?Mathematical formulae have been encoded as MathML and are displayed in this HTML version using MathJax in order to improve their display. Uncheck the box to turn MathJax off. This feature requires Javascript. Click on a formula to zoom.
?Mathematical formulae have been encoded as MathML and are displayed in this HTML version using MathJax in order to improve their display. Uncheck the box to turn MathJax off. This feature requires Javascript. Click on a formula to zoom.abstract
Multispecific proteins, such as bispecific antibodies (BsAbs), that bind to two different ligands are becoming increasingly important therapeutic agents. Such BsAbs can exhibit markedly increased target binding and target residence time when both pharmacophores bind simultaneously to their targets. The cross-arm binding efficiency (χ) describes an increase in apparent affinity when a BsAb binds to the second target or receptor (R2) following its binding to the first target or receptor (R1) on the same cell. χ is an intrinsic characteristic of a BsAb mostly related to the binding epitopes on R1 and R2. χ can have significant impacts on the binding to R2 for BsAbs targeting two receptors on the same cell. JNJ-61186372, a BsAb that targets epidermal growth factor receptor (EGFR) and c-Met, was used as the model compound for establishing a method to characterize χ. The χ for JNJ-61186372 was successfully determined via fitting of in vitro cell binding data to a ligand binding model that incorporated χ. The model-derived χ value was used to predict the binding of JNJ-61186372 to individual EGFR and c-Met receptors on tumor cell lines, and the results agreed well with the observed IC50 for EGFR and c-Met phosphorylation inhibition by JNJ-61186372. Consistent with the model, JNJ-61186372 was shown to be more effective than the combination therapy of anti-EGFR and anti-c-Met monovalent antibodies at the same dose level in a mouse xenograft model. Our results showed that χ is an important characteristic of BsAbs, and should be considered for rationale design of BsAbs targeting two membrane bound targets on the same cell.
Abbreviations
| BsAbs | = | bispecific antibodies |
| c-Met | = | hepatocyte growth factor receptor |
| EGFR | = | epidermal growth factor receptor |
| mAbs | = | monoclonal antibodies |
| NSCLC | = | non-small cell lung cancer |
| pAKT | = | phosphorylated protein kinase B |
| pERK | = | phosphorylated extracellular signal-regulated kinase |
Introduction
Extracellular signal molecules that bind to specific receptors on the cell surface can affect signaling cascades that initiate responses in the target cell, such as proliferation,Citation1 migrationCitation2 or apoptosis.Citation3,4 In multicellular organisms, dysregulation of this receptor-initiated signaling can lead to uncontrolled cell proliferation and cancer.Citation5,6 Monoclonal antibodies (mAbs) and antibody-like molecules, which have emerged as the leading class of protein therapies, build upon the functionally relevant characteristics of natural antibodies, particularly, their highly selective and strong antigen binding.Citation7,8 The two Fab binding arms of conventional mAbs are the same and target the same antigen.Citation9 However, the need to inhibit multiple targets, either due to resistance or the fact that many tumors are driven by multiple growth factor pathways, has led to increased interest in the development of multispecific therapeutic agents.Citation10 Over 50 molcular formats have been engineered for the creation of bispecific molecules, which attests to the excitement and potential therapeutic development opportunities offered by these designs. Citation11,12 Among them, bispecific antibodies (BsAbs), a family of engineered antibody derivatives that recognize two different target antigens (e.g., HER2xHER3,Citation13 HER3xIGF-1RCitation14 and EGFRxHER3Citation15 for pathway blockage, or EpCAMxCD3Citation16 and CD3xCD19 Citation17 for immune effector cell re-direction), are of particular interest. BsAbs can be generated by a variety of approaches.Citation18-21 One promising method is to generate BsAbs using the controlled Fab-arm exchange (cFAE) process, which involves combining two parental antibodies that each harbor a separate mutation in the CH3 domain. Upon a reduction and oxidation process, favorable heterodimer formation occurs.Citation22
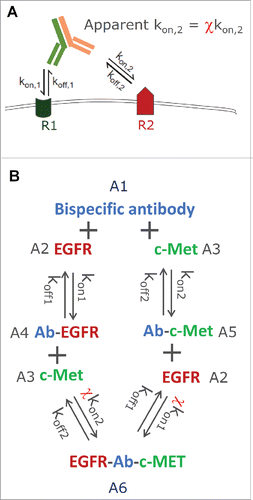
For BsAbs that target two membrane bound receptors on the same cell, once one arm of the antibody is bound to the first target (R1), the second arm is restricted to a narrow region above the plasma membrane (e.g., 100 Å), and is thus concentrated near the cell surface.Citation10 This can result in a much faster secondary binding event to the second receptor (R2), driven by the geometric reach of a cell-surface tethered antibody (). Citation23 Citation10 This acceleration of the secondary binding event can be different among various bivalent antibodies, depending on the epitope and the structural arrangement of the parts on antibody and antigen that interact. Citation24,25 Harms et al.Citation26 introduced the term “cross-arm binding efficiency” (χ) to describe this apparent acceleration of the secondary binding event. χ incorporates the increase of local antibody concentration and restriction of rotational, torsional, and bending freedom that is inherent in the geometry required for cross-arm binding which is epitope- and IgG format-dependent. Citation27 The concept of χ is similar to the concept of avidity, which is commonly referred to as a functional macroscopic affinity describing the accumulated strength of multiple affinities summed up from multiple microscopic binding interactions. Citation25 However, it is not widely appreciated that avidity can be different among different bivalent antibodies depending on the epitopes they bind to. For clarity, we used the term “cross-arm binding efficiency,” or χ, throughout our work here. When Harms et al.Citation26 introduced the concept of χ, it was first used to describe a conventional bivalent mAbs against a single receptor (epidermal growth factor receptor (EGFR), which characterized the monovalent interaction between one binding moiety followed by the bivalent interaction with the same receptor. It was later also used to characterize the bivalent binding ability of HER2 and HER3 scFv antibody fragments joined by a human serum albumin linker.Citation27 The results indicated that χ can have a significant effect on the binding to R2, i.e., the receptor that the second arm of a bivalent antibody binds after the first arm has already bound its target. Citation27 Importantly, it has also been demonstrated that, for conventional bivalent mAbs targeting EGFR, χ can be determined from in vitro cell binding data with an array of EGFR-expressing cell lines using a mathematical model integrating cross-arm binding reaction between antibody and receptor. Citation26
Figure 1. (A) Schematic representation of the cross-arm binding efficiency (χ), (B) the ligand-binding model structure for JNJ-61186372, an EGFR x c-Met bispecific antibody.
