1D11 is a skewed or biased toward TGFβ3. This clearly translates to an in vitro potency differential as 1D11 is most potent at inhibiting TGFβ3 function in vitro. These data suggest that in the tumor microenvironment where each of the TGFβ ligands is expressed, 1D11 would preferentially inhibit TGFβ3 function. In contrast, XPA.42.681 has a similarly high binding affinity for each of the TGFβ ligands which translates into a more uniform inhibition of TGFβ activity in vitro. These data suggest that in the tumor microenvironment, XPA.42.681 would more uniformly suppress TGFβ isoform function. Second, there is a significant improvement in the affinity of XPA.42.681 for all 3 of the isoforms compared to 1D11 which translates to an improved specific activity for the drug in vitro. This is important as the clinical trials conducted with fresolimumab were dosed as high as 15mg/kg before some benefit was achieved. Significant improvements in affinity would allow clinicians to achieve an effective therapeutic dose with less antibody. In addition, it affords the opportunity to dose higher where the amounts of antibody delivered are limiting. Both of these differences have significant implications from a clinical therapy perspective and provide opportunities for improved therapeutic utility.
In conclusion, neutralization of TGFβ function provides an attractive therapeutic approach for the treatment of advanced metastatic cancers as well as fibrosis. We have generated high-affinity fully human monoclonal antibodies capable of neutralizing various isoforms of TGFβ. These antibodies both show reductions in tumor growth in a pharyngeal carcinoma xenograft model and potent neutralization of the receptor-ligand interaction both biochemically and at the cellular level.
Materials and methods
Recombinant proteins and antibodies
Recombinant human TGFβ1, TGFβ2, and TGFβ3 were obtained from PeproTech (Rocky Hill, NJ; Cat# 100-21, 100-35B, and 100-36E, respectively) and R&D Systems (Minneapolis, MN). TGFβ-RI, TGFβ-RII, TGFβ-RIII, and recombinant human TGFβ1 Latency Associated Peptide (rhLAP) were purchased from R&D Systems. Anti-TGFβ human monoclonal IgG antibodies and the anti-KLH control IgG were made by XOMA Corp. (Berkeley, CA). The monoclonal mouse IgG1 1D11 antibody was obtained from R&D Systems (Cat# MAB1835). Recombinant mouse IL-2 and mouse IL-4 were obtained from R&D Systems (Cat# 402-ML and 404-ML, respectively).
Biotinylating TGFβ antigens
The TGFβ antigens were biotinylated with NHS-PEG4-Biotin (Pierce, Rockford, IL). Briefly, TGFβ stored in a low pH buffer was neutralized by the addition of 20x phosphate-buffered saline (PBS) to bring the pH to ∼6.0. A 30-fold molar excess of the pre-activated NHS-PEG4-Biotin was added and the mixture was incubated at room temperature for 20 minutes. After incubation, an equal volume of 10 mM Glycine, pH 3.0 (GE Healthcare, Piscataway NJ) was added and the sample was put immediately into dialysis using a 6-8 kDa cut-off dialysis unit (Calbiochem, Billerica, MA) against a pH 3.5 10 mM citrate buffer for storage (Sigma Aldrich, St. Louis, MO).
Panning using XOMA phage libraries XFab1 and XscFv2
XFab1 and XscFv2 phage libraries were separately panned using solution-based panning against each isoform of TGFβ. In the first selection round, 50X library equivalent was incubated with 200 pmoles of biotinylated TGFβ captured on streptavidin-coated magnetic Dynabeads® M-280 (Life Technologies, Carlsbad, CA) and washed 3 times with PBS with 0.1% Tween (Teknova) followed by 3 PBS washes with 2 minute incubation between washes. Bound phage were eluted from the beads with 100 mM TEA (trimethylamine; Fisher Scientific), neutralized with the addition of an equal volume of 1 M Tris-HCl, pH 7.4 (Teknova) and used to infect log-growing TG1 bacterial cells (OD600 ∼0.5) (Agilent). In the second selection round, 1×1011 cfu (colony forming units) of eluted and amplified phage from the previous round were incubated with 100 pmoles of biotinylated TGFβ and washed following same wash conditions as the previous round. Round 3 selection used 50 pmoles of biotinylated TGFβ and 3 PBS with 0.1% Tween washes with 6 minutes of incubation time between washes followed by 3 PBS washes 4 to 6 minutes in length.
Phage were prepared for use as input in subsequent panning rounds as follows: 100X of the previous round output was rescued by superinfection using MK07 helper phage. Cells were grown at 37°C until cells reached the log-growing phase, and were infected with MK07 (New England Biolabs, Ipswich, MA) at a multiplicity of infection of ∼20 for 30 minutes at 37°C without shaking followed by 30 minutes with shaking. After infection cells were transferred to new 2YT media supplemented with 50 μg/mL Kanamycin and 100 μg/mL carbenicillin (2YTCK) and grown overnight at 25 °C. Phages were separated from cells and debris by centrifugation.
