ABSTRACT
Efforts to develop effective antibody therapeutics are frequently hampered by issues such as aggregation and nonspecificity, often only detected in late stages of the development process. In this study, we used a high throughput cross-reactivity assay to select nonspecific clones from a naïve human repertoire scFv library displayed on the surface of yeast. Most antibody families were de-enriched; however, the rarely expressed VH6 family was highly enriched among nonspecific clones, representing almost 90% of isolated clones. Mutational analysis of this family reveals a dominant role of CDRH2 in driving nonspecific binding. Homology modeling of a panel of VH6 antibodies shows a constrained β-sheet structure in CDRH2 that is not present in other families, potentially contributing to nonspecificity of the family. These findings confirm the common decision to exclude VH6 from synthetic antibody libraries, and support VH6 polyreactivity as a possible important role for the family in early ontogeny and cause for its overabundance in cases of some forms of autoimmunity.
Introduction
Although intended to bind to only a specific antigen, some monoclonal antibodies (mAbs) can bind nonspecifically to other proteins and surfaces, albeit with a much lower affinity. This nonspecificity can lead to off-target effects or accelerated clearance rates due to nonspecific tissue binding.Citation1,Citation2 While not desired in affinity-matured antibodies, polyspecificity may exist in the natural antibody repertoire as a means for broad antigen recognition.Citation3 Studies on this pool of polyreactive, or “natural,” antibodies reveal often elongated CDR H3 regions, which are hypothesized to allow for greater flexibility in antigen recognition.Citation4,Citation5 Long, flexible CDRs are also found in many broadly neutralizing antibodies that target viruses such as influenza and the human immunodeficiency virus, and it is believed that many of these antibodies target specific proteins on the virus surface and engage in nonspecific binding to the virus coat.Citation6-8 Beyond the understanding of polyreactive antibodies in natural repertoires, some studies have implicated the importance of arginine and tryptophan residues, although these were in the context of a synthetic library design.Citation9,Citation10
A greater understanding of nonspecific binding will improve the isolation and development of mAbs using display technologies such as phage and yeast display, which can suffer from poor biophysical characteristics. Multiple naïveCitation11 or semi-syntheticCitation12-14 libraries based on the naïve repertoire are regularly used for the isolation of candidate antibodies, and as such a better understanding of nonspecificity in these libraries could lead to more efficient selections and downstream development.
Traditional methods of assessing specificity include measuring binding of antibody to tissuesCitation15 or a panel of representative non-cognate antigens.Citation10 More recently it has been discovered that complex protein extracts can also serve as nonspecificity reagents. Binding of antibodies to baculovirus particles in an ELISA format correlates remarkably well with a phenotype as complex as clearance rate in humansCitation16 and a high throughput assay using a soluble membrane preparation (SMP) from HEK cells on the surface of yeast also serves as an effective predictor of nonspecificity in displayed antibodies and clearance rates in mice.Citation17,Citation18
In this study, we utilize a yeast surface displayed single-chain variable fragment (scFv) library to assess nonspecificity in the natural repertoire. This library, created from amplified VH and VL segments from pooled human spleen and lymph node RNA, samples the human antibody repertoire, with some differences in antibody frequency and the possibility for non-natural VH/VL pairing.Citation11 To isolate nonspecific clones, a modified version of the SMP assay was applied to sorting, revealing a predominance of nonspecific clones from the VH6 subfamily. Further analysis of these clones revealed CDR H2 as the dominant source of the nonspecificity of these clones. All other families were de-enriched in sorted populations, suggesting their preferred applicability to further naïve or synthetic library designs.
Results
Naïve library panning reveals enrichment of the VH6 family
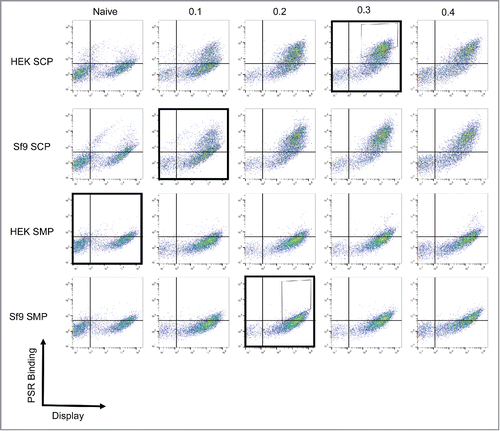
To identify nonspecific clones in the human repertoire, we used a nonimmune human antibody yeast surface display library previously constructed by Feldhaus et al.Citation11 The library was screened against 4 nonspecificity reagents: a preparation of soluble membrane or cytosolic proteins from human embryonic kidney (HEK293) or Spodoptera frugiperda (Sf9) cells. These reagents were chosen to maximize orthogonality between preparations and avoid isolation of specific binders to components in one particular reagent. Populations were positively or negatively selected for binding against the 4 reagents sequentially to maximize round-to-round differences in selection reagent (, Fig. S1). After 4 rounds of selection, the resulting populations were validated by assessing binding to all 4 reagents. This selection process was able to isolate a positive, nonspecific population that displayed robust binding to all reagents and a negative, clean population that bound to none of the reagents (Positive; , Negative; Fig. S1). Sorts were repeated in duplicate to ensure consistency of sorted families.
