ABSTRACT
Active pharmaceutical substances require an International Nonproprietary Name (INN) assigned by the World Health Organization (WHO) to obtain market authorization as a medicinal product. INNs are selected to represent a unique, generic name for a drug enabling unambiguous identification by stakeholders worldwide. INNs may be requested after initiating clinical development of an investigational drug. Pharmaceutical classes are indicated by a common stem or suffix. Currently, INNs for monoclonal antibody-based drugs are recognized by the suffix, -mab, preceded by a source infix such as -xi- (chimeric), -zu- (humanized) or -u- (human) designating the species from which the antibody was derived. However, many technological advances have made it increasingly difficult to accurately capture an antibody's source in its name. In 2014, the WHO and the United States Adopted Names (USAN) Council approached this challenge by implementing changes to antibody source infix definitions. Unfortunately, gaps and ambiguities in the definitions and procedures resulted in inconsistent source category assignments and widespread confusion. The Antibody Society, extensively supported by academic and industry scientists, voiced concerns leading to constructive dialog during scheduled consultations with WHO and USAN Council representatives. In June 2017, the WHO announced that use of the source infix will be discontinued for new antibody INNs effective immediately. We fully support this change as it better aligns antibody INNs with current and foreseeable future innovations in antibody therapeutics. Here we review the changes implemented. Additionally, we analyzed antibody INNs recently assigned under the previous 2014 definitions and provide recommendations for further alignment.
Introduction
“The best laid schemes o' mice an' men / Gang aft a-gley.” This line of the well-known poem by Robert BurnsCitation1 eloquently expresses the notion that things, even though carefully planned, can often go wrong. In fact, this is what happened with well-intended changes to the definitions used to assign the source infix (substem) for (generic) INN and USAN for antibody therapeutics (see appendix for details). Specific concerns with respect to the changes in the INN and USAN source designations have previously been discussed in detail elsewhere.Citation2
Contemporary INNs lack transparency and consistency in source infix designations
The WHO and the USAN Council are not, to our knowledge, planning to change recommended INNs previously issued under the 2014 definitions. Therefore, we considered it important to reinvestigate nomenclature practices for contemporary INNs to identify and highlight shortcomings. We systematically analyzed all INN for antibody therapeutics as they occur in the most recent 2017 INNs Recommended List (RL77).Citation3 The results summarized in reinforce our previous concerns,Citation2,4 and show that multiple inconsistencies occur for antibodies with a chimeric or humanized source designation. The four antibodies at the top of all received a chimeric (-xi-) or mixed (-xizu-) INN designation; the latter referring to antibodies containing both a chimeric and a humanized heavy or light chain. Dinutuximab beta is based on mouse variable (V) domains fused to human constant (C) domains and therefore represents a genuine chimeric antibody generated via classic domain exchange.Citation5 The other three antibodies categorized as chimeras or mixed by their INNs, in contrast, were humanized using common methods.Citation6-8 The chimeric designation of andecaliximab highlights the drawbacks of using linear sequence homology to categorize therapeutic antibodies by source. For this antibody, a humanization procedure was used that employs non-contiguous human framework regions (FRs) aimed at minimizing T cell epitope content and reducing immunogenicity risks.Citation6,9 Next, rozanolixizumab represents the INN with the least transparent source designation in RL77. Despite its mixed source -xizu- designation and its annotation as a “humanized and chimeric antibody,” the INN description (i.e. additional information published by WHO),Citation3 only shows alignments of rozanolixizumab's variable region sequences with Homo sapiens VH and VL reference genes with 86.5 and 76% homology, respectively (ref. Citation3 and ). The IMGT/DomainGapAlign searches saved for rozanolixizumab in IMGT/mAb-DBCitation10,11 also exclusively show the same human reference genes. So if the reference genes are human, then what is the mixed source designation based on? Notably, rerunning the IMGT reference gene database search as described in Methods reveals that the light chain aligns more closely to macaque VL genes with 7 Macaca mulatta reference alleles showing slightly greater homology compared with the first human VL hit (i.e., 77–79% versus 76%, respectively; ). Differences are subtle, however, with rozanolixizumab VL showing 20 amino acid changes in FR1- FR3 compared with both the top macaque (i.e., IGKV1-13*01 with 9 changes in FR1-3 and 11 in complementary-determining region (CDR)1–2) as well as the top human (i.e., IGKV1–17*01 with 8 changes in FR1–3 and 12 in CDR1–2) reference allele hit in IMGT/DomainGapAlign. The lower score for the human references allele can therefore be attributed to a slightly greater dissimilarity of the CDR-L3 which, in fact, was grafted from the parental rat antibody during humanization. Rozanolixizumab's light chain, therefore, reasonably should also have obtained a humanized source designation. Unexpectedly, it was assigned a mixed chimeric stem instead which, in addition, is at odds with the documentation provided by WHO about this particular INN.Citation3 Multiple inconsistencies were also found for 10 of 14 antibodies with a humanized INN designation upon redoing the IMGT/DomainGapAlign search (). In most cases, we obtained a macaque reference gene as a top hit instead of the human gene listed, although in 2 instances mouse reference genes were also found.
Table 1. Antibody names in Recommended INN: List 77.
Table 2. Inconsistencies in source infix designations analyzed for antibodies from RL77.