Here, we applied the concept of χ to characterize the interactions between JNJ-61186372, an anti-EGFR x c-Met BsAb, to their binding targets. JNJ-61186372 prevents binding of EGF and HGF to their respective receptors, EGFR and c-Met. In this study, the “cross-arm binding efficiency” (χ) of JNJ-61186372 was determined via fitting of in vitro flow cytometry data to a ligand binding model that incorporated χ. The model-derived χ value was verified by comparing model-predicted receptor binding to respective receptor phosphorylation inhibition. The effect of χ on EGFR/c-Met binding and tumor growth inhibition were further assessed in a mouse xenograft model.
Results
Step 1: Determination of the monovalent binding affinities of JNJ-61186372 to EGFR and c-Met on cell surfaces
To enable the estimation of χ, the monovalent binding affinities for JNJ-61186372 to cell surface EGFR and c-Met receptors were first determined.
The monovalent binding affinities of JNJ-61186372 and its EGFR and c-Met monovalent parent (gp120 x EGFR and gp120 x c-Met) to the purified EGFR and c-Met extracellular domains (ECD) had been determined by surface plasmon resonance using ProteOn.Citation28,29 The monovalent EGFR and c-Met binding affinity values for JNJ-61186372 were comparable to that of its monovalent parent gp120 x EGFR BsAb and gp120 × c-Met BsAb (kon,EGFR = 0.028 nM−1 min−1; koff,EGFR = 0.039 min−1 and kon,c-Met = 0.040 nM−1 min−1; koff,c-Met = 0.0028 min−1, respectively).Citation29 In these antibody constructs, the “null binding arm,” making the antibody monovalent against EGFR or c-Met receptors, is made with an arm binding to gp120, an epitope that is not present on the cells we used.
To examine whether the affinity values with purified proteins are similar to those with cell-surface EGF and c-Met receptors, the binding curves for gp120 × EGFR and gp120 × c-Met to multiple non-small cell lung cancer (NSCLC) cell lines with known EGFR and c-Met densities were simulated using these kon and koff values. To account for the variability between experiments, the binding of mAbs at each drug concentration was described by the fraction of maximal binding, which was calculated by dividing the observed median fluorescence intensity (MFI) by the MFI value at plateau in the same experiment.
The simulated binding curves for EGFR and c-Met monovalent parent were compared against the observed curves. For c-Met, using the kon and koff values determined by ProteOn, the simulated curve agreed reasonably well with the curve determined by flow cytometry, while for EGFR, the simulated and observed curves showed greater disparities (data not shown). This suggested that the affinity for c-Met determined using purified protein is reasonably close to the binding affinity to the cell surface receptor. Therefore, the kon and koff values for c-Met were fixed at the ProteOn-determined values in subsequent model fitting. For EGFR, kon was also fixed at the ProteOn-determined value while koff was estimated by model fitting.
The next step was to simultaneously fit the cell-binding data of the gp120 x EGFR and gp120 x c-Met BsAbs to 8 NSCLC cell lines with known EGFR and c-Met cell densities (). Cell density was set as a model input parameter. Although this parameter was kept the same for EGFR and c-Met binding to the same cell line as they were measured in the same experiment, cell density values were allowed to float among the different cell lines. A ligand binding model simultaneously fitted the cell-binding data of gp120 x EGFR and gp120 x c-Met BsAb to the 8 NSCLC cell lines with varying EGFR/c-Met densities. This model estimated the single monovalent binding affinity to cell-surface EGFR, as well as the cell densities of all 8 cell lines. The observed fraction of maximum binding over BsAb concentration profiles for all 8 cell lines, as well as the model fitting results were shown in .
Figure 2. Simultaneous fitting of the EGFR and c-Met monovalent parent (gp120 x EGFR Ab and gp120 x c-Met Ab) binding data to 8 NSCLC cell lines, model fitting (line) vs. observed data (symbols).
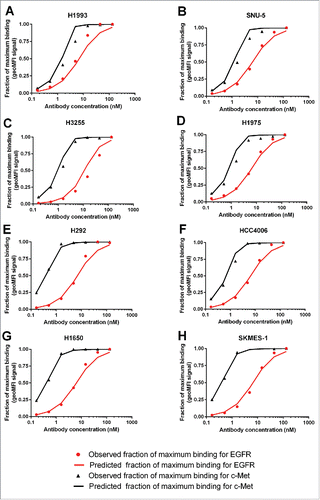
Table 1. EGFR and c-Met receptor densities and model derived cell densities of the 8 NSCLC cell lines used in flow cytometry staining.
The koff value for monovalent EGFR binding determined by flow cytometry in the cell binding assay was estimated to be 4.23-fold of the in vitro koff,EGFR based on ProteOn.Citation29 The model-derived cell densities, based on model estimated EGFR baseline receptor concentrations () for all 8 cell lines were 4.9- to 19.6- fold (mean ± SD = 10.6 ± 6.1) of the cell densities pre-specified (1×106/mL) in the flow cytometry experiment protocol (). Of note, the cell densities were not measured vigorously at the time of experiment, as it was not regarded as an important variable then. In addition, the model-estimated “cell-density” can also be understood as a semi-empirical parameter that accounted for the potential inaccuracies in not only the cell counts, but also in receptor density determination, heterogeneous nature of cell populations, and various model approximations.
Table 2. Model input parameter and model-estimated parameter values.
Step 2: Determination of χ for JNJ-61186372
Binding of JNJ-61186372 was characterized together with that of the gp120 x EGFR and gp120 x c-Met BsAbs in the same set of experiments. Since the cell surface binding of JNJ-61186372 is driven by its monovalent EGFR/c-Met binding affinities and χ, χ was estimated from JNJ-61186372 binding curves using the monovalent EGFR/c-Met binding affinities and the cell density values determined in Step 1.
