 ?Mathematical formulae have been encoded as MathML and are displayed in this HTML version using MathJax in order to improve their display. Uncheck the box to turn MathJax off. This feature requires Javascript. Click on a formula to zoom.
?Mathematical formulae have been encoded as MathML and are displayed in this HTML version using MathJax in order to improve their display. Uncheck the box to turn MathJax off. This feature requires Javascript. Click on a formula to zoom.ABSTRACT
Remsima™ (infliximab) is the first biosimilar monoclonal antibody (mAb) approved by the European Medical Agency and the US Food and Drug Administration. Remsima™ is highly similar to its reference product, Remicade®, with identical formulation components. The 2 products, however, are not identical; Remsima™ has higher levels of soluble aggregates, C-terminal lysine truncation, and fucosylated glycans. To understand if these attribute differences could be amplified during forced degradation, solutions and lyophilized powders of the 2 products were subjected to stress at elevated temperature (40–60°C) and humidity (dry-97% relative humidity). Stress-induced aggregation and degradation profiles were similar for the 2 products and resulted in loss of infliximab binding to tumor necrosis factor and FcγRIIIa. Appearances of protein aggregates and hydrolysis products were time- and humidity-dependent, with similar degradation rates observed for the reference and biosimilar products. Protein powder incubations at 40°C/97% relative humidity resulted in partial mAb unfolding and increased asparagine deamidation. Minor differences in heat capacity, fluorescence, levels of subvisible particulates, deamidation and protein fragments were observed in the 2 stressed products, but these differences were not statistically significant. The protein solution instability at 60°C, although quite significant, was also similar for both products. Despite the small initial analytical differences, Remicade® and Remsima™ displayed similar degradation mechanisms and kinetics. Thus, our results show that the 2 products are highly similar and infliximab's primary sequence largely defines their protein instabilities compared with the limited influence of small initial purity and glycosylation differences in the 2 products.
Introduction
Looming patent expirations for lucrative protein drugs have led to a surge in development, regulatory filings, and approvals for biosimilar products.Citation1 Infliximab was the first biosimilar monoclonal antibody (mAb) to receive approval by the European Medical Agency and, recently, by the US Food and Drug Administration.Citation2,3 The approved biosimilar Remsima™, or Inflectra™, is developed by Celltrion (Incheon, South Korea) and is a copy of Remicade®, marketed by Janssen. Infliximab is approved for various indications, including rheumatoid arthritis, ankylosing spondylitis, and inflammatory bowel diseases such as ulcerative colitis and Crohn's disease.Citation4 Detailed analytical comparison of Remicade® and Remsima™ shows significant product similarity, apart from differences in glycan composition, as well as minor differences in levels of soluble aggregates and basic charged variants.Citation2,5-7 These product variances are not surprising since the biosimilar protein is produced using a different clone of Sp2/0 murine hybridoma cell line, a different manufacturing processes, and in different facilities.Citation2,6,8 Additionally, while the formulations of the 2 products are identical, the final lyophilization process as well as the sources for formulation excipients are likely different. These process and excipient differences could result in various levels of reactive species, such as reduced sugars and peroxide impurities of polysorbates, capable of protein modification during product storage. These factors may all contribute to product quality, safety and efficacy, and ultimately make demonstrating biosimilarity challenging.
One strategy to evaluate biosimilarity is to compare biosimilar and reference products in forced degradation studies using thermal, mechanical, or chemical stressors.Citation9-11 Forced degradation studies are usually performed in the development cycle of biotherapeutics to select formulation excipients, develop dosage forms, and determine product shelf life.Citation9,11 These studies are also used during manufacturing process validation to define hold times, for process intermediates, and in analytical method development to define resolving abilities of various assays. The stress testing conditions used for these analyses are far more extreme than the actual physical and thermal stresses that the product vials are exposed to during manufacturing, transport, storage, and handling by physicians or patients. The results from stress testing likely over-exaggerates the extent of product difference and should, thus, be interpreted with caution. However, stress conditions accelerate protein degradation, enrich impurity levels, and thereby improve analytical resolution to aid with analytical development. Ultimately, examination of protein products under stressed conditions give insights into the mechanisms of how these proteins may unfold, lose efficacy, aggregate, and become immunogenic. Forced degradation studies generate valuable analytical data, in relatively short periods of time, to compare biosimilar and reference products and to help link protein structural information with product quality, safety and efficacy.Citation9,11
In this study, we use Remicade® and Remsima™ as a model reference and biosimilar product pair to investigate whether small variances in initial product purity and manufacturing process between the 2 products could influence protein instability upon stress degradation. Comparisons between batch release data of biosimilar and reference product provide the most important information for assessment of product safety, efficacy and biosimilarity. However, minor structural differences between products may be amplified upon stressing, and such stress testing could provide additional information for biosimilarity evaluation. When protein products are stressed, their degradation pathways and the extent of modifications are determined by multiple factors, including protein sequence, physical state, formulation, excipients, and the levels of initial impurities. When formulations of innovator and biosimilar products are different, forced degradation studies could be particularly important in determining biosimilarity. While the primary sequence, physical state and formulations are identical for Remsima™ and Remicade®, the levels of various impurities such as soluble aggregates, charged isoforms, misassembled light chain-heavy chain impurities and glycation levels are different.Citation2,5 These initial impurities could serve as initiators to facilitate further protein unfolding and degradation upon stress. Hence, it could be reasonably expected that a stress-testing based strategy that compares reference and biosimilar products could help to both accentuate product differences and delineate between protein degradative pathways that are defined by protein structure versus product impurities. Consequently, we expect the stress degradation studies to provide further insight into similarity and differences between the 2 products.
To test our concept, we used elevated humidity and temperature incubation to assess how multiple lots of Remsima™ and Remicade® compared. Rigorous analytical characterization was performed to monitor structural (aggregation, hydrolysis, unfolding) and chemical (oxidation, deamidation) changes brought on due to stressing, to understand infliximab's degradation pathways, and identify modification “hot spots” in the sequence. Lastly, the effect of these modifications on biologic activity was quantified using tumor necrosis factor (TNF) binding ELISA and FcγRIIIa binding assays. Collectively, our results provide both a biosimilarity comparison and information about degradation mechanisms for infliximab in the lyophilized powder form.
Results
Protein stress: Monomeric changes and particulate formation
For forced degradation studies, the powders of the 2 products were subjected to incubation at 40°C, at various humidity levels (dry to 97% relative humidity (RH)). A schematic of the study design is depicted in . Both Remsima™ and Remicade® were supplied as lyophilized powders in vials containing 100 mg of infliximab, 500 mg of sucrose, 0.5 mg Tween 80, and 8 mg phosphate buffer salts.Citation4,12 Overall, the protein content of the powder cake was ∼16% (w/w). The powder melt temperatures, determined by modulated-differential scanning calorimetry (mDSC), were similar, with values of 135.8°C for Remicade® and 138.1°C for Remsima™. Product vials were opened, and powders were aliquoted into Eppendorf tubes and the open tubes were incubated for 0, 1, 2 or 4 weeks at 40°C and elevated humidity. At all incubation conditions, powders visually appeared moist, and those incubated at 97% RH deliquesced.
Figure 1. Schematic of stress study design. Humidity/thermal stress of infliximab samples were performed by incubating the drug powders at 40°C at different %RH for 0–4 weeks, followed by reconstitution in WFI and analysis.