There may be multiple explanations for the observed inconsistencies. First, there is no precise definition of what constitutes “closer to human than to other species” in the reference database search. Presumably, an INN examiner may determine that a sequence aligns most closely to a human reference gene even if non-human genes score slightly higher but the observed homology is of similar magnitude (see appendix). This may be of particular relevance if the result is affected by differential alignment in CDR sequences. Macaque reference genes, for example, may obtain a higher IMGT/DomainGapAlign score through a closer homology with rodent CDRs or, due to an artifact of the local alignment algorithm by having no identity or similarity in the V gene termini (CDR3 and FR3), even though alignment for the corresponding region in the human counterpart is better.Citation2 However, such subjectivity in assigning an appropriate source category is highly problematic as discussed above (). Second, results may vary over time as the composition of the IMGT gene reference database changes due to additional genome sequences becoming available in which specifically the addition of macaque germline reference genes with high allelic variation is of concern.Citation2 Finally, inconsistencies will occur when an INN for an antibody contained in a novel compound was issued before the 2014 change in source definitions. This is exemplified by gemtuzumab ozogamicin. The INN for the antibody portion of this antibody-drug conjugate (ADC) was issued in 2001 without its sequence being made available.Citation12 In fact, sequences have only been systematically disclosed in the INN description since RL57 released in 2007.Citation13 The subjectivity and time-dependence of antibody INNs creates undesired uncertainties with respect to predicting and interpreting INN source categories.
As noted, the USAN definition differs from INN by using an 85% sequence cut-off definition for distinguishing chimeric from humanized antibodies (appendix). When using the USAN definition, only 8 of 19 antibody heavy chains and 9 of 19 light chains would have obtained a humanized designation. Interestingly, rozanolixizumab's nomenclature would be consistent with USAN's definitions for a mixed source antibody. This antibody however has not been assigned a USAN,Citation14 so the pre-existence of a USAN cannot explain the discrepancies described.
Examining the 8 human antibodies in RL77, we observe no discrepancies, which is as expected due to the absence of a definition (; appendix). Ironically, the only antibody in RL77 derived from an immune response in a human individual (i.e., the second Fab in the bispecific DVD lutikizumab was cloned from a patient with auto-immune diseaseCitation15,16) did not earn the antibody a (mixed) human INN designation. The wide range of technologies used to access human sequences for generating therapeutic antibodies is noteworthy. These technologies include mRNA-display, multiple distinct phage-display and several distinct transgenic mice platforms (), and exemplify that therapeutic antibodies can have many origins (). Additional similar technologies, often used in conjunction, are being used to fill early pharmaceutical development pipelines. INNs requests for such antibodies can be expected for submission in the near future.
Figure 1. The INN source substem for therapeutic antibodies. Antibody INNs issued until June 2017 (with the exception of the first antibody INN muromonab-CD3) contain a source infix designating the species. The antibody's origin determined the source infix until 2014. For antibody INN issued between 2014 and early 2017, the source infix was determined using a sequence alignment procedure, which led to inconsistent source infix designations for chimeric and humanized antibodies. No definitions to determine a human source existed (see appendix).
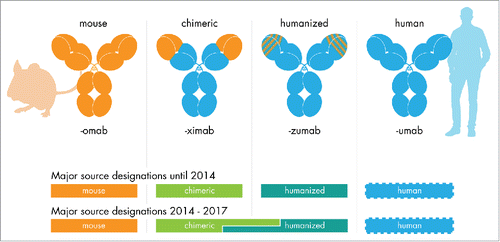
Figure 2. An expanding toolbox for the generation of therapeutic antibodies that meet modern biopharmaceutical requirements. Therapeutic antibodies can be generated in many ways and capturing an antibody's source in a single syllable is therefore no longer possible.
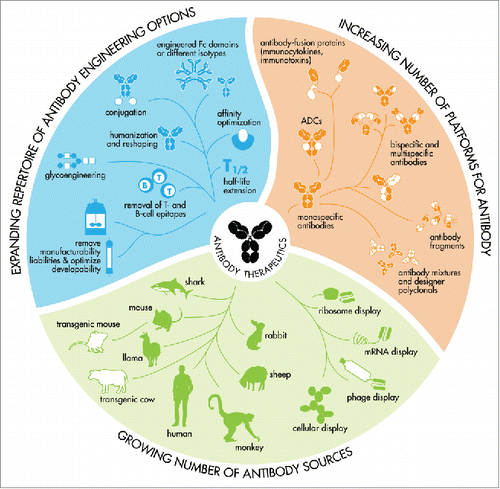
In summary, therapeutic antibody INNs as well as the accompanying description published in the INN Recommended List RL77 lack consistency and transparency in source infix designations for chimeric and humanized antibodies.
Fixing the source infix
On behalf of its members and scientists who signed an online petition, The Antibody Society engaged in discussions with the WHO INN expert group and representatives from the USAN Council and Food and Drug Administration (FDA) during the 62nd open consultation on INN for Pharmaceutic Substances in April 2016 and an ad hoc meeting on Biologicals in September 2016.Citation4,17 The Antibody Society, in collaboration with key stakeholders, developed proposals to revise the INN system to provide scientifically sound, distinguishing names for therapeutic antibodies in current and future development.