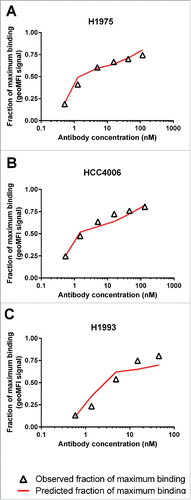
A sensitivity analysis showed that the shape of the JNJ-61186372 binding curves is most sensitive to the change of χ when EGFR and c-Met densities are similar (data not shown). Therefore, data from the 3 cell lines with the most similar levels of EGFR and c-Met (HCC4006, H1975 and H1993) were used for the determination of χ. Using the model-estimated monovalent binding affinities to cell-surface EGFR, c-Met and model-estimated cell densities for these 3 cell lines, the χ for JNJ-61186372 was estimated to be 104 (relative standard error% = 37%) via simultaneously fitting of the JNJ-61186372 binding data (). The simulated JNJ-61186372 binding curves also agreed well with the observed data for the other 5 cell lines, using the model-estimated χ, and the monovalent EGFR/c-Met binding affinities and the cell density values determined in Step 1 (data not shown).
Step 3: Verification of χ value for JNJ-61186372
Model-predicted EGFR/c-Met binding vs. observed EGFR/c-MET phosphorylation inhibition by JNJ-61186372 in cell lines
When JNJ-61186372 binds to cell lines expressing both EGFR and c-Met, the observed binding curve is a mixture of EGFR or c-Met binding events. Using the model-estimated χ ( = 104) for JNJ-61186372, the binding curves for JNJ-61186372 to individual EGFR and c-Met can now be simulated. The results for H1993 and H292, the 2 NSCLC cell lines where phosphorylation of both EGFR and c-Met had been investigated, are shown in . Inhibition of ligand-induced phosphorylation of both EGFR and c-Met was identified as an important mechanism of action that contributes to the anti-tumor activity of JNJ-61186372. Citation28 The IC50 values for EGFR and c-Met phosphorylation inhibition by JNJ-61186372 for H1993 and H292 had been determined. Citation29 The concentrations of JNJ-61186372 that were required to occupy 50% of EGFR or c-Met were plotted against the concentrations of JNJ-61186372 that inhibited 50% of pEGFR and pMet in . Not surprisingly, the observed IC50 values for c-Met phosphorylation inhibition by JNJ-61186372 was about 9-fold higher for H1993 (7.8 nM) compared with H292 (0.86 nM), in line with the over 9-fold higher c-Met density value in H1993 (5.61 × 105) compared with H292 (6.12 × 104) (). However, it is interesting to note the ∼8-fold lower IC50 values for EGFR phosphorylation inhibition by JNJ-61186372 in the H1993 cell line (3.8 nM) compared with that in the H292 cell line(29 nM), despite their similar EGF receptor densities (3.25 × 105 and 3.57 × 105, respectively) was observed (). It suggested that the c-Met arm binding served as an anchor to facilitate EGFR binding, and higher c-Met density in H1993 can lead to lower EC50 values for EGFR binding and thus lower IC50 values for EGFR phosphorylation inhibition by JNJ-61186372, i.e. an apparent impact of χ.
Figure 4. (A) Model-predicted binding of JNJ-61186372 to EGFR and c-Met, respectively, on H1993 and H292; (B) Correlation between the model-predicted EC50 for EGFR and c-Met binding and the observed IC50 values for EGFR and c-Met phosphorylation inhibition in H1993 and H292 cell lines.
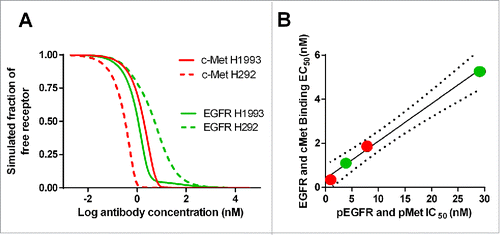
The results showed that the model-simulated EC50 value of EGFR/c-Met binding based on the estimated χ agreed well with the observed IC50 value of pEGFR and pMet inhibition (p = phosphorylation) (), which provided confidence to our model-estimated χ value for JNJ-61186372.
Evaluation of the in vivo role of χ on tumor growth inhibition of JNJ-61186372 in a mouse xenograft model
The in vivo EGFR/c-Met receptor occupancy for the H1975/HGF mouse xenograft model across a wide range of JNJ-61186372 and a combination of EGFR/c-Met monospecific mAb concentrations were simulated (). For JNJ-61186372, χ was set to be 104, and, for the combination of single monovalent antibodies, χ was set to be 0. The EGFR and c-Met receptor densities in the H1975/HGF cell line were used to estimate the in vivo receptor occupancies for EGFR and c-Met.
Figure 5. Simulated profiles of the free fraction of EGFR and c-Met in the H1975/HGF mouse xenograft model for JNJ-61186372 and the combination of both monovalent parent antibodies (gp120 x EGFR Ab + B21M x c-Met Ab).
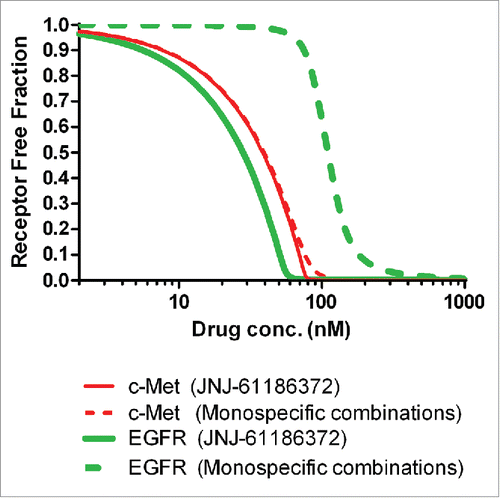
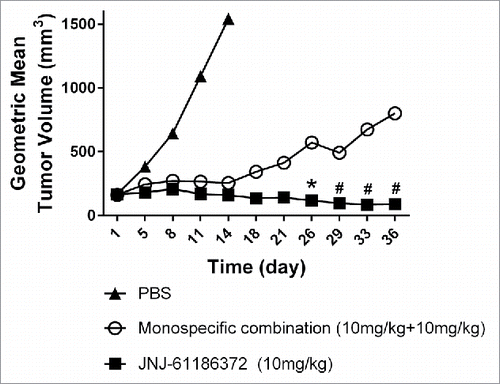
Following 10 mg/kg twice weekly intraperitoneal (i.p.) dosing for 3 weeks, the antibody serum concentrations at 15 days post the last dose were expected to be between 200 and 600 nM (data not shown). Considering the anticipated lower antibody concentration in tumors (∼6% of serum levels based on an in-house dataset), the EGF receptor occupancy could be significantly lower for the monovalent mAb combination than JNJ-61186372 following the 10 mg/kg twice weekly i.p. dosing. The slightly higher expression of c-Met and ∼20-fold higher c-Met binding affinity make c-Met the anchoring receptor for JNJ-61186372 binding. Therefore, the c-Met binding profiles of JNJ-61186372 and the gp120 x c-Met BsAb in the combination therapy are expected to be similar. The actual receptor occupancy values in vivo will be impacted by other factors such as tumor heterogeneity, antibody distribution and receptor internalization rate, but the simulation results in clearly suggested that, following 10 mg/kg twice a week (BIW) i.p. dosing, JNJ-61186372 could be more effective than the combination therapy of anti-EGFR and anti-c-Met monovalent antibodies in a H1975/HGF mouse xenograft model.