After incubation, water for injection (WFI) was added to the samples and levels of protein aggregation were examined by size-exclusion chromatography (SEC). Analysis revealed a time- and humidity-dependent loss of native monomer and formation of soluble aggregate. The kinetics of monomer loss were similar for the 2 products (, ). The rates of monomer loss were calculated using linear regression and are summarized in . The fastest rate of protein aggregation was observed at 97% RH with 0.42% monomer loss per day for Remicade® and 0.44% per day for Remsima™, and virtually no monomer loss was observed for the dry samples. Samples incubated for 4 weeks were further characterized by non-reducing SDS-PAGE (). The gel showed increased levels of aggregates with increasing relative humidity, but also the presence of mAb fragments not previously detected by SEC. Further characterization was performed using reducing SDS-PAGE (). The absence of some aggregate bands after reduction suggests the aggregate formation was partially mediated by disulfide bonds. Jung et al. reported ∼0.12 free –SH mol/mol IgG for infliximab, which may play a role in disulfide bond shuffling.Citation5
Figure 2. Characterization of protein aggregation in stressed samples. A. Representative SEC chromatograms of infliximab following 97% RH/40°C incubation for 0, 1, 2 and 4 weeks. B. SDS PAGE of Remicade® and Remsima™ samples stressed for 4 weeks at 0 (dry), 50, 75 and 97% RH. C. Kinetics of monomer loss at various humidity for Remicade® (solid) and Remsima™ (dashed) as detected by SEC (n = 4 ± SEM). D. Reducing SDS PAGE gel of Remicade® and Remsima™ humidity stressed samples for 4 weeks at 0 (dry), 50, 75 and 97% RH. Molecular weights (MW) are annotated on the ladder in kDa.
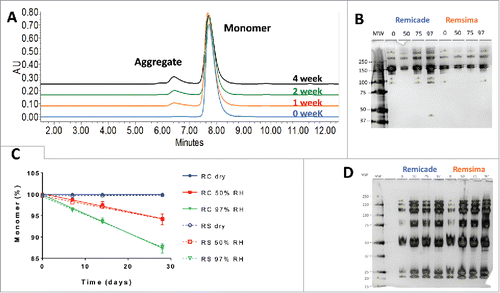
Table 1. Fitted rates of monomer loss for Remicade® and Remsima™ samples after incubation at 40°C and various relative humidity levels.
Particulate size for the reconstituted samples was measured using nanoparticle tracking analysis (NTA). Particulates from 50–350 nm were observed for both products, as shown in and . Remicade® had fewer observed particulates than Remsima™, both initially and after humidity stressing. Although the counts exceed several millions, the particle sizes measured were quite small, and, overall, the particulate counts showed large variability (). Further characterization of these samples was performed using nano-differential scanning calorimetry (nDSC), and . The 3 characteristic melt temperatures were determined (Tm1: 67°C, Tm2: 72°C, and Tm3: 86°C) and were similar for the 2 products, in agreement with previously published values.Citation5 No thermal shifts in the melt temperatures were observed for either product following stress; however, a decrease in overall heat capacity was noticed for the stressed samples. Since the amount of material was held constant, this suggests less energy is required for unfolding after stressing.
Figure 3. Nanoparticle tracking analysis of unstressed (light) and incubated at 97% RH/40°C for 4 weeks (dark) samples of Remicade® (A) and Remsima™ (B). DSC thermal melts of unstressed (dashed) and incubated at 97% RH/40°C for 4 weeks (solid) samples of Remicade® (C) and Remsima™ (D).
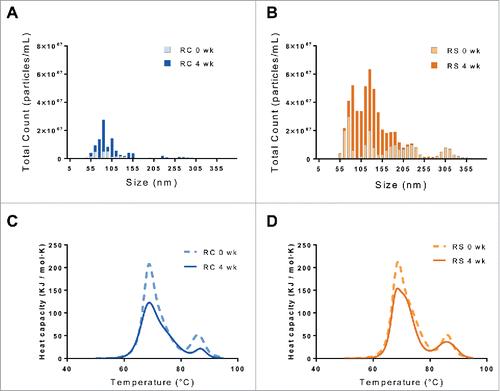
Table 2. Impurity profile of Remicade® and Remsima™ before and after 4-week incubation at 40°C and 97% relative humidity.
Additional forced degradation studies were performed to evaluate antibody stability in solution, and detailed results are in the Supplemental Information. Briefly, at 60°C, both products exhibited rapid aggregation and precipitation within 2 hours, and the kinetics of aggregate formation examined by SEC appeared to be similar for Remicade® and Remsima™ (Fig. S1A). Remicade® samples analyzed by NTA following 2-hour incubation appear on average to have slightly larger aggregates relative to Remsima™ (Fig. S1C), but this difference is not statistically significant due to high sample and measurement variabilities.
Characterization of protein aggregates and hydrolysis products
To augment SEC and SDS-PAGE analyses, unstressed and 97% RH/4 week incubated samples for both products were further characterized by ion mobility mass spectrometry (IM-MS). The IM-MS analysis is capable of examining antibodies under native conditions with minimal handling of samples compared with other MS-based methods. Relatively weak electric fields are used to separate gas-phase protein ions accordingly to their orientationally averaged collision cross sections (CCSs) and charge. IM-MS spectra displayed similar IM drift times for Remicade® and Remsima™ with discrete positions in drift time vs. m/z space for antibody fragments, monomers, dimers, and trimers (). The IM-MS results suggested that small initial levels of antibody impurities increase significantly after humidity and temperature stressing ( and ). For example, the presence of dimer and trimer protein aggregates, as well as 50 and 100 kDa antibody fragments, were observed in stressed samples. Appearance of these species was evident from the different drift times relative to protein monomer drift time, as observed in IM-MS spectra (). While the initial levels of aggregates were slightly higher for Remsima™ (0.35% by SEC, 2.0% by IM-MS) than for Remicade® (0.08% by SEC, 0.8% by IM-MS), these differences did not result in faster aggregation upon stress for Remsima™. The large initial dimer differences between the 2 products as measured by IM-MS may be attributed to method variability () and should be interpreted with caution, as only 2 batches of each product were analyzed. The levels of dimer were similar in stressed samples for Remsima™ (12.4% by SEC and 3.5% by IM-MS) and Remicade® (12.7% by SEC and 3.0% by IM-MS), and the presence of trimers was detected at 0.1% in both products by IM-MS.
Figure 4. Ion mobility spectra and corresponding mass to charge spectrograms of Remicade® (A) and Remsima™ (B) before and after 4 weeks incubation at 97% RH/40°C, Remicade® (C) and Remsima™ (D). Fragment, monomer, dimer and trimer species annotated in ion mobility spectra.
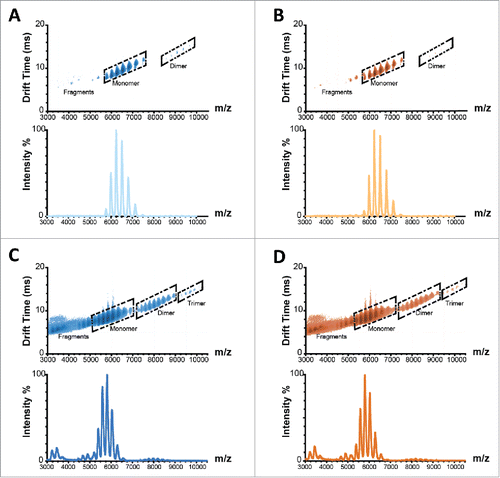
The levels of dimers in stressed samples measured by IM-MS appear to be lower than the levels measured by SEC, indicating that possibly some of the newly formed dimers dissociate during mass spectrometry analysis or during sample preparation, e.g., buffer exchange before mass analysis. Prominent fragment sizes of 50 and 100 kDa were observed by IM-MS and may be antibody fragments corresponding to 1 heavy chain or 2 linked heavy chains, respectively, whereas fragments of 125 kDa by SDS PAGE suggest the loss of one light chain from the mAb. These modifications confirm the susceptibility of infliximab to undergo inter-chain disulfide rearrangements upon humidity stressing. While the levels of protein fragments appear to be higher for Remsima™ than for Remicade® following forced degradation, there is high variability in the measurements and a limited number of samples were available for analysis. Overall, while a significant degree of infliximab aggregation and hydrolysis was observed after the 4-week stress, there were no statistically significant differences in the levels of impurities between the 2 stressed products.