Preparation of soluble fragment antibodies used in primary screening for TGFβ binders
Periplasmic extracts (PPE) containing soluble antibody fragments were prepared following same methods described in Schwimmer et al.Citation34 Briefly individual colonies were picked and 1 mL cultures were grown in 96 well plates At the appropriate density (OD600nm of 0.5) 1 mM isopropyl-β-D-thiogalactopyranoside (Calbiochem, Calbiochem, Billerica, MA, USA) was added to induce antibody fragment expression of cultures at 25°C for 16 to 18 hours. PPE were prepared from the resulting cell pellets by resuspension with 75uls of PPB (potassium phosphate buffer; Teknova) plus protease inhibitor cocktail (Roche) and 225 uL of H20. The plates were centrifuged and supernatants containing soluble antibody fragments were collected.
PPE binding screen by surface plasmon resonance
Primary screening of the soluble antibody fragments were performed by a direct binding SPR assay on a Biacore 4000 system (GE Healthcare). In this assay, a CM5 sensor chip was prepared via standard amine coupling chemistry using the Biacore Amine Coupling kit (GE Healthcare, Piscataway, NJ). Immobilization running buffer was HEPES buffered saline (HBS-EP+) containing 10 mM Hepes, 150 mM sodium chloride, 3 mM EDTA, and 0.05% polysorbate 20 (Teknova). The chip surface was activated with freshly mixed 1:1 solution of 0.1 M N-hydroxysuccinimide (NHS) and 0.4 M 1-ethyl-3-(3-dimethylaminopropyl) carbodiimide hydrochloride (EDC) (GE Healthcare). TGFβ1, TGFβ2, and TGFβ3 were each diluted to 6 μg/mL in acetate, pH 4.0 (GE Healthcare), and immobilized on different spots for 7-10 minutes each; the surface was then deactivated with 1 M ethanolamine (GE Healthcare). This immobilized between 3400 and 4800 response units (RU) of each TGFβ. PPE were diluted 1:1 with HBS-EP+ containing 2 mg/mL BSA (Sigma), filtered through a 0.2 μm Millex GV filter plate (Millipore, Billerica MA), and injected at 30 μL/minute for 240 seconds with a 30 second dissociation. Regeneration after each PPE injection was 10 seconds of 100 mM HCl (Fisher Scientific). The stability early report point was used to evaluate PPE binding levels. Binding level cut-offs were determined for each isoform independently as being visually above background level. RU cutoffs were 245, 175, and 125 for TGFβ1, TGFβ2, and TGFβ3, respectively.
XPA.42.068 light chain shuffling
The XPA.42.068 light chain library was constructed by shuffling the light chain variable regions of XFab1 library with XPA.42.068 heavy chain variable region, creating a library containing clones with same heavy chain variable region but diverse light chains. The following method was used to create the XPA.42.068 light chain sub-library. pXHMV-US2-L.Citation34 Fab DNA was digested simultaneously with 20 U/ug DNA Nco1-HF and Not1-HF enzymes (NEB) for 2 hours at 37°C. pXHMV-US2-K.Citation34 Fab was digested consecutively with 20 U/ug DNA Not1-HF enzymes (NEB) for 1 hour at 37°C followed by 20 U/ug DNA Nco1-HF for 1 hour at 37°C. Digested DNA separated on 0.8% agarose gel; DNA digested product ran at 5240 bp, which is the pXHMV-US2-L or pXHMV-US2-K minus the heavy chain variable and Fd region was excised from agarose gel and purified using Qiaquick Gel Extraction kit (Qiagen).
To isolate XPA.42.068 heavy chain variable and Fd region, pXHMV-XPA.42.068 DNA was digested simultaneously with Nco1-HF and Not1-HF. Digested product was separated on a 2% agarose gel, and XPA.42.068 heavy chain variable and Fd region DNA product, which ran at 684 bp, was excised from the gel and purified using Qiaquick Gel Extraction kit (Qiagen).
XPA.42.068 heavy chain variable and Fd region (684 bp digested and purified product) were ligated into pXHMV-US2-K and pXHMV-US2-L minus the heavy chain variable and Fd region (5240 bp digested and purified product) at a vector-to-insert ratio of 1:5 and 1:3, respectively, using T4 ligase. Ligated products were transformed into TG1 cells and plated on 2YTCG bioassay plates to make pXHMV-US2-L and pXHMV-US2-K libraries of 1×108 library size each.
Panning of XPA.42.068 light chain shuffled libraries
Two rounds of solution phase panning using XPA.42.068 light chain lambda library were performed, each using 1×1012 cfu/ml of starting phage. 50 pmoles of biotinylated TGFβ2 or TGFβ3 was used as antigen for the first panning round and 1 pmoles or 0.5 pmoles of TGFβ3 was used for the second panning round.