To test the hypothesis, JNJ-61186372 (10 mg/kg, dosing = BIW × 3 weeks) and the combination therapy of anti-EGFR and anti-c-Met monovalent antibodies, each at 10 mg/kg, were evaluated in a mouse H1975/HGF xenograft model. Inhibition of both EGFR and c-Met pathways are important for this xenograft model because either anti-EGFR or anti-c-Met treatment alone can only partially inhibit the tumor growth (unpublished data). As shown in , JNJ-61186372 was significantly more effective than the combination therapy of anti-EGFR and anti-c-Met monovalent mAbs. The geometric mean and median tumor volumes in the JNJ-61186372 treated group were consistently lower than those in the combination group throughout the study duration. On the last study day for the combination group (day 36), the tumor volumes in the JNJ-61186372-treated group were ∼90% smaller than those in the monovalent combination treatment groups.
Discussions
Due to the complexity and heterogeneity of many diseases, simultaneous engagement of multiple mechanisms of actions and interference with multiple pathogenic pathways may strengthen therapeutic potential.Citation30 Bi- or multi-specific antibodies capable of blocking multiple growth and survival pathways have a potential to better meet the challenge of blocking cancer growth, and indeed many of them are advancing in clinical development.Citation31,32 Given the roles of membrane-bound receptors in the stimulus recognition and upstream regulation of cell signaling, mechanism-based mathematical models that facilitate the understanding of the binding and inhibition of such receptors can play a critical role in the rational design and development of novel therapeutic strategies. Citation33,34
Here, we characterized cross-arm binding efficiency (χ), the increase in apparent affinity when a bivalent antibody binds to the second target (R2) following its binding to the first receptor (R1) on the same cell, for JNJ-61186372, an EGFR x c-Met BsAb. The χ for JNJ-61186372, was successfully determined via fitting of in vitro flow cytometry data to a ligand-binding model that incorporated χ via a step-wise approach. The model-derived χ value was used to predict the binding of JNJ-61186372 to individual EGFR and c-Met receptors, and the results agreed well with the observed IC50 values for EGFR and c-Met phosphorylation inhibition in two NSCLC cell lines. Among the two cell lines examined, H1993 and H292, the IC50 value differences for c-Met phosphorylation inhibition by JNJ-61186372 is consistent with their difference in receptor densities. A prominent difference in the IC50 value for EGFR phosphorylation inhibition by JNJ-61186372 was observed despite the similar levels of EGFR densities between H1993 and H292.Citation29 The magnitude of IC50 shift corroborates with the anchoring effect of c-Met quantified using the model-estimated χ value (). These results provided confidence for the model-estimated χ for JNJ-61186372.
The pharmacological role of χ for JNJ-61186372 on downstream signaling was further demonstrated by showing that JNJ-61186372 is more efficacious than the combination therapy of anti-EGFR and anti-c-Met monovalent antibodies in the H1975/HGF xenograft mouse model, where both EGFR/c-Met are involved in tumor growth/survival. Our simulations suggested that, as the drug concentrations declined after dosing, the EGFR target occupancy in the monotherapy combination group could be significantly lower compared to the JNJ-61186372 group, especially in the tumor compartment (). On the other hand, the simulated c-Met target occupancy was only slightly higher in the JNJ-61186372 treatment group compared with that in the monotherapy combination group mostly due to much higher monovalent affinity of the c-Met arm. This result is also consistent with our previous observation that JNJ-61186372 is more potent in inhibiting the phosphorylation of ERK (55-65-fold lower IC50 value), a downstream effector of c-Met and EGFR, than the combination of anti-EGFR/c-Met monovalent antibodies in H1975 cell-based assays in the presence of HGF.Citation35
Given that χ is an important characteristic of BsAbs targeting two different membrane bound targets on the same cell, it is of little doubt that χ should be considered for rational design of such antibodies. The challenge is that even though χ is believed to be mostly related to the binding epitopes on R1 and R2 for a bivalent antibody, it cannot be predicted a priori. We demonstrated here that as for conventional mAbs, χ for BsAbs can be determined from in vitro experimental data. The observed cell-binding curve of an antibody is the result of a mixture of monovalently and bivalently bound species. At low concentrations, cell binding is dominated by bivalent binding of antibody molecules to both receptors, whereas the monovalent binding would increase as the concentrations of antibody increase. As shown in , this transition of predominantly bivalent binding to the increasing contribution of monovalent binding, resulted in a unique “shoulder” shape when the fraction maximal binding was plotted against BsAb concentration, and this “shoulder” is driven by the cell surface receptor densities, monovalent binding affinities of the antibodies and the χ value. For JNJ-61186372, the BsAb concentration where the “shoulder” occurs is higher for H1993 (∼5 nM) with high receptor density compared with that for HCC4006 and H1975 (<2 nM) with low receptor densities. This observation is consistent with the findings by Harms et al. Citation26 where the observed “shoulder” concentration for anti-EGFR bivalent mAbs increases (e.g., 0.1 nM to above 2 nM) with increasing EGFR density values (5.8 × 104, 3.6 × 105 and 2 × 106 receptors/cell). Model simulation showed that the “shoulder” would be more apparent when the 2 receptor density values are similar across a wide range of hypothetical χ values. Indeed, we successfully estimated χ for JNJ-61186372 using cell binding data from 3 cell lines with similar EGFR and c-Met receptor density values.