Structural characterization
To better understand the structural changes induced by humidity stressing, selected samples were probed using bulk-averaging spectroscopic techniques. Intrinsic fluorescence and near UV circular dichroism (CD; 250–320 nm) were used to gather information about the tertiary structure, while far UV CD (190–250 nm) measurements were used to probe the secondary structure. Intrinsic fluorescence examines changes in the local environment surrounding aromatic amino acids and it assesses how this local hydrophobicity alters the emission profile upon protein stress.Citation13 The intrinsic fluorescence curves for Remicade® and Remsima™ samples incubated for 0, 2 and 4 weeks at 97% RH are shown in and . Both products had a maximum intensity at 335 nm and they showed no substantial differences upon humidity stressing, apart from a slight decrease in maximum relative fluorescence intensity. Linear drop-offs in the maximum intensity (λ335) are observed with increasing incubation times, suggestive of partial unfolding, aggregation, or monomer loss. Our results, which show high homology between the 2 products, also demonstrate comparable intrinsic fluorescence profiles to published results for other IgG1s.Citation14-16
Figure 5. Biophysical characterization of humidity stressed samples of Remicade® (A, C, E, solid) and Remsima™ (B, D, F, dashed) incubated at 97% RH/40°C for 0 (blue), 2 (red) and 4 (green) weeks. Intrinsic fluorescence spectra ((A)and B), circular dichroism far UV spectra ((C)and D) and near UV spectra ((E)and F).
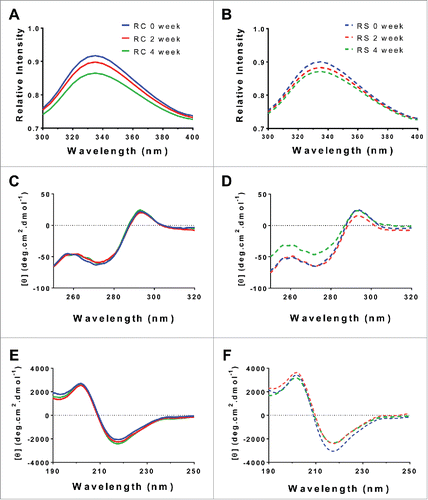
Near UV CD spectrums were collected to confirm the intrinsic fluorescence findings, as shown in and . The Remicade® spectra did not change substantially with the increased duration of protein incubation. The Remsima™ samples showed little change over 2 weeks, but showed a large deviation for the 4-week sample at 250–280 nm not observed for Remicade®. The initial near UV spectrums look similar to published results for infliximab.Citation5 A small shift in absorbance was observed at the near UV region between 280 nm and 300 nm corresponding to the aromatic residues, similar to the earlier drops in the intrinsic fluorescent spectra. Regarding the deviation in the near UV spectrum between 250 nm and 280 nm in the 4-week Remsima™ sample, the spectral pattern suggests a change in the sulfide linkage profile for this sample, in agreement with the earlier findings from IM-MS and SDS-PAGE. It is unclear, however, why similar shifts were not observed for other samples, which should have also undergone similar disulfide rearrangements as suggested by previous data.
We next assessed the far UV CD to capture any secondary structural changes that might have occurred due to humidity stressing. Far UV CD from 190 nm to 250 nm has been used extensively to study secondary structure of proteins and antibodies.Citation17-19 As shown in and , the far UV CD spectra showed considerable similarity between the 2 products. In addition, Remicade® samples displayed small time-dependent deviations in their secondary structure. By contrast, Remsima™ samples showed a slight increase in positive signal at 218 nm, suggesting a greater anti-parallel β structure. Yet, the overall shapes of spectra were similar for the 2 products, indicating that secondary structural differences between Remicade® and Remsima™ following a 4-week incubation were negligible. The variability of native protein concentration in stressed samples somewhat limits the utility of these intrinsically low resolution spectroscopic methods for biosimilarity characterization.
Individual amino acid modifications
To further characterize how humidity stress leads to chemical modifications of the individual amino acids, the proteins were enzymatically digested and analyzed by LC-MS. As shown earlier by LC-MS, both products exhibit similar but significant degrees of oxidation and deamidation, as well as rather different distributions of N-linked glycans.Citation7 Here we applied LC-MS methodology to examine whether initial small differences in chemical modifications can be amplified upon stress, as well as to identify infliximab's “hot spots,” i.e., the amino acid residues easily susceptible to modifications upon elevated humidity and temperature stress.
Chemical modifications of the stressed samples were analyzed by LC-MS and depicted in . The asparagine deamidation levels were initially similar for Remsima™ and Remicade® at light chain residues N-137 and N-158, and at heavy chain residues N-31, N-57, N-318, N-364, N-387, N-392, and N-424. After humidity stress, deamidation levels for both Remsima™ and Remicade® increased at all measured residues (). The highest levels of deamidation after 4-week incubations were observed for LC N-158 (7.5%), HC N-57 (18–19%) and HC-N-392 (10–11%). The deamidation of conserved Asn 384 (N-387, 384+3 due to different numbering schemes) and 389 (N-392), located in the Fc region, that is responsible for antibody binding to Fcγ and neonatal Fc (FcRn) receptors, might be expected to be significant. While we observed significant increases in deamidation of N-387 and N-392 residues, the extent of deamidation was similar between Remicade® and Remsima™. It also has been shown for other antibodies that deamidation of Fc region residues did not significantly alter Fc receptor binding.Citation20-23 The Fc receptor binding is primarily dictated by glycosylation of mAb, specifically the levels of afucosylated species.
Figure 6. Chemical modifications of humidity stressed (97% RH) samples. Deamidation (A), oxidation (B) and N-glycosylation (C) of Remicade® (blue) and Remsima™ (orange) for native proteins and following 97% RH/40°C stress for 2 and 4 weeks as assayed by LC-MS (n = 2 lots ± SEM).
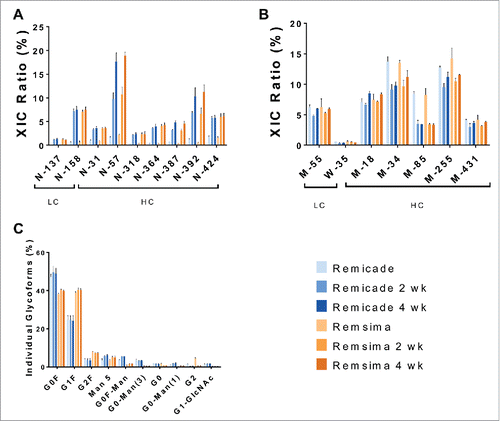
In contrast, deamidation of HC N-57 could be significant due to the residue's location in the complementarity-determining region (CDR); thus, deamidation of HC-N-57 could potentially reduce infliximab's binding to TNF. Other antibodies have been shown to exhibit reduced binding abilities to their respective antigens upon chemical modifications of amino acids in the CDR.Citation20,23,24 Interestingly, while deamidation appears to increase with the duration of incubation for some asparagine residues (N57, N318, and N392), others exhibit the same deamidation levels after 2 and 4 weeks. This provides information regarding the conformational flexibility of the mAbs and the propensity and accessibility of select residues to deamidation. Overall, levels of deamidation appear to be very similar between Remicade® and Remsima™ products both initially and following stressed degradation.
The levels of initial and stress-induced methionine oxidation were analyzed as well (). Our results indicate significant initial oxidation levels for M-55 at 6.3% (LC) and M-18 (∼7%), M-34 (∼13%), M-85 (8.2%), M-255 (∼12%) and M-431 (∼4%) on the heavy chain. However, oxidation levels appeared to be unchanged following forced degradation. Overall, the levels of oxidation are very similar for the 2 products. The oxidation of methionine residues in the Fc region of IgG1 (M-252 and M-431) is known to reduce binding to FcRn and decrease circulation in vivo.Citation25 In addition, oxidation of these methionine residues leads to reduced IgG1 binding to Protein-A column during purification.Citation20-23 While we also observe oxidation of the critical methionine residues HC M-255 (252+3) and HC M-431 (428+3), the levels are similar in the 2 products and do not appear to increase during powder temperature/humidity stressing.