XPA.42.068 affinity maturation PPE binding screen by SPR
Screening of the affinity-matured clones was performed in a way similar to the primary screen, but with much lower levels of immobilized TGFβ to allow for more accurate off-rate analysis. The Biacore A100 was used with a CM5 sensor chip with 200 - 450 RU of each TGFβ immobilized. PPE were diluted 1:1 with HBS-EP+ with 2 mg/mL BSA, filtered through a 0.2 μm Millex GV filter plate, and then injected at 30 μL/minute for 240 seconds with a 600 second dissociation for off-rate measurement. Reference subtracted data was plotted and clones that appeared to have either greater stability or higher binding levels to TGFβ3 were selected for further evaluation and characterization.
Antibody affinity characterization
Immobilized antibody kinetics method
A CM4 sensor chip was used on the Biacore 2000 system. Amine coupling was used as above with immobilization of 136 RU XPA.42.681 on Fc2, 123 RU of XPA.42.068 on Fc3, and 260 RU of XPA.42.089 on Fc4. Fc1 was the activated and deactivated control. Kinetic analysis was performed as above with each TGFβ (TGFβ1, TGFβ2, TGFβ3) injected at 10 nM, 2 nM, 0.4 nM, and 0.08 nM with blanks bracketing each concentration series and quadruplicate injections. Regeneration was then performed with 3 injections of 30 seconds each of 100 mM HCl in 3 M MgCl2. These data were analyzed using Scrubber2 (BioLogic Software, Campbell, Australia) and double referenced by subtracting data from both the blank flow cell and the averaged bracketing blank injections. The rate constants for on-rates (ka) and off-rates (kd) were estimated by fitting these referenced data using a 1:1 binding interaction model.Citation55 The equilibrium dissociation constant (KD) was then calculated as KD = kd/ka.
Immobilized TGFβ affinity method
A CM1 sensor chip (GE Healthcare), which has a planar–COOH surface, was used on a Biacore 2000 system (GE Healthcare). TGFβ proteins were immobilized using standard amine chemistry. The chip surface was activated with 4 minute injections at 20 μL/minute of NHS/EDC. A 0.1 μg/mL solution of TGFβ in acetate, pH 4.0, was injected for several minutes to achieve a target density of ∼30 RU for each TGFβ; the surface was then deactivated with 1 M ethanolamine HCl, pH8.5. For kinetic analysis, the running buffer was switched to a thoroughly degassed form of the HBS-EP+ buffer supplemented with 1 mg/mL BSA. Antibodies were diluted in running buffer to 33.33 nM, 6.67 nM, 1.33 nM, 267 pM, and 53 pM. They were then injected for 4 minutes at 50 μL/minute with a 900 second dissociation time. Regeneration was performed with a 12 μL (14.4 second) injection of 100 mM HCl at 50 μL/minute. Injections were across all flow cells simultaneously and all samples and controls were run in quadruplicate. The data were analyzed as above.
Competition assays
Receptor competition method
A CM5 sensor chip was used on the Biacore 2000 system. Standard amine coupling was used to immobilize the TGFβ receptors. 5 μg/mL of TGFβ-RII or TGFβ-RIII in acetate, pH 4.5, was injected at 20 μL/minute for 4 minutes and resulted in 1000 - 4000 RU of TGFβ receptor immobilized. Fc1 was the activated and deactivated control. The competition assay was performed using HBS-EP+/BSA running buffer as described above. TGFβ was used in all injections except blank controls at 100 ng/mL (4 nM) to 40 ng/mL (1.6 nM) and was prepared with 10 μg/mL (66.6 nM) of competitor and control antibodies. Samples were allowed to come to equilibrium for 40 minutes at room temperature before the Biacore run was started. Equilibrated samples were then injected at 10 μL/minute for 2 minutes. Regeneration was performed every cycle with one 10-second injection of pH 2.5 glycine. Samples were run in at least duplicate and analyzed for level of TGFβ bound.
rhLAP competition
A CM5 sensor chip was used on the Biacore 2000 system. Standard amine coupling was used; 2 μg/mL of rhLAP in acetate, pH 4.5, was injected for 4 minutes and resulted in 400 RU of rhLAP immobilized. Fc1 was the activated and deactivated control. The rhLAP competition assay was performed using HBS-EP+/BSA running buffer as described above. TGFβ1 was used in all injections except blank controls at 0.25 μg/mL (10 nM) and was prepared with 10 μg/mL (66.6 nM) of competitor and control antibodies. Samples were allowed to come to equilibrium for 40 minutes at room temperature before the Biacore run was started. Equilibrated samples were then injected at 40 μL/minute for 2 minutes over the control and the rhLAP surface. Regeneration was performed every cycle with 2 10-second injections of 100 mM HCl. Samples were run in duplicate and analyzed for level of TGFβ1 bound.
Stoichiometry evaluation
A C1 sensor chip was used on the Biacore 2000 system and standard amine coupling chemistry was used to immobilize between 240-270 RU of anti-TGFβ antibodies. Flow cell 1 was an activated and deactivated control, and the other flow cells had either the XPA.42.681, XPA.42.089, or the 1D11 antibodies. Stoichiometry analysis was performed using HBS-EP+/BSA running buffer as above. Each antibody was injected to verify that there was no binding to the surface in the absence of TGFβ. TGFβ sample injections were performed at 1 μg/mL (40 nM) followed by a 10 μg/mL (66.6 nM) antibody injection. Regeneration was performed with pH 2.0 glycine for 12 seconds. Binding levels of TGFβ were measured and compared to binding levels of the injected antibody.