Two potential solutions were discussed. The first was to drop the use of the source infix and sequence alignments to categorize antibodies altogether. The second was to improve the current system, for example by generating a new expanded ‘engineered’ source infix which should take current and future developments in antibody generation technologies into account. Although no general consensus was reached at the workshop, dropping the source infix was a favored solution.Citation17 Removing the source infix would, as a side effect, create more flexibility in the assignment of INNs. This is important as it was noted that, due the large increase in applications for biologicals, it is becoming increasingly difficult to design new distinguishable INNs.Citation17
After considering all options, the WHO announced it was decided at the 64th consultation on INN for Pharmaceutical Substances held April 4–7, 2017 in Geneva, Switzerland that they will discontinue the use of the source infix in antibody INNs.Citation18 The make-up of previous and new antibody INN nomenclature rules are summarized in .
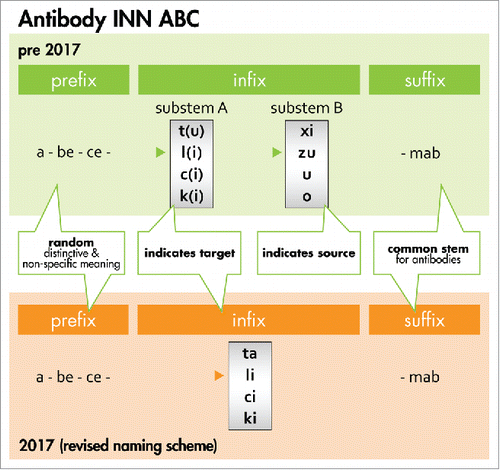
Figure 3. Antibody INN ABC. The general naming scheme for antibody INN before 2017 is compared with the new system. Prior to 2017, the random prefix was followed by a target infix (substem A) of which -t(u)- for tumor, -l(i)- for immunomodulatory, -c(i)- for cardiovascular, and -k(i)- for interleukin represented major classes. The source infix (substem B) indicated the source of which -xi- for chimeric, -zu- for humanized and -u- for human represented major classes (see the Bioreview (2014)Citation27 for complete listing). In the new scheme, the source infix designating the species has been discontinued as recommended by the INN expert group during the 64th INN Consultation.Citation18, 60 To avoid confusion with earlier schemes, -ta- now designates tumor antigen. Furthermore, -ba- designates bacterial, -ami- serum amyloid protein(SAP)/amyloidosis, -ci- cardiovascular, -fung- fungal, -gros- skeletal muscle mass-related growth factors and receptors, -ki- interleukin, -li- immunomodulating, -ne- neural, -os- bone, -toxa- toxin and -vi- viral. The source infix -vet- for veterinary use antibodies is retained and added to the ‘target’ infix list. The suffix -mab represents the common stem for antibody therapeutics.Citation18
Antibody INNs: Beyond the source infix
The target infix (substem A) is determined by the target (molecule, cell or organ) class.Citation17 The information provided by a single syllable can only in very general terms describe the intended target for an antibody, especially as an antibody's target molecule is often expressed on multiple cells in multiple organs. In addition, since therapeutic antibodies may be developed for additional or different indications after the INN has been assigned, the INN is not always consistent with the disease target. For example, rituximab includes the -tu- target infix consistent with its oncology but not inflammatory disease indications. Discontinuation of the target infix was discussed as a means of generating further flexibility in generating clearly distinct INNs for antibodies.Citation17 Overall, arguments for discontinuing the target infix are less compelling and urgent than for the source infix. Optimizing or potentially discontinuing the use of the target infix should therefore be addressed in future discussions.
Antibody INNs may sometimes contain a second wordCitation17 that is added to conjugates of antibodies with other molecules such as a toxin, chelator or chemical (e.g., the ADCs shown in ). If the antibody also contains a radiolabel, the isotope is listed at the start of the INN (i.e., name of the isotope, element and isotope number followed by the generic antibody name). The INN does not contain a specific designation to specify that a substance is a fusion protein, although this is currently under discussion. Antibodies with identical amino acid sequence (or containing minor processing differences) but distinct glycosylation are provided with a new INN containing a Greek letter as a second word, e.g., daclizumab beta (). This antibody was generated from a new cell line using a distinct manufacturing process generating carbohydrate species that are more homogenously fucosylated than the originator product Zenapax®(daclizumab), thereby reducing IgG Fc receptor IIIa binding and antibody-dependent cell-mediated cytotoxicity.Citation17,19 Finally, the second word ‘pegol’ indicates PEGylated antibodies, e.g., certolizumab pegol.
The stem -mab has been used in INNs for all antibody-containing substances. However, the intention to introduce INNs for antibody-fusion proteins consisting of a single word containing the stem -fusp has been discussed. Although general consensus was not reached, it was decided to test the -fusp stem on 2 outstanding requests.Citation17
For recombinantly expressed polyclonal antibodies (also known as designer polyclonals), each antibody in the mixture will usually require a separate INN. However, this may not be appropriate for recombinant polyclonal antibodies manufactured using single batch production strategies. In a USAN, the suffix -pabCitation20 may then be used, such as in rozrolimupab, which comprised 25 different recombinant anti-rhesus D antibodies.Citation21 A suffix for such recombinant polyclonal drug substances is not available for INNs, but in view of future development should likely be considered.
Beyond INN
In some cases, a prefix may be added to specific antibody products to avoid medication errors and facilitate pharmacovigilance.Citation22 The FDA, for example, included the prefix ado- to the ADC ado-trastuzumab emtansine (Kadcyla) to distinguish it from the non-drug conjugated trastuzumab. This is to mitigate the risk that the name is misread or mislabeled and to avoid administration of the wrong drug, which could lead to serious adverse events.Citation22,23 Further use of prefixes for specific drugs should be considered as additional conjugates with the same antibody are being developed (e.g., trastuzumab duocarmazine ()).