The results of our exploratory infliximab solution thermal stability study indicated significantly increased HC M-255 oxidation levels after incubation at 60°C (Fig. S1D-E), highlighting the importance of this residue as a “hot-spot” for infliximab degradation pathways. In addition, increases in tryptophan di-oxidation were observed in heat-stressed protein solutions, specifically the di-oxidation of HC W-33, W-47 and W-161, which increased from <0.5% to ∼2–4%. We also observed an increase in deamidation, especially for HC N-387, N-392 and N-424.
In addition to oxidation and deamidation, we examined conserved N-glycosylation of the Fc domain by LC-MS. Glycosylation occurs during recombinant production of infliximab and is defined to a large degree by the specific cell line clone as well as cell culture conditions.Citation26 Both products were produced using murine hybridoma cell line Sp2/0, but using different infliximab expressing clones generated by the innovator and biosimilar companies.Citation2,8 Thus, it is not surprising that a biosimilar product like Remsima™ exhibits different distributions of glycans relative to the reference product Remicade®.Citation2,6,7 We also confirmed initial differences in glycan distribution, with G0F, G0F-Man, G0F-Man(3) and G0F-Man(1) being more abundant in Remicade® while G1F, G2F and G2 more prevalent in Remsima™ (). These initial differences are important, as it appears that overall levels of mannose and afucosylated glycans are higher in Remicade® than in Remsima™, and this is known to affect Fc receptor binding, antibody dependent cell-mediated cytotoxicity (ADCC), circulation time and immunogenicity.Citation27 We also observed that glycan distribution remained virtually unchanged for both Remicade® and Remsima™, as expected. Proteins are glycosylated enzymatically during production by the host cell; thus, it is unlikely that glycosylation could be altered during stressed stability studies apart from possible selective aggregation/degradation of a specific glycan type.
In vitro efficacy
In addition to the analytical characterization of the humidity stressed samples, we also performed in vitro bioactivity assays to gauge how stressing affects infliximab's abilities to bind TNF and FcγRIIIa (). We expected that stressed-induced individual amino acid modification in the CDR could reduce infliximab's binding to TNF, while modification of the Fc domain could alter FcγRIIIa interactions. In addition, the reduction of intact infliximab monomer content over the duration of the forced degradation study could lead to further reduction in antigen and receptor binding.
Figure 7. Binding of Remicade® and Remsima™ samples after stressing at 97% RH/40°C. A. TNF binding as measured by ELISA. B. FcγRIIIa binding as measured by BLITZ (n = 2 lots ± SEM; * P< 0.05 ** P<0.01 compared with unstressed).
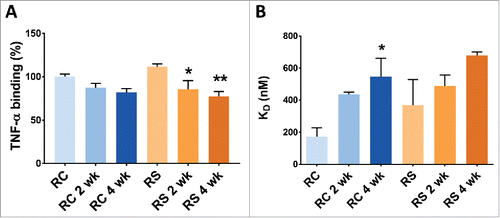
According to regulatory filings, Remicade® and Remsima™ exhibit similar initial ability to bind and neutralize TNF.Citation2,6 The 90% confidence interval for the mean difference between Remicade® and Remsima™ TNF binding affinity measured by ELISA falls entirely within the equivalence margin.Citation6 Our measurements, derived from only 2 lots for each product, indicated that the TNF binding affinity was slightly higher for Remsima™, at 111.7% of the initial Remicade® value, but this result was not statistically significant (). Following 4 weeks of forced degradation, TNF neutralization decreased to 81.8% (Remicade®) and 77.2% (Remsima™) of the initial value for unstressed Remicade® standard. This decrease could be attributed to increased deamidation levels of HC-N-57 or reduced infliximab monomer content or both. However, there was no statistically significant difference in TNF binding affinity between 2 stressed products at the corresponding time points.
The differences in FcγRIIIa binding corresponding to the lower levels of afucosylation for Remsima™ relative to Remicade® were reported previously.Citation7 In the regulatory filing, decreased afucosylation was reported, and corresponded to a 20% lower binding efficiency to FcγRIIIa and 20% lowered ADCC for Remsima™ relative to Remicade®.Citation6 The initial unstressed samples of Remicade® showed tighter binding to FcγRIIIa relative to Remsima™ with the respective KD of 173 ± 56 nM and 368 ± 160 nM, as measured by biolayer interferometry (). The receptor binding progressively weakened following stress degradation of the 2 products, and the KD increased to 545 ± 117 nM for Remicade® and 680 ± 22 nM for Remsima™ after 4 weeks at elevated humidity and temperature. Since no significant changes in glycosylation between the 2 products were detected upon incubation, the reduction in Fc receptor binding is likely attributed to the progressive increase in aggregation and fragmentation of the 2 antibodies, in conjunction with the chemical modifications, thus reducing the total amount of bioactive monomer available to bind receptors. Additionally, the initial differences in Fc binding between the 2 products appear to largely diminish upon stressing, highlighting the importance of structural integrity over glycosylation patterns for bioactivity. Overall, no statistically significant differences in FcγRIIIa binding were observed throughout the entire study, which was possibly due to the small number of lots tested (n = 2).
Discussion
In this study, humidity- and temperature-induced forced degradation was used to analytically compare a biosimilar mAb, Remsima™, with its reference product, Remicade®. Despite the minor differences in initial product profiles (glycosylation pattern, levels of dimer and basic variants), as well as differences in the manufacturing processes of 2 mAbs,Citation2,5,6 the 2 products behaved remarkably similarly in the forced degradation studies. Very similar rates of degradation, along with similar types and levels of impurities were detected in the 2 stressed products. Hence, for products with high analytical similarity and identical formulations, such as Remsima™ and Remicade®, the degradation mechanisms appear to be defined primarily by protein sequence and structure rather than by minor initial product differences. However, other biosimilar products may be formulated differentlyCitation28,29 or produced in different expression systems compared with their reference products, both potentially leading to greater product differences and varying mechanisms of protein instability in forced degradation studies.
Our results revealed the main infliximab degradation products to be protein aggregates and antibody fragments of 50 and 100 kDa ( and ). The rate of monomer loss was nearly identical for the 2 products under the various humidities tested (), and higher initial levels of dimers in Remsima™ did not result in faster protein aggregation. The actual levels of formed aggregates and antibody fragments varied slightly when measured by SDS-PAGE, IM-MS and SEC. An increase in the level of subvisible particulates was observed for both products by NTA after stressing. Each analytical method has inherent limitations due to variations in the sample handling, e.g., concentration/dilution, buffer exchange, mixing with gel loading buffer and binding to column matrix, which is known to affect aggregate before the detection.Citation30 Hence, use of orthogonal methods allows for a more complete picture of protein aggregation and hydrolysis.