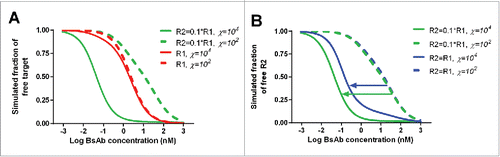
Additional model simulation was conducted to identify scenarios where χ would have more of an effect on R2 (e.g.,, the non-anchoring receptor) binding. The results showed that the effect of χ on R2 binding is more apparent when: 1) the monovalent binding affinity of the BsAb to R2 is lower; 2) the receptor density for R2 is lower; 3) the relative difference in binding affinities between R1 and R2 is bigger; and 4) the relative difference in receptor densities between R1 (e.g., the anchoring receptor) and R2 is bigger. The simulation results for the first 2 scenarios are shown in , respectively. Note that in contrast to the binding curve shape change, which is more sensitive to χ when R1 and R2 densities are similar, χ would lead to bigger shift in R2 binding when the relative difference in receptor densities between R1 and R2 is bigger. Harms et al. Citation27 previously reported that conventional bivalent mAbs targeting different epitopes on the same receptor can demonstrate different χ, such that the parameter χ can differ by several orders of magnitude (χ ∼102–105). The χ for JNJ-61186372 between the EGFR and c-Met arm seems to be in the lower range of the χ values reported for conventional mAbs. Both monovalent target binding affinity and cross-arm binding efficiency χ are engineerable properties of a mAb.Citation36,37 Although we cannot yet predict χ a priori, we can quantitatively evaluate χ and use it for candidate selection. Other bispecific entities in which the two targeting binding units are connected by a flexible linker may be engineered more easily compared with a rigid format of an IgG like BsAb to obtain enhanced cross-arm binding, as was demonstrated by the candidate selection for MM-141, a tetravalent bispecific antibody antagonist of IGF-IR and HER3. Citation27
Figure 7. Model simulation to show the effect of χ on the binding of a bispecific antibody (BsAb) to receptors (A) χ has more significant effect on the binding of a BsAb to the second receptor (R2), than the anchor receptor (R1, which is assumed to have 10-fold higher abundance and 10-fold higher affinity); (B) The effect of χ on R2 binding is more apparent when the R2 baseline density is lower (R1 is assumed to have 10-fold higher affinity).
In summary, kinetic computational models that capture protein-protein interactions using mass action kinetics are a valuable tool for understanding the binding properties of antibodies to their targets. This simple parameterization of antibody cross-arm binding allows the model to successfully describe and predict antibody binding curves across a wide variety of experimental conditions, including variations in target receptor densities and target-binding affinities. Our results showed that χ is an important characteristic of BsAb that can be determined from in vitro cell binding data. It should be considered for rational design of BsAbs targeting two membrane bound targets on the same cell. In addition, insights from our work can help to predict the effect of antibody properties such as affinity and χ on drug potency. More thorough examination of the structure–activity relationships, and likely more sophisticated network models, however, will be required to fully understand the link between cell surface receptor binding, downstream signaling events and the therapeutic outcomes of antibodies targeting cell surface receptors.
Materials and methods
Materials
Human IgG1 BsAbs (JNJ-61186372 (EGFR x cMet), gp120 x c-Met Ab, gp120 x EGFR Ab, B21M x c-Met), were produced by Janssen R&D, LLC (Spring House, PA) using the method reported by Labrijn et al. Citation22 The anti-human immunodeficiency virus (gp120) and anti-respiratory syncytial virus (B21M) Fab arms do not recognize any known target on the tumor cell lines and serve as the ‘inert’ arms.
The NSCLC cell lines used in this study, including H292, SKMES-1, H1975, H3255, H1650, HCC4006, H1993, and SNU-5, were obtained from the American Type Culture Collection (ATCC; Manassas, VA, USA) and cultured as recommended by ATCC.
Flow cytometry staining and determination of cell-surface receptor density
The cell-surface density of EGFR and c-Met in a panel of NSCLC tumor cell lines was determined using flow cytometry and QuantiBRITE™ PE Beads as described previously.Citation38 Parental bivalent EGFR and c-Met antibodies and their corresponding monovalent BsAbs with ‘inert’ arms were conjugated to R-phycoerythrin (R-PE)Citation39 and used for receptor quantitation studies. Briefly, cells were resuspended and kept at 4°C to prevent receptor internalization. Cells were incubated on ice for 1 h with serial dilutions of R-PE-labeled antibodies. Cells were washed and resuspended in stain buffer containing 1:50 diluted DRAQ7 live/dead stain. Samples were read on either a BD FACSCalibur with the nonspecific bindings being subtracted using a PE-labeled isotype-matching control Ab (BD Biosciences, San Jose, CA) or Miltenyi MACSQuant flow cytometer (Miltenyi Biotec, Auburn, CA).
Receptor density values are reported as the antibody binding capacity (ABC). ABC values were derived from standard curves generated with QuantiBRITE™ PE Beads (BD Biosciences). Citation29 ABC values presented in are the specific antibody binding capacity (sABC) calculated by subtracting the ABC values from an isotype matched control from the ABC of the EGFR or c-Met mAbs.
EGFR and c-Met phosphorylation assays
The inhibition of ligand-induced receptor phosphorylation by JNJ-61186372 in NSCLC tumor cell lines was characterized using Phospho-Met (Tyr1349) and Phospho-EGFR (Tyr1173) Assay Whole Cell Lysate Kit (Meso Scale Discovery). Details of the assay were described elsewhere. Citation29 In brief, following serum starvation, H1993 and H292 cells were incubated for 1 h with antibody in 50 μL of starvation media. Either 50 ng/mL EGF or 100 ng/mL HGF was then added to antibody-treated and control wells. Cells were incubated for 15 min, then treatment was removed and cells were lysed in lysis buffer. The lysates were transferred to pre-coated, pre-blocked MSD 96-well MultiArray plates for Phospho-Met and Phospho-EGFR detection. IC50 values were calculated in GraphPad Prism 6, and data from multiple experiments were used to calculate mean IC50 ± SEM.