The addition of a Biological Qualifyer (BQ) to the INN has also been extensively discussed by WHO, the USAN Council and the FDA. It is envisioned that each biologic product, including originator products, related biologic products and biosimilars, would obtain an additional suffix. The WHO proposed the use of a random 4-letter code supplemented with a 2-digit checksum as a BQ, which should be used in conjunction with the INN to uniquely identify an antibody product to promote world-wide pharmacovigilance.Citation24 The BQ would furthermore help the accurate identification of antibody products by health care providers and patients and avoid unintended substitution. Although the BQ is considered an important issue, details are still being worked out while a pilot program with regulators is being discussed.Citation17 The FDA recently released an Industry Guidance document proposing the use of a 4-letter suffix as a biologic qualifier.Citation22,25 Three biosimilar antibody products have so far been issued such a suffix, these include adolimumab-atto (Amjevita), infliximab-dyyb (Inflectra) and infliximab-abda (Renflexis). A further alignment between regulatory authorities will be essential to achieve consistency and maximal leverage of the approach.
Future perspectives
The WHO decision to discontinue the use of the source infix in antibody INNs is an important step forward and addresses the concerns and objections raised by The Antibody Society on behalf of many academic and industry scientists in the antibody field.Citation2 The new naming scheme accepted at the 64th Consultation has swiftly been implemented by WHO as we have learned that it has already been adopted in INNs currently proposed to applicants. Since the shortcomings of the 2014 source definitions extend to the Additional Information provided in the INN description (e.g., as in RL77 discussed here), we urge the WHO INN expert group to completely retire the use of categorizing therapeutic antibodies for source by using sequence alignments. The WHO announced that the INN description is envisioned to contain more extensive information on the antibody's origin and that information regarding the species on which the antibody's sequence is based will continue to be included in the definition for antibody INN.Citation18 We fully support the inclusion of more precise information of the antibody's origin and generation in the INN description. However, continuing the practice of describing antibodies as chimeric, humanized and human in the INN description on the basis of ambiguous and inconsistent definitions, in spite of discontinuing the source infix in the actual INN, would be a very poor solution that should be reconsidered. Going forward, we request the WHO to consider omitting the source infix from antibody INNs currently under discussion (e.g., INNs in the 2016 Proposed List PL116),Citation26 as well as to review anomalies in previously assigned INNs. Most importantly, discontinuing the use of sequence alignments to determine an antibody's origin in the INN description would allow the most complete resolution of the issue.
Methods
Database analysis of antibody INNs
The antibody names in the Recommended INN: List 77Citation3 were examined. The assigned VH and VL reference genes or alleles were searched in IMGT/mAb-DBCitation34,35 using the INN as “General Query” and examined in the IMGT/2D structure-DB card for the INNs using the link provided in the Table on the respective IMGT/mAb-DB result page. The saved IMGT/DomainGapAlign results were accessed using the links provided at the bottom in the box designating the V-domain of the heavy and light chain, respectively. This link provides the top 5 “Closest reference gene and allele(s) from the IMGT V domain directory.” The top hit corresponds to the reference gene and species listed in RL77.Citation3 The top hit and percentage homology are noted in in this manuscript. Next, a new IMGT/DomainGapAlign search against the current databased was performed by scrolling to the top of the page and executing “Align and IMGT -gap my sequence(s).” Searches were performed against the database available in weeks 17 and 18 (2017). Antibodies for which a different top hit relative to the species stated in RL77 was found are highlighted in red in and further details are provided in . Patent applications and patents can be accessed via https://worldwide.espacenet.com/.
Web-based materials
All links and searches in this manuscript were checked for accuracy at the time of writing. Since links may become inoperative or linked information may be retired or changed, relevant copies are being kept on file at The Antibody Society and may be accessed in the “member's only” area on The Antibody Society's website (http://www.antibodysociety.org) or directly requested from the authors.
Disclosure of potential conflicts of interest
The authors are employees (PWHIP and PJC) or are associated (AP) with companies that have a commercial interest in therapeutic antibody products and antibody engineering technologies.
Acknowledgments
We thank Dennis Burton, Janine Schuurman, Isidro Hötzel, Andrew Popplewell, Max Vasques, Tim Jones and Matthew Baker for insightful discussions and for critically reviewing the manuscript. We thank Joost Bakker for preparing the graphics.