Structural changes in the 2 products were compared spectroscopically. The near- and far-UV spectral results for the initial samples are in agreement with the published results for the native protein and small changes in spectra were observed in stressed samples.Citation5 No thermal shifts were observed in stressed samples by nDSC, but lower enthalpy indicated minimal structural changes. Further analysis suggests these products slightly unfold, but undergo significant deamidation at specific “hot spots” upon stressing, as measured by LC-MS (). Overall, structural and chemical modifications were comparable between the products. The TNF neutralizing ability of both antibodies was reduced significantly upon stress, by ∼20–30% relative to the unstressed protein levels (). This reduction could be attributed to individual amino acids modifications in the CDR domain (LC-M-55, HC-M-34, HC-N-31 and HC-N-57), as well as to the decrease in functional protein monomer content. Nonetheless, regardless of mechanism, the 2 products exhibited a similar magnitude of reduction in TNF binding. Additionally, post-stress FcγRIIIa binding was reduced significantly for both products, by 2–3-fold, with slightly larger mean decreases for Remsima™ binding at each time point. However, there were no statistically significant differences between the reference and biosimilar products. Interestingly, using a larger number of lots for Remicade® and Remsima™, we showed that the level of afucosylation was significantly higher in unstressed Remicade, which corresponded to stronger FcγRIIIa binding relative to Remsima™. Such differences in glycosylation, along with the reduction in FcγRIIIa binding and ADCC were reported in the regulatory filings for Remsima™, resulting in the deferral of biosimilar approval for inflammatory bowel disease indications by Health Canada.Citation2,6,31,32 While glycosylation differences between products did not change following forced degradation, the FcγRIIIa binding decreased similarly for the 2 products (, ). A several fold reduction in the FcγRIIIa binding after forced degradation could be attributed to individual amino acid modifications in the Fc domain, as well as to extensive aggregation and hydrolysis of the 2 mAbs. However, this finding shows that forced degradation could mute rather than amplify initial differences between biosimilar and reference product, which was surprising and contradictory to the initial expectations for this study.
Stress testing is often used to identify instabilities in proteins, select optimal formulations to reduce protein degradation during stability testing, and to define shelf-life and storage conditions. Antibodies, particularly IgG1s, have been stressed in a variety of ways, including thermally, physically (e.g., stirring, shaking), and chemically (e.g., oxidation, pH).Citation16,33 Overall, different mechanisms of stressing have yielded different by-products; however, in general, IgG1s are prone to aggregation and chemical modification of several conserved residues identified as “hot-spots.”Citation20 Most stress conditions lead to aggregate formation, either insoluble, soluble, or both.Citation9,11 Additionally, structural studies have characterized changes in secondary and tertiary features of mAbs that may initiate aggregation.Citation14,18 Chemical stressing has also identified several oxidation hot-spots on mAbs and accessibility of various residues.Citation16,20,34 Aggregates formed via different stressors are heterogeneous and express differing immunogenic patterns test in various immunogenicity models.Citation16,35,36 In general, physical stressing (thermal, agitation) leads to greater particulate formation and yields a stronger immune response than chemically stressed samples.Citation35,36 Stress-induced particulate formation highlights a need for characterizing how process parameters, excipients and formulation differences in biosimilars affect protein aggregation, as they may ultimately affect product immunogenicity and safety.
To demonstrate biosimilarity, the 2 products should have no “clinically meaningful” differences. Thus, comparison of lot release data for biosimilar and reference products are the most critical for determining biosimilarity. However, the rates and mechanisms of degradation of reference and biosimilar products can also be compared during forced degradation studies. These comparisons are particularly useful if the biosimilar is formulated differently compared with the reference product, as it could result in new types of degradation products observed in the biosimilar that are not present in the reference product. The presence of new types of degradation products raises additional questions if these degradants could potentially affect product efficacy and safety.
While forced degradation studies allow rapid generation of a large volume of analytical comparability data, the mechanism of protein instability and type of degradation products formed are largely determined by the stress conditions. Often the conditions of forced degradation studies are more extreme than the actual stresses that product vials are exposed to during transport, storage, and clinical administration. In this study, to accelerate protein degradation, the vials were opened and products were exposed to much higher temperatures and humidity than the environment inside sealed and refrigerated vials. For traditional biosimilarity comparisons the analyses are performed using several lots of each product. In our constrained study, only 2 lots of each product were subjected to the forced degradation, which limits our ability to perform rigorous statistical analysis. We observed large deviations in product modifications in the stressed samples, which, in addition to limited sample size, may also arise from the heterogeneous nature of degradant formation, especially at the primary chemical levels. Due to the limited amount of protein, we did not examine the effects of oxidative, mechanical, and chemical stress on reconstituted infliximab solutions for every time point. Furthermore, we have not characterized the type, shapes, and sizes of formed protein aggregates and have not examined the levels of protein glycation in this study, which may be of interest for immunogenicity characterizations.
The immunogenic propensity of biologics is a key safety parameter identified by regulatory agencies. To date, several studies have used a variety of stresses to form immunogenic products, and characterized their immunogenicity in various in vitro and in vivo models.Citation16,35,36 General correlations between characteristics of stressed protein (type, shape and size of protein aggregates) and the immunogenicity of these degradation products were developed.Citation20,33-36 Thus, forced degradation could provide valuable predictive data regarding the immunogenic propensity of the biosimilar product, and this data could ultimately be used in product regulatory licensing applications. The challenge in applying this stress testing strategy is the lack of universal conditions/guidelines to perform the studies, which leads to different setup conditions and, thus, difficulty in extrapolating results. Ideally, the solution would be to adopt guidelines to perform these tests, but this is confounded by the diverse nature of biotherapeutics, each of which may display different behaviors. Given these concerns, it is recommended that stress studies be designed with specific ends in mind, and that several orthogonal robust analytical techniques be used to ensure confidence in the final outcomes.
Biosimilar approvals are poised to bring substantial positive change to healthcare, yet establishing biosimilarity is challenging. Forced degradation studies provide a unique approach to examine the appearance of any minor differences that may have clinical safety, immunogenicity, and efficacy implications. In this study, we used thermal and humidity stress testing on both reference, Remicade®, and biosimilar, Remsima™, infliximab products. Our results show similar levels of aggregate formation, structural variation, and chemical modifications to support the notion that the products are biosimilar. We anticipate stress testing will be used widely for biosimilar assessment as patents for more biologic products expire and new biosimilar products permeate the markets, and this work will help guide future studies.
Materials and methods
Remicade® was purchased from the University of Michigan Hospital Pharmacy (Ann Arbor, MI) and Remsima™ was acquired from Celltrion (Incheon, South Korea). Both Remsima™ and Remicade® are supplied as lyophilized powders in vials containing 100 mg of infliximab, 500 mg of sucrose, 0.5 mg Tween 80, and 8 mg phosphate buffer salts.Citation4,12 The lot numbers and expiry dates of the products used in this study are listed in Supplemental Table 1. All forced degradation studies were conducted within the expiry dates of the samples. Samples were reconstituted using pure WFI (Thermo-HyClone) to a concentration of 1 mg/mL unless specified otherwise. All chemical reagents were of analytical grade or purer and were purchased from either Fisher or Sigma Aldrich.
Protein stress study set-up
Powders of Remicade® and Remsima™ contain ∼16% of protein by weight. Vials of both products were opened and aliquoted in 1.5 mL Eppendorf tubes (∼6.25 mg of powder or 1 mg of infliximab in each tube). Saturated solutions of NaBr and K2SO4 in distilled water were prepared to simulate 53% and 97% RH, respectively.Citation37 Desiccant was used to simulate dry conditions. The infliximab powders in open tubes were placed in desiccators at a specific RH and incubated at 40°C for 1, 2, or 4 weeks. Samples were removed from the desiccators and reconstituted with WFI to 1 mg/mL. Reconstituted samples were further aliquoted for the various analytical assays and stored at either 4°C or −80°C until analysis. Reconstituted proteins were also subject to thermal stress and are detailed in the supplemental information.
Size exclusion chromatography
SEC was performed using a Waters Binary HPLC pump 1525 equipped with Waters auto-sampler 2707 and UV/visible detector 2489. TSK Gel 3000 SWxl column (Tosoh 7.8 mm × 30 cm, 5 µm). The mobile phase, phosphate-buffered saline (PBS; pH 7.4), was delivered at 1 mL/min. Protein samples were filtered through 0.45 μm filter (Millipore) before injection. A 25 μL injection volume was used and the UV signal was monitored at 210 and 280 nm. The area under the curve was used to calculate the percentage of monomer, aggregates and fragments. The monomer content for unstressed samples served as the 100% reference and the loss of monomer content at each stressed time point was calculated by decrease of monomer peak area.