Xenograft mouse study
The in vivo effect of different treatments was evaluated in female nude mice implanted with human lung H1975 tumor cells engineered to express human HGF (H1975/HGF).Citation40 It is known that both EGFR and c-Met pathways are involved in tumor growth in the H1975/HGF xenograft model. Citation41 Once tumors reached a mean size of 180-185 mm3, animals were dosed with either PBS vehicle control, JNJ-61186372, or a combination of anti-EGFR and anti-c-Met monovalent antibodies (10 mg/kg, BIW × 3 weeks, n = 8 animals/group). Tumor volumes were measured twice weekly during dosing and after treatment. Mice were sacrificed when their tumor volume exceeded 1500 mm3.
Model structure for BsAb-receptor interactions
The dual targeting ligand-receptor binding model () was described using the following ordinary differential equations (ODEs):(Eq. 1)
(Eq. 1)
(Eq. 2)
(Eq. 2)
(Eq. 3)
(Eq. 3)
(Eq. 4)
(Eq. 4)
(Eq. 5)
(Eq. 5)
(Eq. 6)
(Eq. 6)
The parameters in the above equations are defined as the following:
| kON1: | = | the on rate for the monovalent binding of the BsAb to R1 |
| kOFF1: | = | the off rate for the monovalent binding of the BsAb to R1 |
| kON2: | = | the on rate for the monovalent binding of the BsAb to R2 |
| kOFF2: | = | the off rate for the monovalent binding of the BsAb to R2 |
| χ*kON1: | = | the on rate for R2-BsAb binding to R1, forming R2-BsAb-R1 |
| χ*kON2: | = | the on rate for R1-BsAb binding to R2, forming R1-BsAb-R2 |
| kU1: | = | the off rate for R2-BsAb to disassociate from R2-BsAb-R1, which was set to be the same as KOFF1 |
| kU2: | = | the off rate for R1-BsAb to disassociate from R1-BsAb-R2, which was set to be the same as KOFF2 |
| VMEDIA: | = | the volume of media in the in vitro cell binding experiment |
Since the differences in χ for different Abs is believed to be driven by the binding epitopes on both R1 and R2, i.e., steric hindrance for the two arms of an antibody to bind to targets simultaneously, the same χ was used regardless of which arm of the antibody binds the target first. The experiment data did not support implementing 2 χ values for R1 and R2 either. Similarly, it is not possible to separate the impact of χ on kon or koff from the experimental data, χ was only applied to kon of the second binding event, where theoretically the impact of χ should be most apparent. The initial values of A(2) and A(3) are the baseline receptor concentrations (nM) of EGFR and c-Met for each cell line, respectively, and the initial values of A(1), A(4), A(5) and A(6) all equal to 0.
No internalization of receptor complex was confirmed by conducting the in vitro cell binding experiments at 4°C, and thus no such process was incorporated in the current model.
The target (receptor), i.e., target (receptor) accessible to BsAb binding, concentration was calculated as(Eq. 7)
(Eq. 7)
Citation42The in vitro target cell density was obtained by dividing the total number of target-carrying cells by the media volume (L) and was assumed to remain the same throughout the experiment. The receptors are assumed to be uniformly distributed in the media. Citation43 For the monovalent monospecific mAbs of each receptor, the full model was simplified to the ligand-receptor binding model without the inclusion of χ.
Model fitting and simulations were performed in NONMEM® (v 7.2.0, ICON Development Solutions) with PsN (v 3.6.2) and Pirana (v 2.8.2). First-order conditional estimation with interaction (FOCEI) method was used. ADVAN13 and TOL = 6 were used for SUBROUTINE while NSIG = 3 and SIGL = 6 were implemented in ESTIMATION. The objective function value computed by NONMEM® was used in a log-likelihood ratio test for the comparison of hierarchical models. The addition of a structural or variance parameter was considered statistically significant when the objective function value dropped by at least 3.84 (p < 0.05 for 1 degree of freedom). Successful minimization, completion of the covariance step, goodness-of-fit plots, etc. were used to evaluate model performance. Additional data fitting (e.g.,, IC50 of receptor phosphorylation inhibition) and visualization were performed in GraphPad Prism 6 (GraphPad Software).
Disclosure of potential conflict of interest
No potential conflicts of interest were disclosed.
Acknowledgment
The authors would like to thank Drs. Sylvie Laquerre and Donald Heald, employees of Janssen R&D, LLC, as well as Drs. Joost Neijssen and Aran Labrijn, employees of Genmab for their critical scientific reviews of this manuscript.
Funding
All authors are full time employees of Janssen R&D, LLC, which supported the study financially.