References
- Burns R. To a mouse, on turning her up in her nest with the plough. 1785. The Complete Works of Robert Burns. http://www.robertburns.org/works/75.shtml
- Jones TD, Carter PJ, Plückthun A, Vasquez M, Holgate RG, Hotzel I, Popplewell AG, Parren PW, Enzelberger M, Rademaker HJ, et al. The INNs and outs of antibody nonproprietary names. MAbs 2016; 8:1-9; PMID:26716992; https://doi.org/10.1080/19420862.2015.1114320
- World Health Organization. International non-proprietary names for pharmaceutical substances (INNs). RECOMMENDED international nonproprietary names: List 77. WHO Drug Inform 2017; 31(1): 1-150. http://www.who.int/medicines/publications/druginformation/issues/77_INN_Recommended_List.pdf?ua=1
- World Health Organization. 62nd Consultation on International Nonproprietary Names for Pharmaceutical Substances. Geneva, 12–15 April. 2016. INN Working Doc. 16.395. July 2016. http://www.who.int/medicines/services/inn/62nd_Executive_Summary.pdf?ua=1
- Gillies SD, Lo KM, Wesolowski J. High-level expression of chimeric antibodies using adapted cDNA variable region cassettes. J Immunol Methods 1989; 125:191-202; PMID:2514231; https://doi.org/10.1016/0022-1759(89)90093-8
- Di Paolo J, Breckenridge D, Bates J, Tumas D. Combinations of the BTK inhibitor GS-4059 with inhibitors selected from a JAK, ASK1, BRD and/or MMP9 inhibitor to treat cancer, allergic disorders, autoimmune diseases or inflammatory diseases. 2017. WO 2017/059252.
- Reilly EB, Phillips AC, Buchanan FG, Kingsbury G, Zhang Y, Meulbroek JA, Cole TB, DeVries PJ, Falls HD, Beam C, et al. Characterization of ABT-806, a humanized tumor-specific anti-EGFR monoclonal antibody. Mol Cancer Ther 2015; 14:1141-51; PMID:25731184; https://doi.org/10.1158/1535-7163.MCT-14-0820
- Finney HM, Lawson ADG, Shaw SG, Smith BJ, Tyson K, Kevorkian L. Anti-FcRn antibodies. 2014. WO2014/019727 A1.
- Holgate RG, Weldon R, Jones TD, Baker MP. Characterisation of a novel anti-CD52 antibody with improved efficacy and reduced immunogenicity. PLoS One 2015; 10:e0138123; PMID:26372145; https://doi.org/10.1371/journal.pone.0138123
- Ehrenmann F, Kaas Q, Lefranc MP. IMGT/3Dstructure-DB and IMGT/DomainGapAlign: A database and a tool for immunoglobulins or antibodies, T cell receptors, MHC, IgSF and MhcSF. Nucleic Acids Res 2010; 38:D301-7; PMID:19900967; https://doi.org/10.1093/nar/gkp946
- Ehrenmann F, Lefranc MP. IMGT/DomainGapAlign: IMGT standardized analysis of amino acid sequences of variable, constant, and groove domains (IG, TR, MH, IgSF, MhSF). Cold Spring Harb Protoc 2011; 2011:737-49; PMID:21632775; https://doi.org/10.1101/pdb.prot5636
- World Health Organization. International nonproprietary names for pharmaceutical substances (INNs). RECOMMENDED international nonproprietary names (Rec. INNs): List 45. WHO Drug Inform 2001; 15(1): 40. http://www.who.int/medicines/publications/druginformation/innlists/RL45.pdf?ua = 1
- World Health Organization. International nonproprietary names for pharmaceutical substances (INNs). RECOMMENDED international nonproprietary names: List 57. WHO Drug Inform 2007; 21:53-83. http://www.who.int/medicines/publications/druginformation/innlists/RL57.pdf?ua=1
- American Medical Association. United States Adopted Name (USAN) Drug Finder. 2017. https://searchusan.ama-assn.org/finder/usan/search/*/relevant/1
- Lacy SE, Wu C, Ambrosi DJ, Hsieh CM, Bose S, Miller R, Conlon DM, Tarcsa E, Chari R, Ghayur T, et al. Generation and characterization of ABT-981, a dual variable domain immunoglobulin (DVD-Ig(TM)) molecule that specifically and potently neutralizes both IL-1alpha and IL-1beta. MAbs 2015; 7:605-19; PMID:25764208; https://doi.org/10.1080/19420862.2015.1026501
- Garrone P, Djossou O, Fossiez F, Banchereau J. Human monoclonal antibodies against human cytokines and methods of making and using such antibodies. 1995. WO95/14780.