Figure 1. Isolation of nonspecific antibodies. A nonimmune scFv library was subjected to 4 rounds of yeast display-based selections. To ensure nonspecificity, the antigen used in each round was changed, with the following order used: HEK SMP, Sf9 SCP, Sf9 SMP, HEK SCP (Outlined boxes). The first 2 rounds were completed using magnetic-activated cell sorting and the final 2 rounds were completed using fluorescence-activated cell-sorting with the indicated sort gates (Thin boxes). The populations were validated after each round by measuring binding to each of the 4 reagents. As expected, the positively selected panel displays binding to all reagents.
We sequenced the resulting populations and analyzed the distribution of the various germline families (). Examination of negatively sorted sequences shows there was an enrichment in sequences from the VH1 family, with some depletion of sequences from the VH4 and VH6 families. More strikingly, an average of 92% of the positively sorted antibodies originated from the VH6 family, with depletions in all other families. This rare germline family, represented by a single V-gene, represents approximately 1% of the expressed germline antibody pool in humans.Citation19 It is worth noting that this family was already overrepresented in the original library, present in 40% of sequences, which likely contributed to the overall dominance of this family in the sequencing output.
Figure 2. Antibody germline family distribution. Variable heavy (A) and light (B) chain sequences were aligned against human germline genes using IMGT V-QUEST and grouped into major families. Abundances of each family are displayed for the natural human repertoire (Blue), naive library (Red), negatively sorted pool (Green), and positively sorted population (Purple). Bars represent the average percentage of clones among 2 sorts and error bars indicate standard error.
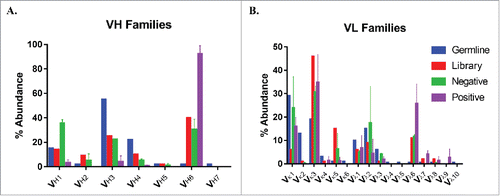
The repertoire of light chain segments was enriched for Vκ1 segments and depleted for Vκ3 segments in both positive and negative cases, while there was a considerable enrichment of sequences from the Vλ6 subfamily among positively sorted sequences. The enrichment in Vκ1 and Vκ3 could be attributed to the fact that the initial library sequencing was completed on the naïve, unselected population and we have selected all clones for successful display, which likely subtly increases their abundance as these segments both tend to be well behaved. With respect to the Vλ6 subfamily, this germline family was overrepresented in the original library (0.5% vs. 11%), but was enriched 2.3-fold among nonspecific sequences.
Nonspecific binding is driven by CDR H2
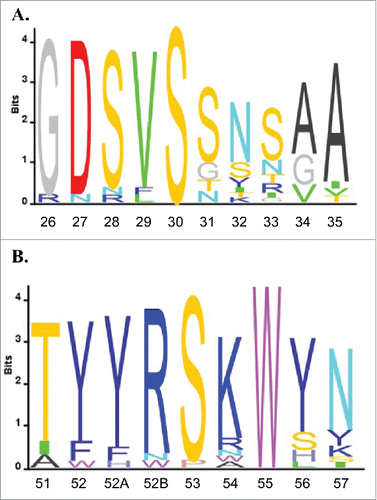
We next investigated what sequences drive nonspecificity within the VH6 germline family, focusing first on the conserved V gene segment, which contains the sequences of both CDR H1 and H2. Aligning all positively selected VH6 sequences we see a high level of homology in both the H1 and H2 sequences (). It has been suggested that positive charge and hydrophobic regions can lead to nonspecific behavior.Citation9,Citation10 This led us further investigate the H2 sequence, which contains both two positively charged residues and one tryptophan residue.