References
- Pardee AB. G1 events and regulation of cell proliferation. Science 1989; 246:603-8; PMID:2683075; http://dx.doi.org/10.1126/science.2683075
- Lauffenburger DA, Horwitz AF. Cell migration: a physically integrated molecular process. Cell 1996; 84:359-69; PMID:8608589; http://dx.doi.org/10.1016/S0092-8674(00)81280-5
- Xia Z, Dickens M, Raingeaud J, Davis RJ, Greenberg ME. Opposing effects of ERK and JNK-p38 MAP kinases on apoptosis. Science 1995; 270:1326-31; PMID:7481820; http://dx.doi.org/10.1126/science.270.5240.1326
- Sordella R, Bell DW, Haber DA, Settleman J. Gefitinib-sensitizing EGFR mutations in lung cancer activate anti-apoptotic pathways. Science 2004; 305:1163-7; PMID:15284455; http://dx.doi.org/10.1126/science.1101637
- Olayioye MA, Neve RM, Lane HA, Hynes NE. The ErbB signaling network: receptor heterodimerization in development and cancer. EMBO J 2000; 19:3159-67; PMID:10880430; http://dx.doi.org/10.1093/emboj/19.13.3159
- Plati J, Bucur O, Khosravi-Far R. Dysregulation of apoptotic signaling in cancer: Molecular mechanisms and therapeutic opportunities. J Cell Biochem 2008; 104:1124-49; PMID:18459149; http://dx.doi.org/10.1002/jcb.21707
- Spreter Von Kreudenstein T, Lario PI, Dixit SB. Protein engineering and the use of molecular modeling and simulation: The case of heterodimeric Fc engineering. Methods 2014; 65:77-94; PMID:24211748; http://dx.doi.org/10.1016/j.ymeth.2013.10.016
- Slamon DJ, Leyland-Jones B, Shak S, Fuchs H, Paton V, Bajamonde A, Fleming T, Eiermann W, Wolter J, Pegram M, et al. Use of chemotherapy plus a monoclonal antibody against HER2 for metastatic breast cancer that overexpresses HER2. N Engl J Med 2001; 344:783-92; PMID:11248153; http://dx.doi.org/10.1056/NEJM200103153441101
- Hudson PJ. Recombinant antibody constructs in cancer therapy. Curr Opin Immunol 1999; 11:548-57; PMID:10508712; http://dx.doi.org/10.1016/S0952-7915(99)00013-8
- Fitzgerald J, Lugovskoy A. Rational engineering of antibody therapeutics targeting multiple oncogene pathways. mAbs 2011; 3:299-309; PMID:21393992; http://dx.doi.org/10.4161/mabs.3.3.15299
- Kontermann R. Dual targeting strategies with bispecific antibodies. Mabs 2012; 4(2):182-197; PMID:22327426; http://dx.doi.org/10.4161/mabs.4.2.19000
- Garber K. Bispecific antibodies rise again. Nat Rev Drug Dis 2014; 13:799-801; PMID:25359367; http://dx.doi.org/10.1038/nrd4478
- McDonagh CF, Huhalov A, Harms BD, Adams S, Paragas V, Oyama S, Zhang B, Luus L, Overland R, Nguyen S, et al. Antitumor activity of a novel bispecific antibody that targets the ErbB2/ErbB3 oncogenic unit and inhibits heregulin-induced activation of ErbB3. Mol Cancer Therapeut 2012; 11:582-93; PMID:22248472; http://dx.doi.org/10.1158/1535-7163.MCT-11-0820
- Fitzgerald JB, Johnson BW, Baum J, Adams S, Iadevaia S, Tang J, Rimkunas V, Xu L, Kohli N, Rennard R, et al. MM-141, an IGF-IR- and ErbB3-directed bispecific antibody, overcomes network adaptations that limit activity of IGF-IR inhibitors. Mol Cancer Therapeut 2014; 13:410-25; PMID:24282274; http://dx.doi.org/10.1158/1535-7163.MCT-13-0255
- Kamath AV, Lu D, Gupta P, Jin D, Xiang H, Wong A, Leddy C, Crocker L, Schaefer G, Sliwkowski MX, et al. Preclinical pharmacokinetics of MEHD7945A, a novel EGFR/HER3 dual-action antibody, and prediction of its human pharmacokinetics and efficacious clinical dose. Cancer Chemother Pharmacol 2012; 69:1063-9; PMID:22203367; http://dx.doi.org/10.1007/s00280-011-1806-6
- Seimetz D, Lindhofer H, Bokemeyer C. Development and approval of the trifunctional antibody catumaxomab (anti-EpCAM x anti-CD3) as a targeted cancer immunotherapy. Cancer Treat Rev 2010; 36:458-67; PMID:20347527; http://dx.doi.org/10.1016/j.ctrv.2010.03.001
- Topp MS, Kufer P, Gokbuget N, Goebeler M, Klinger M, Neumann S, Horst HA, Raff T, Viardot A, Schmid M, et al. Targeted therapy with the T-cell-engaging antibody blinatumomab of chemotherapy-refractory minimal residual disease in B-lineage acute lymphoblastic leukemia patients results in high response rate and prolonged leukemia-free survival. J Clin Oncol 2011; 29:2493-8; PMID:21576633; http://dx.doi.org/10.1200/JCO.2010.32.7270
- Kontermann RE. Recombinant bispecific antibodies for cancer therapy. Acta Pharmacologica Sinica 2005; 26:1-9; PMID:15659107; http://dx.doi.org/10.1111/j.1745-7254.2005.00008.x
- Rouet R, Christ D. Bispecific antibodies with native chain structure. Nat Biotechnol 2014; 32:136-7; PMID:24509759; http://dx.doi.org/10.1038/nbt.2812
- Lewis SM, Wu X, Pustilnik A, Sereno A, Huang F, Rick HL, Guntas G, Leaver-Fay A, Smith EM, Ho C, et al. Generation of bispecific IgG antibodies by structure-based design of an orthogonal Fab interface. Nat Biotechnol 2014; 32:191-8; PMID:24463572; http://dx.doi.org/10.1038/nbt.2797
- Chan AC, Carter PJ. Therapeutic antibodies for autoimmunity and inflammation. Nature Rev Immun 2010; 10:301-16; PMID:20414204; http://dx.doi.org/10.1038/nri2761
- Labrijn AF, Meesters JI, de Goeij BE, van den Bremer ET, Neijssen J, van Kampen MD, Strumane K, Verploegen S, Kundu A, Gramer MJ, et al. Efficient generation of stable bispecific IgG1 by controlled Fab-arm exchange. Proc Natl Acad Sci U S A 2013; 110:5145-50; PMID:23479652; http://dx.doi.org/10.1073/pnas.1220145110
- Lauffenburger DA, Linderman J. Receptors: Models for Binding, Trafficking, and Signaling Oxford University Press, Oxford, United Kingdom
- McCloskey N, Turner MW, Goldblatt TD. Correlation between the avidity of mouse-human chimeric IgG subclass monoclonal antibodies measured by solid-phase elution ELISA and biospecific interaction analysis (BIA). J Immunol Methods 1997; 205:67-72; PMID:9236916; http://dx.doi.org/10.1016/S0022-1759(97)00059-8
- Rudnick SI, Adams GP. Affinity and avidity in antibody-based tumor targeting. Cancer Biother Radiopharmaceut 2009; 24:155-61; PMID:19409036; http://dx.doi.org/10.1089/cbr.2009.0627
- Harms BD, Kearns JD, Su SV, Kohli N, Nielsen UB, Schoeberl B. Chapter four - Optimizing Properties of Antireceptor Antibodies Using Kinetic Computational Models and Experiments. In: Wittrup KD, Gregory LV, eds. Methods Enzymol: Academic Press, 2012:67-87.