- World Health Organization. 63rd Consultation on International Nonproprietary Names for Pharmaceutical Substances. Geneva, 18–21 October. 2016. INN Working Doc. 17.408. January 2017. http://www.who.int/medicines/services/inn/63rd_Executive_Summary.pdf?ua=1
- World Health Organization. Revised monoclonal antibody (mAb) nomenclature scheme. INN Working Doc. 17.416. May 2017. http://www.who.int/medicines/services/inn/Revised_mAb_nomenclature_scheme.pdf?ua=1
- European Medines Agency. Assessment report. Zinbryta. International non-proprietary name: daclizumab. 2016. EMA/458317/2016. Committee for Medicinal Products for Human Use (CHMP). http://www.ema.europa.eu/docs/en_GB/document_library/EPAR_-_Public_assessment_report/human/003862/WC500210601.pdf
- American Medical Association. Monoclonal Antibodies. 2017. https://www.ama-assn.org/about/monoclonal-antibodies
- Robak T, Windyga J, Trelinski J, von Depka Prondzinski M, Giagounidis A, Doyen C, Janssens A, Alvarez-Román MT, Jarque I, Loscertales J, et al. Rozrolimupab, a mixture of 25 recombinant human monoclonal RhD antibodies, in the treatment of primary immune thrombocytopenia. Blood 2012; 120:3670-6; PMID:22915649; https://doi.org/10.1182/blood-2012-06-438804
- Food and Drug Administration. Nonproprietary Naming of Biological Products. Guidance for Industry. 2017. U.S. Department of Health and Human Services. Food and Drug Administration. Center for Drug Evaluation and Research (CDER). Center for Biologics Evaluation and Research (CBER). January 2017 Labeling. https://www.fda.gov/downloads/Drugs/GuidanceComplianceRegulatoryInformation/Guidances/UCM459987.pdf
- Institute for Safe Medication Practices. The National Alert Network. Confusion regarding the generic name of the HER2-targeted drug KADCYLA (ado-trastuzumab emtansine). 2013. https://www.ismp.org/NAN/files/20130417.pdf
- World Health Organization. Biological Qualifier An INN Proposal. Programme on International Nonproprietary Names (INNs). 2015. INN Working Doc. 14.342. Rev. Final October 2015. http://www.who.int/medicines/services/inn/WHO_INN_BQ_proposal_2015.pdf?ua=1
- Department of Health and Human Services. Food and Drug Adminstration. Nonproprietary naming of biological products; guidance of industry; availability. Federal Register 2017; 82(9):4345-7. https://www.gpo.gov/fdsys/pkg/FR-2017-01-13/pdf/2017-00694.pdf
- World Health Organization. International Nonproprietary Names for Pharmaceutical Substances (INN). Proposed INN: List 116. WHO Drug Inform 2016; 30(4):605-710. http://www.who.int/medicines/publications/druginformation/innlists/PL116.pdf?ua=1
- World Health Organization. International Nonproprietary Names (INN) for biological and biotechnological substances (a review). 2014. WHO/EMP/RHT/TSN/2014.1. http://www.who.int/medicines/services/inn/BioRev2014.pdf?ua=1
- World Health Organization. The use of stems in the selection of International Nonproprietary Names (INN) for pharmaceutical substances. 2013. WHO/EMP/RHT/TSN/2013.1. http://www.who.int/medicines/services/inn/StemBook_2013_Final.pdf?ua=1
- Reichert JM. Therapeutic monoclonal antibodies approved or in review in the European Union or the United States. 2017. http://www.antibodysociety.org/news/approved-antibodies/
- World Health Organization. International non-proprietary names. WHO Drug Inform 2009; 23:193-9. http://www.who.int/medicines/publications/druginformation/issues/DrugInfo09_Vol23-3.pdf?ua=1
- World Health Organization. Guidelines on the Use of INNs for Pharmaceutical Substances. 1997. WHO/PHARMS/NOM 1570. http://apps.who.int/iris/bitstream/10665/63779/1/WHO_PHARM_S_NOM_1570.pdf
- American Medical Association. Naming Biologics. 2017. https://www.ama-assn.org/about/naming-biologics
- American Medical Association. New Monoclonal Antibody Rules. 2017. https://www.ama-assn.org/sites/default/files/media-browser/public/usan/2015-mab-rules_0.pdf
- The International Immunogenetics Information System. IMGT/mAb-DB query page. 2017. http://www.imgt.org/mAb-DB/query
- Poiron C, Wu Y, Ginestoux C, Ehrenmann F, Duroux P, Lefranc MP. IMGT/mAb-DB: The IMGT® database for therapeutic monoclonal antibodies. 2010. http://www.imgt.org/IMGTposters/SFI2010_IMGTmAb-DB.pdf
- Yuan J, Qu X, Lin J, Ye X, Cao G, Tao W, Zhang L, Zhang L, Yang L. PD-1 antibody, antigen-binding fragment thereof, and medical application thereof. 2016. US2016/0376367.
- Rollins S, Alvarez R, Rother R, Kawar Z, Mcever R. Anti-P-selectin antibodies and methods of their use and identification. 2012. WO2012/088265.
- Bhinder TK, Ford SK, Gemaschewski V, Lewis AP, Pepys MB. Antigen binding proteins specific for serum amyloid P component. 2011. WO2011/107480.
- Russo AF, Kaiser E, Recober A, Kuburas A, Raddant AC, Kovacevich BR, Latham JA, Smith JTL, Garcia-Martinez LF. Use of anti-CGRP or anti-CGRP-R antibodies or antibody fragments to treat or prevent chronic and acute forms of diarrhea. 2012. WO2012/162253.
- Poulsen KT, Shelton DL, Zeller J, Machin I, Corradini L. Methods for treating chronic pain. 2016. US 9,328,167 B1.
- Hamann PR, Hinman LM, Hollander I, Beyer CF, Lindh D, Holcomb R, Hallett W, Tsou HR, Upeslacis J, Shochat D, et al. Gemtuzumab ozogamicin, a potent and selective anti-CD33 antibody-calicheamicin conjugate for treatment of acute myeloid leukemia. Bioconjug Chem 2002; 13:47-58; PMID:11792178; https://doi.org/10.1021/bc010021y
- Bebbington CR, Yarranton GT, Palath V. Treatment of leukemias and chronic myeloproliferative diseases with antibodies to EPHA3. 2010. WO2010/102244.
- Gurney AL, Bond CJ. RSPO3 binding agents and uses thereof. 2014. WO2014/012007.