The Rmax kinetics experiment was also performed to address stoichiometry. A CM5 sensor chip was used on the Biacore 2000 system to immobilize 3000 RU of anti-human Fc capture antibody (GE Healthcare). Human antibodies were captured at 105-120 RU, TGFβ2 was injected in duplicates of 20 nM, 4 nM, and 0.8 nM for 4 minutes, and regeneration was then performed with 3 M MgCl2. The data were analyzed using Scrubber2 software and were double referenced by subtracting both the blank flow cell data and the averaged bracketing blank injections. The on-rate (ka) and off-rate (kd) rate constants were estimated by fitting these data using a 1:1 binding interaction model, and the RUmax from the fit was reported.
HT-2 assay
An HT-2 proliferation assay was adapted from an assay previously described by Reugemer et al.Citation35 HT-2 murine T cells (ATCC, Manassas, VA) were maintained by splitting every 2-3 days at 1.5 × 104 – 2.5 × 104 cells/mL in RPMI medium containing 10% FBS, 10 mM Hepes, 2 mM glutamine, 50 μM 2-ME (Sigma-Aldrich). Recombinant mouse IL-2 was freshly added at 200 IU/mL to each flask from a concentrated stock. On day 1, cells were washed in media to remove IL-2 and dispensed into opaque 96-well plates at 10,000 cells/well with 2000 IU/mL recombinant mouse IL-4. TGFβ1, -2, or -3 was added after 1 hour pre-incubation with or without antibodies across a titration series. After 48 hours of incubation at 37°C, ATP was measured as a readout for metabolically active cells (i.e. viability) on a MDS Flexstation3 using the Cell Titer GloR assay (Promega, Madison, WI), according to the manufacturer's recommendations.
IL-11 release assay
The IL-11 release assay was adapted from an assay previously described by Rapoza et al.Citation61 in which A549 lung carcinoma cells were pre-incubated with neutralizing or control antibodies and then treated with or without TGFβ. A549 cells were grown in F12 + 10% serum. The day prior to treatment, cells were detached with versene solution (to retain receptor expression) and seeded at 40,000 cells/well into a 96-well flat-bottom plate. The next day, TGFβ1, -2, or -3 at EC80 were pre-incubated for 1 hour with or without neutralizing or control antibodies across a dilution series prior to addition to the cells. As controls, TGFβ alone, TGFβ + anti-KLH IgG2 or media alone were added to the plates. After 24 hours at 37°C, IL-11 levels in cell culture supernatants were measured by ELISA using the IL-11 DuoSet® ELISA kit (R&D Systems, Cat# DY218), according to the manufacturer's recommendations.
pSMAD2 assay
Detroit 562 (human pharyngeal carcinoma) cells were maintained in IMDM + 10% FBS. Cells were detached with versene solution and plated at 500,000 cells/well into a 6-well dish. The next day, cells were cultured in serum-free IMDM for 3 hours and pre-incubated for 1 hour with or without antibodies (50 ng/mL) prior to addition of TGFβ1, -2, or -3 (5 ng/mL). After 30 minutes at 37°C, cells were lysed in cell lysis buffer (Cell Signaling, Cat# 9803) containing 1 mM freshly added PMSF. After rocking for 5 minutes at 4°C, cells were scraped off and dispensed into a 96-deep-well plate to lyse on ice for 20 minutes. Lysates were spun down at 3000 rpm for 10 minutes at 4°C. Lysates were diluted and phospho-SMAD2 (pSMAD2) and total SMAD2 levels were measured by ELISA according to the manufacturer's recommendations (Cell Signaling, Cat# 7348 and Cat# 7244, respectively). Levels of pSMAD2 were normalized to total SMAD2 and percent inhibition was calculated for each clone relative to the anti-KLH control.
Detroit 562 xenograft study
A tumor xenograft study using Detroit 562 cells was performed to evaluate dose-related inhibition of tumor growth progression in nude mice. Detroit 562 cells were implanted subcutaneously at 5 × 106 cells per animal mixed 1:1 with matrigel (BD Biociences, San Jose, CA) into the lower left ventral abdominal region of 8-9-week old female athymic (nu/nu) mice (Charles River Laboratories, Wilmington, MA). All protocols were reviewed by XOMA's Institutional Animal Care and Use Committee and all experiments were conducted under its guidance. The Study Director software application (Studylog Systems, Inc., San Francisco, CA) was used to track body weights, monitor growth of the tumors throughout the study, and randomize animals into groups of 12 mice each. Mice were IP injected twice weekly with 3 mg/kg of XPA.42.068, XPA.42.681, or with control antibody (anti-KLH hIgG2 isotype control) starting 7 days after tumor implantation. Tumors were measured biweekly with a digital caliper and tumor volumes were calculated based on the formula, Volume = L x W2/2. Animals were sacrificed on the day after the last dose (day 30) after 7 doses of antibody treatment. Tumor weights were recorded immediately after harvest. All tumor measurements were taken by personnel blinded to the treatment groups. Statistical significance for all measurements was determined by Student's t-test (one-tailed).