- Harms BD, Kearns JD, Iadevaia S, Lugovskoy AA. Understanding the role of cross-arm binding efficiency in the activity of monoclonal and multispecific therapeutic antibodies. Methods 2014; 65:95-104; PMID:23872324; http://dx.doi.org/10.1016/j.ymeth.2013.07.017
- Moores, SL, Chiu M, Bushey B, Chevalier K, Haytko P, Neijssen, J, Parren, P, Schuurman J, Anderson M, Attar R, et al. Discovery and preclinical pharmacology of JNJ-61186372: A novel bispecific antibody targeting EGFR and cMET. [abstract]. In: Proceedings of the 105th Annual Meeting of the American Association for Cancer Research; 2014 Apr 5–9; San Diego, CA. Philadelphia (PA): AACR; Cancer Res 2014;74(19 Suppl):Abstract nr DDT01-03. doi:10.1158/1538-7445.AM2014-DDT01-03.
- Jarantow SW, Bushey B, Pardinas JR, Boakye K, Lacy E, Sanders R, Sepulveda MA, Moores SL, Chiu ML. Impact of Cell-Surface Antigen Expression on Target Engagement and Function of an EGFR x c-Met Bispecific Antibody. J Biol Chem 2015; 290: 24689-24704; PMID:26260789; http://dx.doi.org/10.1074/jbc.M115.651653
- Schuurman J, W.H.I. Parren P. Mind the gap. Methods 2014; 65:1-4; PMID:24184353; http://dx.doi.org/10.1016/j.ymeth.2013.10.012
- Chames P, Baty D. Bispecific antibodies for cancer therapy: the light at the end of the tunnel? mAbs 2009; 1:539-47; PMID:20073127; http://dx.doi.org/10.4161/mabs.1.6.10015
- Nunez-Prado N, Compte M, Harwood S, Alvarez-Mendez A, Lykkemark S, Sanz L, Álvarez-Vallina L. The coming of age of engineered multivalent antibodies. Drug Dis Today 2015; 20:588-94; PMID:25757598; http://dx.doi.org/10.1016/j.drudis.2015.02.013
- Ruiz-Herrero T, Estrada J, Guantes R, Miguez DG. A Tunable Coarse-Grained Model for Ligand-Receptor Interaction. PLoS Comput Biol 2013; 9:e1003274; PMID:24244115; http://dx.doi.org/10.1371/journal.pcbi.1003274
- Krzyzanski W. Systems pharmacology models for guiding drug design. CPT: Pharmacometrics Systems Pharmacol 2013; 2:e39; http://dx.doi/org/10.1038/psp.2013.15
- Moores S, Chiu M, Bushey B, Chevalier K, Chin D, Chippari S, Haytko P, McCabe F, Neijssen J, Parren P, et al. Bispecific antibody targeting EGFR and cMet demonstrates superior activity compared to the combination of single pathway inhibitors. [abstract]. In: Proceedings of the AACR-NCI-EORTC International Conference: Molecular Targets and Cancer Therapeutics; 2013 Oct 19–23; Boston, MA. Philadelphia (PA): AACR; Mol Cancer Ther 2013;12(11 Suppl): Abstract nr B241.
- Robinson MK, Hodge KM, Horak E, Sundberg AL, Russeva M, Shaller CC, von Mehren M, Shchaveleva I, Simmons HH, Marks JD, et al. Targeting ErbB2 and ErbB3 with a bispecific single-chain Fv enhances targeting selectivity and induces a therapeutic effect in vitro. Br J Cancer 2008; 99:1415-25; PMID:18841159; http://dx.doi.org/10.1038/sj.bjc.6604700
- Dong J, Sereno A, Aivazian D, Langley E, Miller BR, Snyder WB, Chan E, Cantele M, Morena R, Joseph IB, Boccia A, et al. A stable IgG-like bispecific antibody targeting the epidermal growth factor receptor and the type I insulin-like growth factor receptor demonstrates superior anti-tumor activity. mAbs 2011; 3:273-88; PMID:21393993; http://dx.doi.org/10.4161/mabs.3.3.15188
- Iyer SB, Hultin LE, Zawadzki JA, Davis KA, Giorgi JV. Quantitation of CD38 expression using QuantiBRITE beads. Cytometry 1998; 33:206-12; PMID:9773881; http://dx.doi.org/10.1002/(SICI)1097-0320(1998-1001)33:2%3c206::AID-CYTO15%3e3.0.CO;2-Y
- Pannu KK, Joe ET, Iyer SB. Performance evaluation of QuantiBRITE phycoerythrin beads. Cytometry 2001; 45:250-8; PMID:11746094; http://dx.doi.org/10.1002/1097-0320(20011201)45:4%3c250::AID-CYTO10021%3e3.0.CO;2-T
- Yamada T, Matsumoto K, Wang W, Li Q, Nishioka Y, Sekido Y, Sone S, Yano S. Hepatocyte growth factor reduces susceptibility to an irreversible epidermal growth factor receptor inhibitor in EGFR-T790M mutant lung cancer. Clin Cancer Res 2010; 16:174-83; PMID:20008840; http://dx.doi.org/10.1158/1078-0432.CCR-09-1204
- Nakagawa T, Takeuchi S, Yamada T, Nanjo S, Ishikawa D, Sano T, Kita K, Nakamura T, Matsumoto K, Suda K, et al. Combined therapy with mutant-selective EGFR inhibitor and Met kinase inhibitor for overcoming erlotinib resistance in EGFR-mutant lung cancer. Mol Cancer Therap 2012; 11:2149-57; PMID:22844075; http://dx.doi.org/10.1158/1535-7163.MCT-12-0195
- Shah DK, Haddish-Berhane N, Betts A. Bench to bedside translation of antibody drug conjugates using a multiscale mechanistic PK/PD model: a case study with brentuximab-vedotin. J Pharmacokinet Pharmacody 2012; 39:643-59; PMID:23151991; http://dx.doi.org/10.1007/s10928-012-9276-y
- Mac Gabhann F, Yang MT, Popel AS. Monte Carlo simulations of VEGF binding to cell surface receptors in vitro. Biochim Biophys Acta 2005; 1746:95-107; PMID:16257459; http://dx.doi.org/10.1016/j.bbamcr.2005.09.004