Circular dichroism
CD was performed using a Jasco J-815 CD spectrometer equipped with temperature controller (CDF-426S/15) and Peltier cell at 25°C. The samples were diluted to 0.1 mg/mL for near UV and to 0.5 mg/mL for far UV measurements. The samples were measured in quartz cuvettes (Hellma) with a path length of 1 mm for far UV and 1 cm for near UV. The spectra were collected in continuous mode at a speed of 50 nm/min, bandwidth of 1 nm and a DIT of 1 s. The average of 10 scans were reported. Blank buffer without the antibody was subtracted from each spectrum using the Jasco spectra manager software (Version 2.1). The raw data were converted to mean residual ellipticity (MRE) using the following equation:where θλ is the observed ellipticity in degrees at wavelength λ, d is the path length in cm, c is the concentration in g/mL, and mean residual weight (MRW) is 110 for infliximab. Data smoothing was performed using GraphPad Prism Software (Version 6.07) using a 0th order polynomial with 4 neighbors at each point.
Intrinsic fluorescence
Intrinsic fluorescence was performed with a Jasco J-815 spectrometer equipped FMO-427S/15 detector and Peltier controller set at 25°C. The samples were diluted to 0.1 mg/mL and were measured in black window quartz cuvettes (Hellma) with a path length of 1 mm. The spectra were collected in continuous mode with a data pitch of 5 nm, scan speed of 50 nm/min and a DIT of 1sec; data were averages of 5 scans. The excitation wavelength was 280 nm and emission spectra were collected from 300 to 400 nm, with a gain voltage of 850 V. Data smoothing was performed using GraphPad Prism Software (Version 6.07) using a 2nd order polynomial with 4 neighbors at each point.
Gel electrophoresis
Selected samples were analyzed by non-reducing and reducing SDS-PAGE to examine for presence of mAb aggregates and fragments. Samples (∼10 µg) were mixed with NuPAGE LDS sample buffer, at 3:7 sample: loading buffer ratio and denatured at 90°C for 3 minutes before gel loading. For reduced samples, 5% v/v β-mercaptoethanol was added before heat denaturation. Samples were run on Invitrogen PowerEase 500 with NuPAGE 4–12% BisTris gel. BioRad Precision Plus Protein™ All Blue Standards were used for molecular weight controls. Gels were stained using Thermo Pierce Silver Stain Kit and accompanying protocol and analyzed using Fluorchem M (ProteinSimple).
Nanoparticle tracking analysis
NTA (Nanosight NS300, Malvern) was used to quantify subvisible particulates in protein samples. The NS3000 was fitted with a SCMOS camera, a 405 nm blue laser and the sample chamber. The samples were diluted 10-fold before analysis and transferred to sterile syringes (BD) before injection into the sample chamber. The sample chamber was flushed with antibody buffer and then with sterile water in between sample analysis. The sample results were averages of three 60s runs measured at 0.1 mL/min flow rate and were analyzed using the NTA 3.0 software.
Modulated differential scanning calorimetry
Modulated DSC (Discovery, TA Instruments, New Castle, DE) was used to determine the Tg and heat of enthalpy of the lyophilized powders. Sample powders (∼10 mg) were sealed in hermatic aluminum pans. The measurements were performed at a heating rate of 2 °C per min from 0 to 180 °C under a nitrogen gas flow of 25 mL/min. The modulation amplitude was 0.5 °C and the period was 40 s. The Tg and enthalpy were obtained by fitting data with TA Trios software (v4.1.1).
Nano-differential scanning calorimetry
The thermal melt profiles for stressed and unstressed samples of Remicade® and Remsima™ were measured using TA Instruments nDSC equipped with an autosampler. The thermograms were obtained using a scan rate of 1°C/min from 10°C to 100°C. The thermograms were analyzed using TA NanoAnalyze software (v2.4.1) after blank buffer subtraction using multiple scaled 2 state models to determine the 3 transition melt temperatures.
Liquid chromatography-mass spectrometry
Antibody tryptic digests were prepared per manufacturer procedure from the low pH protein digestion kit (Promega, CAS # CS1895A01), designed to prevent non-enzymatic protein modifications during digest. Antibody samples were denatured in 8 M urea, reduced, and alkylated with iodoacetamide. The reactions were diluted 7-fold and incubated with Trypsin Gold and Lys-C (Promega) at 20:1:1 (w/w/w) ratio overnight at 37°C. After digestion was complete, the reactions were acidified with trifluoroacetic acid.
500 ng of each digested sample was analyzed by nano UPLC-MS/MS with a Proxeon EASY-nLC 1000 HPLC system interfaced to a ThermoFisher Q Exactive HF mass spectrometer. Peptides were loaded on a trapping column and eluted over a 75 µm × 50 cm analytical column (Thermo Fisher P/N ES-803) at 300 nL/min by using a 2-hour reverse phase gradient; both columns were packed with PepMap RSLC C18, 2 µm resin (Thermo Scientific). The mass spectrometer was operated in data-dependent mode, with MS and MS/MS performed in the Orbitrap at 70,000 and 17,500 FWHM resolution respectively. The 15 most abundant ions were selected for MS/MS.
Data analysis for LC-MS/MS analysis of digested specimens was performed with Byonic search software (Protein Metrics Inc., San Carlos, CA, USA).Citation38,39 In this instance, the search used the infliximab sequence. Identifications for peptide ions were made by matching the precursor (MS1) mass and expected fragment ion masses (MS2) to infliximab peptides. The search included variable modifications such as mono- and di-oxidation on methionine and tryptophan, deamidation and ammonia loss from asparagine, and a wide range of N-linked glycans.
Quantification of modifications relative to unmodified and other modified peptides was accomplished using the Byologic software (Protein Metrics), which uses a label-free quantification approach with extracted ion chromatogram areas (XIC areas). This software automated the XIC extraction and data organization automatically from the Byonic results or in silico generated lists of potentially observed molecular ions.
Ion mobility mass spectrometry
Reconstituted antibody samples for native MS experiments were buffer exchanged into 100 mM ammonium acetate buffer using Micro Bio-Spin 30 columns (Bio-Rad, Hercules, CA) without further purification. Sample aliquots (∼7 µL) were analyzed by IM-MS on a quadrupole-ion mobility-time-of-flight mass spectrometer (Q-IM-ToF MS) instrument (Synapt G2 HDMS, Waters, Milford, MA).Citation40,41 Antibody ions were generated using a nESI source in the positive mode. Capillary voltages of 1.4 kV-1.6 kV were applied and the sampling cone was operated at 60 V. The trap traveling-wave ion guide was pressurized to 3.4 × 10−2 mbar of argon gas. The traveling-wave ion mobility separator was operated at a pressure of ∼3.5 mbar and used a series of DC voltage waves (40 V wave height traveling at 600 m/s) to generate ion mobility separation. The ToF-MS was operated over the m/z range of 1000–10000 at a pressure of 1.7 × 10−6 mbar.
Mass spectra were calibrated externally using a solution of cesium iodide (100 mg/mL) and processed with Masslynx V4.1 software (Waters, Milford, MA). Exact molecular masses of intact mAb samples were calculated by assigning the charge states based on the set that gives lowest standard deviation for a given average mass assignment.Citation42,43 Relative dimer ratios were calculated from total ion counts of major charge states (30+ to 36+) compared with that of major monomer charge states (20+ to 26+). Peaks were fitted to Gaussian models and integrated using OriginPro 9 software.