Disclosure of potential conflicts of interest
At the time work was conducted, all authors were employees of XOMA Corporation.
Bedinger et al Supplemental Data
Download PDF (545.9 KB)Acknowledgments
The authors thank Diane Wilcock for thoughtful review of the manuscript and insightful comments as well as Donna Combs, Kristen Michelson, Jennifer Green, Yero Espinoza, Angela Lau, and Billy Cheung for their technical assistance.
References
- Korpal M, Kang Y. Targeting the transforming growth factor-β signalling pathway in metastatic cancer. Eur J Cancer 2010; 46:1232–40; PMID:20307969; http://dx.doi.org/10.1016/j.ejca.2010.02.040
- Miyazono K. Transforming growth factor-β signaling in epithelial-mesenchymal transition and progression of cancer. Proc Japan Acad Series B, Phys Biolog Sci 2009; 85:314–23; PMID:19838011; http://dx.doi.org/10.2183/pjab.85.314
- Akhurst RJ, Hata A. Targeting the TGFbeta signalling pathway in disease. Nat Rev Drug Discov 2012; 11:790–811; PMID:23000686; http://dx.doi.org/10.1038/nrd3810
- Wahl SM, Wen J, Moutsopoulos N. TGF-β: a mobile purveyor of immune privilege. Immunol Rev 2006; 213:213–27; PMID:16972906; http://dx.doi.org/10.1111/j.1600-065X.2006.00437.x
- Lee Y, Awasthi A, Yosef N, Quintana FJ, Xiao S, Peters A, Wu C, Kleinewietfeld M, Kunder S, Hafler DA, et al. Induction and molecular signature of pathogenic TH17 cells. Nat Immunol 2012; 13:991–9; PMID:22961052; http://dx.doi.org/10.1038/ni.2416
- van 't Veer LJ, Dai H, van de Vijver MJ, He YD, Hart AA, Mao M, Peterse HL, van der Kooy K, Marton MJ, Witteveen AT, et al. Gene expression profiling predicts clinical outcome of breast cancer. Nature 2002; 415:530–6; PMID:11823860; http://dx.doi.org/10.1038/415530a
- Massague J. TGF-β signal transduction. Annu Rev Biochem 1998; 67:753–91; PMID:9759503; http://dx.doi.org/10.1146/annurev.biochem.67.1.753
- Brown PD, Wakefield LM, Levinson AD, Sporn MB. Physicochemical activation of recombinant latent transforming growth factor-β's 1, 2, and 3. Growth Factors 1990; 3:35–43; PMID:2200452; http://dx.doi.org/10.3109/08977199009037500
- Schultz-Cherry S, Ribeiro S, Gentry L, Murphy-Ullrich JE. Thrombospondin binds and activates the small and large forms of latent transforming growth factor-β in a chemically defined system. J Biol Chem 1994; 269:26775–82; PMID:7929413
- D'Angelo M, Billings PC, Pacifici M, Leboy PS, Kirsch T. Authentic matrix vesicles contain active metalloproteases (MMP). a role for matrix vesicle-associated MMP-13 in activation of transforming growth factor-β. J Biol Chem 2001; 276:11347–53; PMID:11145962; http://dx.doi.org/10.1074/jbc.M009725200
- Derynck R, Jarrett JA, Chen EY, Eaton DH, Bell JR, Assoian RK, Roberts AB, Sporn MB, Goeddel DV. Human transforming growth factor-β complementary DNA sequence and expression in normal and transformed cells. Nature 1985; 316:701–5; PMID:3861940; http://dx.doi.org/10.1038/316701a0
- Gentry LE, Lioubin MN, Purchio AF, Marquardt H. Molecular events in the processing of recombinant type 1 pre-pro-transforming growth factor β to the mature polypeptide. Mol Cell Biol 1988; 8:4162–8; PMID:3185545; http://dx.doi.org/10.1128/MCB.8.10.4162
- Madisen L, Webb NR, Rose TM, Marquardt H, Ikeda T, Twardzik D, Seyedin S, Purchio AF. Transforming growth factor-β 2: cDNA cloning and sequence analysis. DNA 1988; 7:1–8; PMID:3162414; http://dx.doi.org/10.1089/dna.1988.7.1
- Grutter C, Wilkinson T, Turner R, Podichetty S, Finch D, McCourt M, Loning S, Jermutus L, Grutter MG. A cytokine-neutralizing antibody as a structural mimetic of 2 receptor interactions. Proc Natl Acad Sci USA 2008; 105:20251–6; PMID:19073914; http://dx.doi.org/10.1073/pnas.0807200106
- Cheifetz S, Bassols A, Stanley K, Ohta M, Greenberger J, Massague J. Heterodimeric transforming growth factor β. Biological properties and interaction with three types of cell surface receptors. J Biol Chem 1988; 263:10783–9; PMID:2899081