- Cardillo TM, Govindan SV, Sharkey RM, Trisal P, Goldenberg DM. Humanized anti-Trop-2 IgG-SN-38 conjugate for effective treatment of diverse epithelial cancers: Preclinical studies in human cancer xenograft models and monkeys. Clin Cancer Res 2011; 17:3157-69; PMID:21372224; https://doi.org/10.1158/1078-0432.CCR-10-2939
- Wang J, Goetsch L, Tucker L, Zhang Q, Gonzalez A, Vaidya KS, Oleksijew A, Boghaert E, Song M, Sokolova I, et al. Anti-c-Met monoclonal antibody ABT-700 breaks oncogene addiction in tumors with MET amplification. BMC Cancer 2016; 16:105; PMID:26879245; https://doi.org/10.1186/s12885-016-2138-z
- Wang J, Anderson MG, Oleksijew A, Vaidya KS, Boghaert ER, Tucker L, Zhang Q, Han EK, Palma JP, Naumovski L, et al. ABBV-399, a c-Met antibody-drug conjugate that targets both MET-amplified and c-Met-overexpressing tumors, irrespective of MET pathway dependence. Clin Cancer Res 2017; 23:992-1000; PMID:27573171; https://doi.org/10.1158/1078-0432.CCR-16-1568
- Hudziak RM, Lewis GD, Winget M, Fendly BM, Shepard HM, Ullrich A. p185HER2 monoclonal antibody has antiproliferative effects in vitro and sensitizes human breast tumor cells to tumor necrosis factor. Mol Cell Biol 1989; 9:1165-72; PMID:2566907; https://doi.org/10.1128/MCB.9.3.1165
- Zhang L, Liu J, Cao G, Sun P. IL17A binding agent and uses thereof. 2016. US2016/0289321.
- Sommer A, Kopitz C, Schatz CA, Nising CF, Mahlert C, Lerchen HG, Stelte-Ludwig B, Hammer S, Greven S, Schuhmacher J, et al. Preclinical efficacy of the Auristatin-based antibody-drug conjugate BAY 1187982 for the treatment of FGFR2-positive solid tumors. Cancer Res 2016; 76:6331-9; PMID:27543601; https://doi.org/10.1158/0008-5472.CAN-16-0180
- Yamazaki Y, Urakawa I, Yoshida H, Yamashita T, Shimada T, Hasegawa H. anti-FGF23 antibody and a pharmaceutical composition comprising the same. 2011. US 7,883,705 B2.
- Towne JE, Cheng JD, O'Neill JC, Zhang L, Sun Y, Cerne H, Piper DE, Ketchem RR. Human IL23 antigen binding proteins. 2014. US 8,722,033 B2.
- Kock K, Pan WJ, Gow JM, Horner MJ, Gibbs JP, Colbert A, Goletz TJ, Newhall KL, Rees WA, Sun Y, et al. Preclinical development of AMG 139, a human antibody specifically targeting IL-23. Br J Pharmacol 2015; 172:159-72; PMID:25205227; https://doi.org/10.1111/bph.12904
- Shabanzadeh AP, Tassew NG, Szydlowska K, Tymianski M, Banerjee P, Vigouroux RJ, Eubanks JH, Huang L, Geraerts M, Koeberle PD, et al. Uncoupling Neogenin association with lipid rafts promotes neuronal survival and functional recovery after stroke. Cell Death Dis 2015; 6:e1744; PMID:25950474; https://doi.org/10.1038/cddis.2015.109
- Willuda J, Linden L, Lerchen HG, Kopitz C, Stelte-Ludwig B, Pena C, Lange C, Golfier S, Kneip C, Carrigan PE, et al. Preclinical anti-tumor efficacy of BAY 1129980 - a novel Auristatin-based Anti-C4.4A (LYPD3) antibody-drug conjugate for the treatment of non-small cell lung cancer. Mol Cancer Ther 2017; 16:893-904; PMID:28292941; https://doi.org/10.1158/1535-7163.MCT-16-0474
- Salfeld JG, Allen DJ, Hoogenboom HRJM, Kaymakcalan Z, Labkovsky B, Mankovich JA, McGuinness BT, Roberts AJ, Sakorafas P, Schoenhaut D, et al. Human antibodies that bind human TNFα. 2001. US 6,258,562 B2.
- Hsieh C-M, Cuff C, Tarcsa E, Hugunin M. Discovery and characterization of ABT-122, an anti-TNF/IL17 DVD-Ig molecule as potential therapeutic candidate for rheumatoid arthritis. Abstract number 1421. Arthritis Rheumatism 2013; 65: abstract 4676. http://acrabstracts.org/abstract/discovery-and-characterization-of-abt-122-an-anti-tnfil-17-dvd-ig-molecule-as-a-potential-therapeutic-candidate-for-rheumatoid-arth
- Gurnett-Bander A, Perez-Caballero D, Sivapalasingam S, Duan X, MacDonald D. Human antibodies to respiratory syncytial virus F protein and methods of use thereof. 2016. US 9,447,173 B2.