Figure 3. VH6 family sequence alignment. All VH6 antibody sequences were aligned using IMGT V-QUEST and sequence logos were plotted using Matlab for CDR H1 (A) and CDR H2 (B). Residues are numbered using Kabat standard numbering and CDRs are defined using IMGT definitions.Citation41
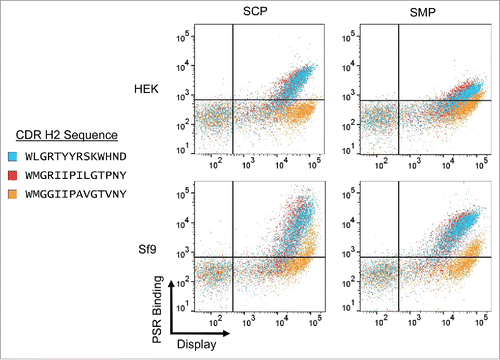
To isolate any effects driven by the H2, we spliced the CDR H2 sequence from two “clean” VH1 antibodies into an example antibody derived from the VH6 family and measured poly-specificity reagent (PSR) binding for all resulting clones. Interestingly, while one of the mutants (VH6.H2.1) displayed similar binding to the original sequence, the other mutant (VH6.H2.2) completely ablated binding to all four nonspecificity reagents (). This suggests that the H2 is indeed driving nonspecificity in the family, but not every “clean” H2 sequence is able to reverse this phenotype. Comparing the sequences of the two VH1 segments used, there are four amino acids that are different, with one of these, Arg50, matching the VH6 sequence. We next mutated these four residues individually in the VH6.H2.1 and measured PSR binding. No individual mutation was able to fully ablate nonspecific binding, but most of the mutations had some marginal effect, with the R50G mutation displaying the greatest drop in PSR binding (Supp. Figure 1). Combined, this suggests a dominant role of Arg50 in driving nonspecificity, with appreciable contributions from other residues in the H2 loop.
Homology modeling of the VH6 CDR H2 segment
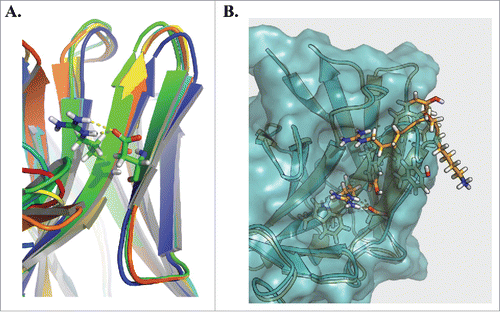
To better understand structurally the importance of Arg50 in the structure of the CDR loop, we used the Rosetta Antibody serverCitation20-22 to create homology models of three representative clones from the VH6 family. All threeclones displayed similar structures in the region surrounding and containing CDR H2, and Arg50 lies at the base of the H2 region (). Interestingly, the exposed surface area of this site is low, and its main effect appears to be in giving added structure to the loop, creating a hydrogen bond with Asp58, which stabilizes a β sheet not widely present in other families of antibodies. This rigid structure allows the tip of the loop to protrude further than other H2 loops, allowing for greater exposure of the other residues in the loop (). While Asp58 is not present in VH6.H2.1, Arg50 is similarly able to form a hydrogen bond with Asn58 that also appears to stabilize the H2 loop. This may explain why VH6.H2.1 retains its nonspecificity, whereas VH6.H2.1, which is unable to make this contact, does not.
Figure 5. Homology modeling of the VH6 CDRH2. Rosetta Antibody was used to homology model 3 representative clones from the VH6 family (A). Alignment of the structures highlights a common constrained β sheet structure stabilized by hydrogen bond interactions between Arg50 and Asp58. Comparing this structure against that of clone VH6.H2.2 (B), we observe the CDRH2 of the VH6 family clone (orange) protrudes farther than that of the mutant (blue).
Discussion
The initial intention of this study was to unveil motifs present in the natural repertoire that may lead to nonspecificity. The sort output was unexpectedly dominated by clones from the usually rare VH6 family, making it difficult to analyze the enrichment of other motifs that may also drive nonspecificity. These clones have conserved CDR H1 and H2 regions, with little to no similarity in the H3 region. Mutation of the H2 region revealed that it was the main driver of nonspecificity in the family. This loop is more structured than other naturally encoded H2 loops, driven by the interaction of Arg50 and Asp58. This added structure causes the H2 loop to protrude farther than in other families, exposing multiple residues.
Of the residues exposed in the loop, in general tyrosine and serine have not been shown to increase nonspecific binding.Citation9,Citation10 However, the positively charged arginine and lysine have been implicated as leading to increased nonspecificity.Citation2,Citation9,Citation10,Citation23-25 While in isolation positively charged residues can be important in establishing charge-charge contacts in inter- and intra-molecular interactions, the small positively charged patch formed by the arginine and lysine residue may contribute to the nonspecific binding profile we see in this work.
In establishment of the human repertoire, the VH6 family is present in early development and in immature B lymphoid cells, but rare to absent in the developed human repertoire.Citation26-28 Interestingly, the VH6 family has been shown to be overrepresented in patients with some autoimmune diseases,Citation29-33 and the broad binding spectrum of autoantibodies from this family has been shown in prior work.Citation26,Citation34 Despite the polyspecificity of the family as a whole, highly specific antibodies have been discovered from this family against influenzaCitation35 and bacterial pathogens.Citation36 The VH6–1 gene segment is also the most JH proximal, which is preferentially used in ontogeny.Citation27 Taken as a whole, it appears that nonspecificity, or polyreactivity, of the VH6 family may be important for early immunologic protection, which is then downregulated as the full repertoire develops. This may explain why a V gene encoding nonspecific antibodies is not completely deleted as a result of evolution, and can help explain why it may be overrepresented in patients with autoimmune disorders.