- Ehrlich M, Gutman O, Knaus P, Henis YI. Oligomeric interactions of TGF-β and BMP receptors. FEBS Lett 2012; 586:1885–96; PMID:22293501; http://dx.doi.org/10.1016/j.febslet.2012.01.040
- Groppe J, Hinck CS, Samavarchi-Tehrani P, Zubieta C, Schuermann JP, Taylor AB, Schwarz PM, Wrana JL, Hinck AP. Cooperative assembly of TGF-β superfamily signaling complexes is mediated by two disparate mechanisms and distinct modes of receptor binding. Mol Cell 2008; 29:157–68; PMID:18243111; http://dx.doi.org/10.1016/j.molcel.2007.11.039
- Rodriguez C, Chen F, Weinberg RA, Lodish HF. Cooperative binding of transforming growth factor (TGF)-β 2 to the types I and II TGF-β receptors. J Biol Chem 1995; 270:15919–22; PMID:7608141; http://dx.doi.org/10.1074/jbc.270.27.15919
- Wrana JL, Attisano L, Wieser R, Ventura F, Massague J. Mechanism of activation of the TGF-β receptor. Nature 1994; 370:341–7; PMID:8047140; http://dx.doi.org/10.1038/370341a0
- Horbelt D, Denkis A, Knaus P. A portrait of Transforming Growth Factor β superfamily signalling: Background matters. Int J Biochem Cell Biol 2012; 44:469–74; PMID:22226817; http://dx.doi.org/10.1016/j.biocel.2011.12.013
- Nickel J, Sebald W, Groppe JC, Mueller TD. Intricacies of BMP receptor assembly. Cytokine Growth Factor Rev 2009; 20:367–77; PMID:19926516; http://dx.doi.org/10.1016/j.cytogfr.2009.10.022
- Ehrlich M, Horbelt D, Marom B, Knaus P, Henis YI. Homomeric and heteromeric complexes among TGF-β and BMP receptors and their roles in signaling. Cell Signal 2011; 23:1424–32; PMID:21515362; http://dx.doi.org/10.1016/j.cellsig.2011.04.004
- Lopez-Casillas F, Wrana JL, Massague J. Betaglycan presents ligand to the TGF β signaling receptor. Cell 1993; 73:1435–44; PMID:8391934; http://dx.doi.org/10.1016/0092-8674(93)90368-Z
- Rotzer D, Roth M, Lutz M, Lindemann D, Sebald W, Knaus P. Type III TGF-β receptor-independent signalling of TGF-beta2 via TbetaRII-B, an alternatively spliced TGF-β type II receptor. EMBO J 2001; 20:480–90; PMID:11157754; http://dx.doi.org/10.1093/emboj/20.3.480
- del Re E, Babitt JL, Pirani A, Schneyer AL, Lin HY. In the absence of type III receptor, the transforming growth factor (TGF)-β type II-B receptor requires the type I receptor to bind TGF-beta2. J Biol Chem 2004; 279:22765–72; PMID:14996829; http://dx.doi.org/10.1074/jbc.M401350200
- Cheifetz S, Hernandez H, Laiho M, ten Dijke P, Iwata KK, Massague J. Distinct transforming growth factor-β (TGF-β) receptor subsets as determinants of cellular responsiveness to three TGF-β isoforms. J Biol Chem 1990; 265:20533–8; PMID:1700790
- Dasch JR, Pace DR, Waegell W, Inenaga D, Ellingsworth L. Monoclonal antibodies recognizing transforming growth factor-β. Bioactivity neutralization and transforming growth factor β 2 affinity purification. J Immunol 1989; 142:1536–41; PMID:2537357
- de Martin R, Haendler B, Hofer-Warbinek R, Gaugitsch H, Wrann M, Schlusener H, Seifert JM, Bodmer S, Fontana A, Hofer E. Complementary DNA for human glioblastoma-derived T cell suppressor factor, a novel member of the transforming growth factor-β gene family. EMBO J 1987; 6:3673–7; PMID:3322813
- ten Dijke P, Hansen P, Iwata KK, Pieler C, Foulkes JG. Identification of another member of the transforming growth factor type β gene family. Proc Natl Acad Sci USA 1988; 85:4715–9; PMID:3164476; http://dx.doi.org/10.1073/pnas.85.13.4715
- Mao XW, Kettering JD, Gridley DS. Immunotherapy with low-dose interleukin-2 and anti-transforming growth factor-β antibody in a murine tumor model. Cancer Biotherapy 1994; 9:317–27; PMID:7719379; http://dx.doi.org/10.1089/cbr.1994.9.317
- Lucas C, Bald LN, Fendly BM, Mora-Worms M, Figari IS, Patzer EJ, Palladino MA. The autocrine production of transforming growth factor-β 1 during lymphocyte activation. A study with a monoclonal antibody-based ELISA. J Immunol 1990; 145:1415–22; PMID:2384664
- Flanders KC, Roberts AB, Ling N, Fleurdelys BE, Sporn MB. Antibodies to peptide determinants in transforming growth factor β and their applications. Biochemistry 1988; 27:739–46; PMID:2450577; http://dx.doi.org/10.1021/bi00402a037