FcγRIIIa binding
The binding of infliximab to FcγRIIIa was tested with biolayer interferometry using a BLITZ instrument (Fortebio, Menlo Park, CA). The procedure used here was adopted from the method reported previously.Citation44,45 Protein G biosensor tips were used and the binding measurement was performed at 25°C. Infliximab samples at 4.6 µM (1 mg/mL) initial concentration in formulation buffer were diluted to 0.8 µM with PBS containing 1 mg/mL casein as a blocking agent (kinetic buffer). Binding studies were performed as follows: First, the protein G biosensor tip was hydrated in PBS for 10 min and then incubated for 30 min in kinetic buffer. Next, an initial baseline (30 s) was established in the kinetics buffer and then the protein G biosensor tips were loaded (120 s) with infliximab samples at a concentration of 0.8 µM to a response level of ∼4 nm. A new baseline (240 s) was then established followed by the association (180 s) and dissociation (360 s) of FcγRIIIa measured by dipping the biosensor into solutions of FcγRIIIa and PBS kinetic buffer, respectively. The biosensor tips were regenerated as described previouslyCitation44 after each assay cycle. To determine the dissociation constant (KD), a range of FcγRIIIa concentrations from 0.4 µM-3.2 µM was evaluated. Data generated from the binding of the receptor to infliximab were collected in triplicate for each incubation time point and product, lot and globally fitted to a 1:1 binding model using BLITZ Pro software.
ELISA for TNF binding
96-well ELISA plates (Nunc Maxisorp) were coated with 1 µg/mL TNF (R&D systems) in PBS (pH 7.4) overnight. The plates were washed for 4 cycles with PBS (pH 7.4) using plate washer (Thermo Wellwash 4 MK-2) and subsequently blocked for 2 h at room temperature with 1% bovine serum albumin (BSA) in PBS (pH 7.4) solution. The plates were washed again and incubated for 1 h at room temperature with infliximab standards (1 ng/mL to 100,000 ng/mL) and diluted samples of equal concentrations. All standard and sample dilutions were prepared in PBST-BSA (PBS (pH 7.4) containing 0.02% Tween 80 and 1% BSA). The plates were once again washed and then incubated with 1000-fold dilution of AP-conjugated anti-human Fc IgG (Sigma-SKU: A9544) in PBST-BSA for 1 h at room temperature. The plate was then washed again to remove residual secondary antibody and incubated with p-nitrophenyl phosphate (pNPP) (Sigma) for 30 min at room temperature for color development. Absorbance at 405 nm was then read using plate reader (Spectra Max M3, Molecular Devices). A standard curve was built using a sigmoidal fit and concentrations of diluted samples were calculated. The sample concentrations were divided by the initial unstressed Remicade® concentration to determine the relative TNF binding activity.
Statistical analysis
Statistical analysis was performed using Prism 6 (Graphpad) suite. 2-way ANOVA hypothesis testing was performed using multiple comparisons relative to initial samples of Remicade® or Remsima™. Corrections for multiple comparisons were performed using Dunnett's method and significance levels were set at 0.05 (95% confidence interval).
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Supplemental_Materials.docx
Download MS Word (64.4 KB)Acknowledgments
The work here is supported by National Institute for Pharmaceutical Technology and Education (NIPTE) FDA U01 FD004275 (AS, SPS, TT), NIGMS R01 GM090080 (TT), NSF CAREER 1253384 (BR), NIGMS R44GM100634 (CB, EC), American Foundation for Pharmaceutical Education Pre-Doctoral Fellowship (KP) and NIH Biotechnology Training Grant 5-T32-GM008359 (SZO). The editorial help of Emily E. Morin and Beth Schachter is greatly appreciated.
References
- Konara CS, Barnard RT, Hine D, Siegel E, Ferro V. The tortoise and the hare: Evolving regulatory landscapes for biosimilars. Trends Biotechnol 2016; 34(1):70-83; PMID:26620970; https://doi.org/10.1016/j.tibtech.2015.10.009
- European Medicines Agency. Assessment report: Inflectra; 2013.
- U.S. Food & Drug Administration. FDA Approves Inflectra, A Biosimilar To Remicade; 2016 Apr 05. [accessed 2016 Apr 16]. http://www.fda.gov/NewsEvents/Newsroom/PressAnnouncements/ucm494227.htm
- Janssen Pharmaceutical. Remicade ® [package insert]; 2015.
- Jung SK, Lee KH, Jeon JW, Lee JW, Kwon BO, Kim YJ, Bae JS, Kim D, Lee SY, Chang SJ. Physicochemical characterization of remsima®. mAbs 2014; 6(5):1163-77; PMID:25517302; https://doi.org/10.4161/mabs.32221
- Celltrion, Inc. FDA Arthritis Advisory Committee. FDA Briefing Document: Arthritis Advisory Committee Meeting BLA 125544: CT-P13 a proposed biosimilar to remicade (infliximab); 2016. U.S. FDA.
- Pisupati K, Tian Y, Okbazghi S, Benet A, Ackermann R, Ford M, Saveliev S, Hosfield CM, Urh M, Carson E, et al. A multidimensional analytical comparison of Remicade and the biosimilar Remsima. Anal Chem 2017; 89(9):4838-46; PMID:28365979; https://doi.org/10.1021/acs.analchem.6b04436
- Celltrion, Inc. CT-P13 (inflximab biosimilar): Briefing Document for the Arthritis Advisory Committee; 2016. U.S. FDA.
- Hawe A, Wiggenhorn M, van de Weert M, Garbe JH, Mahler H, Jiskoot W. Forced degradation of therapeutic proteins. J Pharm Sci 2012; 101(3):895-913; PMID:22083792; https://doi.org/10.1002/jps.22812
- Luo Q, Joubert MK, Stevenson R, Ketchem RR, Narhi LO, Wypych J. Chemical modifications in therapeutic protein aggregates generated under different stress conditions. J Biol Chem 2011; 286(28):25134-44; PMID:21518762; https://doi.org/10.1074/jbc.M110.160440
- Chan CP. Forced degradation studies: Current trends and future perspectives for protein-based therapeutics. Expert Rev Proteomics 2016; 13(7):651-8; PMID:27285431; https://doi.org/10.1080/14789450.2016.1200469
- Celltrion, Inc. Remsima™ [package insert]; 2014.
- Lakowicz JR. Principles of fluorescence spectroscopy. New York City, NY: Springer Science & Business Media; 2013.
- Ohadi K, Legge RL, Budman HM. Intrinsic fluorescence-based at situ soft sensor for monitoring monoclonal antibody aggregation. Biotechnol Prog 2015; 31(5):1423-32; PMID:26137937; https://doi.org/10.1002/btpr.2140
- Garidel P, Hegyi M, Bassarab S, Weichel M. A rapid, sensitive and economical assessment of monoclonal antibody conformational stability by intrinsic tryptophan fluorescence spectroscopy. Biotechnol J 2008; 3(9–10):1201-11; PMID:18702089; https://doi.org/10.1002/biot.200800091
- Filipe V, Jiskoot W, Basmeleh AH, Halim A, Schellekens H, Brinks V. Immunogenicity of different stressed IgG monoclonal antibody formulations in immune tolerant transgenic mice. mAbs 2012; 4(6):740-52; PMID:22951518; https://doi.org/10.4161/mabs.22066
- Cathou RE, Kulczycki A, Haber E. Structural features of -immunoglobulin, antibody, and their fragments. circular dichroism studies. Biochemistry 1968; 7(11):3958-64; PMID:5722265; https://doi.org/10.1021/bi00851a024
- Joshi V, Shivach T, Yadav N, Rathore AS. Circular dichroism spectroscopy as a tool for monitoring aggregation in monoclonal antibody therapeutics. Anal Chem 2014; 86(23):11606-13; PMID:25350583; https://doi.org/10.1021/ac503140j
- Houde DJ, Berkowitz SA. Biophysical characterization of proteins in developing biopharmaceuticals. Boston MA: Elsevier; 2014.