- Fisher TS, Kamperschroer C, Oliphant T, Love VA, Lira PD, Doyonnas R, Bergqvist S, Baxi SM, Rohner A, Shen AC, et al. Targeting of 4-1BB by monoclonal antibody PF-05082566 enhances T-cell function and promotes anti-tumor activity. Cancer Immunol Immunother 2012; 61:1721-33; PMID:22406983; https://doi.org/10.1007/s00262-012-1237-1
- Gearing DP, Virtue ER, Gearing RP, Drew AC. A fully caninised anti-NGF monoclonal antibody for pain relief in dogs. BMC Vet Res 2013; 9:226; PMID:24206926; https://doi.org/10.1186/1746-6148-9-226
- World Health Organization. 64th Consultation on International Nonproprietary Names for Pharmaceutical Substances. Geneva, 4–7 April. 2017. In Preparation. http://www.who.int/medicines/services/inn/meetings/en/
Appendix. The INN source infix explained
The WHO provides International Nonproprietary Names (INNs) to therapeutic antibodies.Citation27,28 A complete and current list of INNs for therapeutic antibodies approved or under regulatory review in the US and EU can be found on The Antibody Society's website.Citation29 The INN is composed of a random, unique prefix of several syllables, a first infix (substem A), which is defined by the target, and second infix (substem B), which is defined by the source, and the suffix (stem) -mab.Citation27 The source infixes were developed during 1991–1993,Citation30 and, although definitions were refined over the years, the delineations between the various categories remained the same until 2014 (see ).Citation28,31 The most common infix indicating the source are:-o- for all mouse sequence-derived antibodies, -u- for all human sequence-derived antibodies, -xi- for antibodies in which the mouse-derived variable region were combined with a human constant region (variable domain exchange) and -zu- for antibodies in which the mouse-derived complementarity determining regions were combined with a human variable region framework and human constant region. The origin of the antibody or the technology used to generate the therapeutic antibody defined the infix unequivocally in the early years.
Progress in antibody technologies, however, has increasingly blurred boundaries between the various source categories, resulting in an antibody landscape consisting of a continuum of sequences. Through these advances, therapeutic antibodies with the characteristics that are required for modern biopharmaceuticals can be generated with sequences derived from a myriad of in vitro and in vivo technologies, distinct animal species and transgenic animals or even fully synthetic sources. These further changes include the introduction of defined point mutations for optimizing binding, the mitigation of manufacturability and developability liabilities (such as replacing amino acids prone to undesired post-translation modifications) and the removal of T-cell epitopes (to lower the antibody's immunogenicity risk profile). Finally, many technologies to optimize the therapeutic antibodies' functionality are being applied (see ). The existing nomenclature system, therefore, was becoming outdated, which was a challenge that the WHO INN expert group attempted to address by developing novel infix definitions.
The WHO updated definitions for the source infix released in 2014Citation27 handled the distinction between chimeric antibodies and humanized antibodies in a novel and unprecedented way. The new definitions included a sequence alignment procedure whereby the source infixes were now defined by the sequence of the end product and not their factual origin. In 2015, the INN expert group clarified that alignments need to be performed by using the IMGT/DomainGapAlign tool.Citation10,11 The -xi- or -zu- infix is determined via the alignment of the amino acid sequence encoded by the V-gene only (i.e., FR1 through FR3 for the heavy chain (VH) and FR1 through CDR3 for the light chain (VL)) to the IMGT sequence reference database. The top hit(s) determine the closest species. For a humanized antibody, the first hit for “Closest reference gene and allele(s) from the IMGT V domain” in the DomainGapAlign result therefore should be a human sequence. A first hit on the list with any other species designates a chimeric antibody. Although this sounds easy in theory, execution is less straightforward. In cases where multiple sequences have similar scores, such as for example where ‘human’ is second or third to ‘macaque’ with only small deviations in the percentage homology score, the designation ‘humanized’ may still be provided by WHO. However, since precise cut-offs have not been disclosed, it is no longer possible for stakeholders to unambiguously predict the source designation that will be provided in the INN. The procedure therefore lacks transparency and consistency. Remarkably, no definitions for determining the human -u- source infix have been provided, and its designation therefore still hinges on the origin of the antibody's variable domains in a human sequence library of some sort, and relies on information provided by the INNs applicant. This creates a necessity for an arbitrary definition regarding whether an antibody with a fully human sequence derived from 1) a library of natural genes, 2) a library of synthetic genes, 3) from transgenic animals, or 4) from human patients would automatically receive the -u- infix, and potentially blurs the boundary between the -u- and -zu- infix.
Although INNs are used world-wide, several countries use a separate system of non-proprietary names, one example of which is the United States Adapted Names (USAN). The USAN council is administered by the American Medical Association (AMA), and includes members of several agencies, e.g., the Food and Drug Administration (FDA). Interestingly, the post-2014 assignment of source infixes has also generated debate between the INN expert group and the USAN Council. In contrast to the INN top hit procedure explained above, the source designation for a USAN is defined by an 85% sequence cut-off (with +85% human sequence content designating a humanized antibody and a <85% content a chimeric).Citation32,33 This discrepancy has further confused the field.Citation2 During the 62nd open consultation on INN, the FDA and INN expert group representatives acknowledged the difference and indicated that harmonization is essential because therapeutic antibodies in development may have either an INN or a USAN and about half have both.Citation4 We note that all USAN for therapeutic antibodies issued in 2017 thus far are also registered with INNs. In contrast, of the 27 INNs published in 2017, only 9 also carry a USAN.Citation3,32
The flaws in determining “humanness” of antibody sequences by alignment approaches were analyzed and debated by us and others elsewhere.Citation2 Overall, the notion that an antibody's origin can be captured in a single syllable has lost its validity due to the increasing complexity of the antibody landscape (). Moreover, the once useful information that was carried in this one syllable can become outright misleading. The cut-off value of a USAN according to human sequence content at precisely 85% is highly questionable, as is an INN assigned according to the top homology hit being a macaque or almost identical human sequence. The source designation in antibody INNs therefore became a highly disputed issue, and its resolution was awaiting an urgent deployment.