- Danielpour D, Dart LL, Flanders KC, Roberts AB, Sporn MB. Immunodetection and quantitation of the two forms of transforming growth factor-β (TGF-β 1 and TGF-β 2) secreted by cells in culture. J Cell Physiol 1989; 138:79–86; PMID:2910889; http://dx.doi.org/10.1002/jcp.1041380112
- Schwimmer LJ, Huang B, Giang H, Cotter RL, Chemla-Vogel DS, Dy FV, Tam EM, Zhang F, Toy P, Bohmann DJ, et al. Discovery of diverse and functional antibodies from large human repertoire antibody libraries. J Immunol Meth 2013; 391:60–71; http://dx.doi.org/10.1016/j.jim.2013.02.010
- Ruegemer JJ, Ho SN, Augustine JA, Schlager JW, Bell MP, McKean DJ, Abraham RT. Regulatory effects of transforming growth factor-β on IL-2- and IL-4-dependent T cell-cycle progression. J Immunol 1990; 144:1767–76; PMID:2407783
- Hoefer M, Anderer FA. Anti-(transforming growth factor β) antibodies with predefined specificity inhibit metastasis of highly tumorigenic human xenotransplants in nu/nu mice. Cancer Immunol Immunother 1995; 41:302–8; PMID:8536276; http://dx.doi.org/10.1007/BF01517218
- Sengle G, Ono RN, Sasaki T, Sakai LY. Prodomains of transforming growth factor β (TGFbeta) superfamily members specify different functions: extracellular matrix interactions and growth factor bioavailability. J Biol Chem 2011; 286:5087–99; PMID:21135108; http://dx.doi.org/10.1074/jbc.M110.188615
- De Crescenzo G, Pham PL, Durocher Y, Chao H, O'Connor-McCourt MD. Enhancement of the antagonistic potency of transforming growth factor-β receptor extracellular domains by coiled coil-induced homo- and heterodimerization. J Biol Chem 2004; 279:26013–8; PMID:15044491; http://dx.doi.org/10.1074/jbc.M400655200
- Van Aarsen LA, Leone DR, Ho S, Dolinski BM, McCoon PE, LePage DJ, Kelly R, Heaney G, Rayhorn P, Reid C, et al. Antibody-mediated blockade of integrin α v β 6 inhibits tumor progression in vivo by a transforming growth factor-β-regulated mechanism. Cancer Res 2008; 68:561–70; PMID:18199553; http://dx.doi.org/10.1158/0008-5472.CAN-07-2307
- Lonning S, Mannick J, McPherson JM. Antibody targeting of TGF-β in cancer patients. Curr Pharm Biotechnol 2011; 12:2176–89; PMID:21619535; http://dx.doi.org/10.2174/138920111798808392
- Kubiczkova L, Sedlarikova L, Hajek R, Sevcikova S. TGF-β - an excellent servant but a bad master. J Trans Med 2012; 10:183; http://dx.doi.org/10.1186/1479-5876-10-183
- Mittl PR, Priestle JP, Cox DA, McMaster G, Cerletti N, Grutter MG. The crystal structure of TGF-β 3 and comparison to TGF-β 2: implications for receptor binding. Protein Sci 1996; 5:1261–71; PMID:8819159; http://dx.doi.org/10.1002/pro.5560050705
- Kingsley DM. The TGF-β superfamily: new members, new receptors, and new genetic tests of function in different organisms. Genes Dev 1994; 8:133–46; PMID:8299934; http://dx.doi.org/10.1101/gad.8.2.133
- Graycar JL, Miller DA, Arrick BA, Lyons RM, Moses HL, Derynck R. Human transforming growth factor-β 3: recombinant expression, purification, and biological activities in comparison with transforming growth factors-β 1 and -β 2. Mol Endocrinol 1989; 3:1977–86; PMID:2628733; http://dx.doi.org/10.1210/mend-3-12-1977
- Roberts AB, Sporn MB. Differential expression of the TGF-β isoforms in embryogenesis suggests specific roles in developing and adult tissues. Mol Repro Dev 1992; 32:91–8; http://dx.doi.org/10.1002/mrd.1080320203
- Gold LI, Jussila T, Fusenig NE, Stenback F. TGF-β isoforms are differentially expressed in increasing malignant grades of HaCaT keratinocytes, suggesting separate roles in skin carcinogenesis. J Pathol 2000; 190:579–88; PMID:10727984; http://dx.doi.org/10.1002/(SICI)1096-9896(200004)190:5%3c579::AID-PATH548%3e3.0.CO;2-I
- Yu L, Border WA, Huang Y, Noble NA. TGF-β isoforms in renal fibrogenesis. Kidney Int 2003; 64:844–56; PMID:12911534; http://dx.doi.org/10.1046/j.1523-1755.2003.00162.x