- Luo Q, Joubert MK, Stevenson R, Ketchem RR, Narhi LO, Wypych J. Chemical modifications in therapeutic protein aggregates generated under different stress conditions. J Biol Chem 2011; 286(28):25134-44; PMID:21518762; https://doi.org/10.1074/jbc.M110.160440
- Bertolotti-Ciarlet A, Wang W, Lownes R, Pristatsky P, Fang Y, McKelvey T, Li Y, Li Y, Drummond J, Prueksaritanont T. Impact of methionine oxidation on the binding of human IgG1 to FcRn and fcγ receptors. Mol Immunol 2009; 46(8):1878-82; PMID:19269032; https://doi.org/10.1016/j.molimm.2009.02.002
- Liu D, Ren D, Huang H, Dankberg J, Rosenfeld R, Cocco MJ, Li L, Brems DN, Remmele Jr RL. Structure and stability changes of human IgG1 fc as a consequence of methionine oxidation. Biochemistry 2008; 47(18):5088-100; PMID:18407665; https://doi.org/10.1021/bi702238b
- Houde D, Peng Y, Berkowitz SA, Engen JR. Post-translational modifications differentially affect IgG1 conformation and receptor binding. Mol Cell Proteomics 2010; 9(8):1716-28; PMID:20103567; https://doi.org/10.1074/mcp.M900540-MCP200
- Khawli LA, Goswami S, Hutchinson R, Kwong ZW, Yang J, Wang X, Yao Z, Sreedhara A, Cano T, Tesar D, et al. Charge variants in IgG1: Isolation, characterization, in vitro binding properties and pharmacokinetics in rats. mAbs 2010; 2(6):613-24; PMID:20818176; https://doi.org/10.4161/mabs.2.6.13333
- Wang W, Vlasak J, Li Y, Pristatsky P, Fang Y, Pittman T, Roman J, Wang Y, Prueksaritanont T, Ionescu R. Impact of methionine oxidation in human IgG1 fc on serum half-life of monoclonal antibodies. Mol Immunol 2011; 48(6):860-6; PMID:21256596; https://doi.org/10.1016/j.molimm.2010.12.009
- Zhu J. Mammalian cell protein expression for biopharmaceutical production. Biotechnol Adv 2012; 30(5):1158-70; PMID:21968146; https://doi.org/10.1016/j.biotechadv.2011.08.022
- Hmiel L, Brorson K, Boyne M, II. Post-translational structural modifications of immunoglobulin G and their effect on biological activity. Anal Bioanal Chem 2015; 407(1):79-94; PMID:25200070; https://doi.org/10.1007/s00216-014-8108-x
- Sandoz. FDA Arthritis Advisory Committee. BLA 761042 GP2015: a proposed biosimilar to Enbrel (etanercept); 2016. U.S. FDA.
- Sandoz. Oncologic Drugs Advisory Committee: ZARXIO (filgrastim); 2015. U.S. FDA.
- Carpenter JF, Randolph TW, Jiskoot W, Crommelin DJA, Middaugh CR, Winter G. Potential inaccurate quantitation and sizing of protein aggregates by size exclusion chromatography: Essential need to use orthogonal methods to assure the quality of therapeutic protein products. J Pharm Sci 2010; 99(5):2200-08; PMID:19918982; https://doi.org/10.1002/jps.21989
- Health Canada. Remsima: Summary basis of decision; 2014.
- Feagan BG, Choquette D, Ghosh S, Gladman DD, Ho V, Meibohm B, Zou G, Xu Z, Shankar G, Sealey DC. The challenge of indication extrapolation for infliximab biosimilars. Biologicals 2014; 42(4):177-83; PMID:24962198; https://doi.org/10.1016/j.biologicals.2014.05.005
- Ahmadi M, Bryson CJ, Cloake EA, Welch K, Filipe V, Romeijn S, Hawe A, Jiskoot W, Baker MP, Fogg MH. Small amounts of sub-visible aggregates enhance the immunogenic potential of monoclonal antibody therapeutics. Pharm Res 2015; 32(4); PMID:25319104; https://doi.org/10.1007/s11095-014-1541-x
- Paul R, Graff-Meyer A, Stahlberg H, Lauer ME, Rufer AC, Beck H, Briguet A, Schnaible V, Buckel T, Boeckle S. Structure and function of purified monoclonal antibody dimers induced by different stress conditions. Pharm Res 2012; 29(8):2047-59; PMID:22477068; https://doi.org/10.1007/s11095-012-0732-6
- Bi V, Jawa V, Joubert MK, Kaliyaperumal A, Eakin C, Richmond K, Pan O, Sun J, Hokom M, Goletz TJ, et al. Development of a human antibody tolerant mouse model to assess the immunogenicity risk due to aggregated biotherapeutics. J Pharm Sci 2013; 102(10):3545-55; PMID:23925953; https://doi.org/10.1002/jps.23663
- Rombach-Riegraf V, Karle AC, Wolf B, Sordé L, Koepke S, Gottlieb S, Krieg J, Djidja M, Baban A, Spindeldreher S, et al. Aggregation of human recombinant monoclonal antibodies influences the capacity of dendritic cells to stimulate adaptive T-cell responses in vitro. PLoS ONE 2013; 9(1):e86322; PMID:24466023; https://doi.org/10.1371/journal.pone.0086322
- Greenspan L. Humidity fixed points of binary saturated aqueous solutions. Journal of Research of the National Bureau of Standards 1977; 81A(1):89; https://doi.org/10.6028/jres.081A.011
- Bern M, Kil YJ, Becker C. Byonic: Advanced peptide and protein identification software. Curr Protoc Bioinformatics 2012; Dec;Chapter 13:Unit 13.20; PMID:23255153; https://doi.org/10.1002/0471250953.bi1320s40
- Bern M, Cai Y, Goldberg D. Lookup peaks: A hybrid of de novo sequencing and database search for protein identification by tandem mass spectrometry. Anal Chem 2007; 79(4):1393-1400; PMID:17243770; https://doi.org/10.1021/ac0617013
- Zhong Y, Hyung S, Ruotolo BT. Characterizing the resolution and accuracy of a second-generation traveling-wave ion mobility separator for biomolecular ions. Analyst 2011; 136(17):3534-41; PMID:21445388; https://doi.org/10.1039/c0an00987c
- Giles K, Williams JP, Campuzano I. Enhancements in travelling wave ion mobility resolution. Rapid Commun Mass Spectrom 2011; 25(11):1559-66; PMID:21594930; https://doi.org/10.1002/rcm.5013
- Tito MA, Tars K, Valegard K, Hajdu J, Robinson CV. Electrospray time-of-flight mass spectrometry of the intact MS2 virus capsid. J Am Chem Soc 2000; 122(14):3550-51; https://doi.org/10.1021/ja993740k
- McKay AR, Ruotolo BT, Ilag LL, Robinson CV. Mass measurements of increased accuracy resolve heterogeneous populations of intact ribosomes. J Am Chem Soc 2006; 128(35):11433-42; PMID:16939266; https://doi.org/10.1021/ja061468q
- Okbazghi SZ, More AS, White DR, Duan S, Shah IS, Joshi SB, Middaugh CR, Volkin DB, Tolbert TJ. Production, characterization, and biological evaluation of well-defined IgG1 fc glycoforms as a model system for biosimilarity analysis. J Pharm Sci 2016; 105(2):559-74; PMID:26869419; https://doi.org/10.1016/j.xphs.2015.11.003
- Alsenaidy MA, Okbazghi SZ, Kim JH, Joshi SB, Middaugh CR, Tolbert TJ, Volkin DB. Physical stability comparisons of IgG1-Fc variants: Effects of N-Glycosylation site occupancy and asp/gln residues at site asn 297. J Pharm Sci 2014; 103(6):1613-27; PMID:24740840; https://doi.org/10.1002/jps.23975