- Mead AL, Wong TT, Cordeiro MF, Anderson IK, Khaw PT. Evaluation of anti-TGF-beta2 antibody as a new postoperative anti-scarring agent in glaucoma surgery. Investigative ophthalmol Visual Sci 2003; 44:3394–401; http://dx.doi.org/10.1167/iovs.02-0978
- Trabeculectomy Study Group, CAT-152 Trabeculectomy Study. Ophthalmology 2007; 114:1950; PMID:17908596; http://dx.doi.org/10.1016/j.ophtha.2007.07.024
- Denton CP, Merkel PA, Furst DE, Khanna D, Emery P, Hsu VM, Silliman N, Streisand J, Powell J, Akesson A, et al. Recombinant human anti-transforming growth factor beta1 antibody therapy in systemic sclerosis: a multicenter, randomized, placebo-controlled phase I/II trial of CAT-192. Arthritis Rheumatism 2007; 56:323–33; PMID:17195236; http://dx.doi.org/10.1002/art.22289
- Trachtman H, Fervenza FC, Gipson DS, Heering P, Jayne DR, Peters H, Rota S, Remuzzi G, Rump LC, Sellin LK, et al. A phase 1, single-dose study of fresolimumab, an anti-TGF-β antibody, in treatment-resistant primary focal segmental glomerulosclerosis. Kidney Int 2011; 79:1236–43; PMID:21368745; http://dx.doi.org/10.1038/ki.2011.33
- Morris JC, Tan AR, Olencki TE, Shapiro GI, Dezube BJ, Reiss M, Hsu FJ, Berzofsky JA, Lawrence DP. Phase I study of GC1008 (fresolimumab): a human anti-transforming growth factor-β (TGFbeta) monoclonal antibody in patients with advanced malignant melanoma or renal cell carcinoma. PloS One 2014; 9:e90353; PMID:24618589; http://dx.doi.org/10.1371/journal.pone.0090353
- Stevenson JP, Kindler HL, Papasavvas E, Sun J, Jacobs-Small M, Hull J, Schwed D, Ranganathan A, Newick K, Heitjan DF, et al. Immunological effects of the TGFbeta-blocking antibody GC1008 in malignant pleural mesothelioma patients. Oncoimmunology 2013; 2:e26218; PMID:24179709; http://dx.doi.org/10.4161/onci.26218
- Khaw P, Grehn F, Hollo G, Overton B, Wilson R, Vogel R, Smith Z. A phase III study of subconjunctival human anti-transforming growth factor β(2) monoclonal antibody (CAT-152) to prevent scarring after first-time trabeculectomy. Ophthalmology 2007; 114:1822–30; PMID:17908591; http://dx.doi.org/10.1016/j.ophtha.2007.03.050
- Myszka DG. Kinetic analysis of macromolecular interactions using surface plasmon resonance biosensors. Curr Opin Biotechnol 1997; 8:50–7; PMID:9013659; http://dx.doi.org/10.1016/S0958-1669(97)80157-7
- Kozack RE, d'Mello MJ, Subramaniam S. Computer modeling of electrostatic steering and orientational effects in antibody-antigen association. Biophysical J 1995; 68:807–14; http://dx.doi.org/10.1016/S0006-3495(95)80257-2
- Torres M, Fernandez-Fuentes N, Fiser A, Casadevall A. The immunoglobulin heavy chain constant region affects kinetic and thermodynamic parameters of antibody variable region interactions with antigen. J Biol Chem 2007; 282:13917–27; PMID:17353196; http://dx.doi.org/10.1074/jbc.M700661200
- Karlsson R, Roos H, Fägerstam L, Persson B. Kinetic and Concentration Analysis Using BIA Technology. Methods 1994; 6:99–110; http://dx.doi.org/10.1006/meth.1994.1013
- De Crescenzo G, Pham PL, Durocher Y, O'Connor-McCourt MD. Transforming growth factor-β (TGF-β) binding to the extracellular domain of the type II TGF-β receptor: receptor capture on a biosensor surface using a new coiled-coil capture system demonstrates that avidity contributes significantly to high affinity binding. J Mol Biol 2003; 328:1173–83; PMID:12729750; http://dx.doi.org/10.1016/S0022-2836(03)00360-7
- Myszka DG. Improving biosensor analysis. J Mol Recognition 1999; 12:279–84; http://dx.doi.org/10.1002/(SICI)1099-1352(199909/10)12:5%3c279::AID-JMR473%3e3.0.CO;2-3
- Rapoza ML, Fu D, Sendak RA. Development of an in vitro potency assay for therapeutic TGFbeta antagonists: the A549 cell bioassay. J Immunol Meth 2006; 316:18–26; http://dx.doi.org/10.1016/j.jim.2006.07.009
- Catriona L, Buchanan Ard, Donna KF, Celia PH, Robert GH, Lutz UJ, Steven R. Ledbetter ANTIBODIES TO TGF-β. US: Optein, Inc., Wilmington, DE (US); Genzyme Corporation, Cambridge, MA (US), 2010:43