Applying these lessons to creating display libraries for the isolation of new therapeutic antibodies, these results caution against the use of the VH6–1 gene segment in naïve human repertoire designs. While it can be possible to derive highly specific antibodies in certain cases, there is still considerable risk that the majority of clones will display nonspecificity and poor development characteristics, including rapid clearance, which are correlated with nonspecificity.Citation2,Citation16,Citation18 The VH6 class was accidentally overrepresented in the library used in this work, either by PCR amplification bias during construction or an abnormal antibody distribution in the donors used, which is likely detrimental for antigen selections. Outside of the VH6 family, all other families were less prevalent in nonspecific clones, with a noticeable enrichment of the VH1 family in negatively selected clones. These families, more prevalent in the developed natural repertoire, appear to be better starting points for both natural repertoire library construction and for use as scaffolds in synthetic library designs.
Materials and methods
Antigen preparation
SMPs were prepared as described previously.Citation17 Briefly, one billion HEK or Sf9 cells were pelleted, washed and homogenized. The homogenate was separated into membrane and cytosolic fractions via centrifugation at 40,000 × g for 1h at 4° C. The supernatant was collected as the soluble cytosolic preparation (SCP) and the pellet was further washed and solubilized to create the enriched membrane fraction (EMF). Both the SCP and EMF were diluted to a concentration of 1 mg/mL and biotinylated using NHS-LC-Biotin (Pierce, Thermo Fisher Cat# 21336). Biotin solution was prepared according to manufacturer's protocol and 20 µL 10 mM biotin reagent was added per 1 mg sample. The reaction mixture was incubated for 3 h at 4 °C with gentle agitation, after which the biotinylated EMF (b-EMF) was pelleted at 40,000 × g for 1 h at 4 °C and the biotinylated SCP (b-SCP) was buffer exchanged into phosphate-buffered saline, pH 7.4 (PBS; Corning) using a Zeba Spin Desalting column (Thermo, 7K MWCO). The b-EMF was subsequently solubilized overnight using a buffer containing 1% n-dodecyl-b-D-maltopyranoside (DDM) and then centrifuged at 40,000 × g for 1h at 4 °C. The resulting supernatant was collected as the final, biotinylated SMP (b-SMP) product.
Yeast display experiments
All experiments were conducted using the naïve human scFv library described by Feldhaus et al.Citation11 using described previously yeast display techniques.Citation37,Citation38 Initial rounds of sorting were conducted using Dynabeads biotin binder beads (Life Technologies), ensuring at least ten-fold coverage of the library or population size in each round. After tworounds, subsequent sorting was completed on a FACSAria III cell sorter (BD Biosciences). Due to concern over selection of scFvs specific to a single protein, the order of antigens was chosen to maximize round-to-round differences. In order, the population was positively screened against: HEK bSMP, Sf9 bSCP, Sf9 bSMP, and HEK bSCP. To ensure no enrichment of binders to biotin, 1 mM biotin (Sigma) was added to sort rounds three and four. In all instances, nonspecificity was evaluated using a 1:10 dilution of each biotinylated nonspecificity reagent as primary and streptavidin AlexaFluor 488 conjugate (1:100, Life Technologies) as secondary. Simultaneously, successful display was evaluated using chicken anti-c-Myc primary (1:250, Gallus Immunotech cat# ACMYC) followed by goat anti-chicken secondary (1:100, Life Technologies cat # A-21449) antibodies.
Clone analysis
Yeast DNA was isolated using a Zymoprep Yeast Plasmid Miniprep II kit (Zymo Research). For most analysis, sequencing of individual clones was completed by Sanger sequencing (Macrogen). For high throughput sequencing efforts, insert DNA was first amplified for Illumina sequencing using the pCTFwdSeq and pCTRevSeq primers (See Supplementary Table 1 for sequences). Illumina libraries were prepared using NexteraXT (Illumina, Inc.) and sequenced on an Illumina Miseq using a 300nt kit (v2). IMGT/V-QUEST was used for all sequence alignment and assignment of closest germline V-genes.Citation39 Both input and output libraries were sequenced, and clones with greater than two-fold enrichment over input and p-value less than 0.01 (Student's t-test, n = 4 replicates) we deemed to be significantly enriched. Motif enrichment and sequence analysis was conducted using Matlab.
Mutant construction
All H2 mutants were constructed via homologous recombination of three DNA fragments in yeast. Designed overlap sequences were located at the two ends of the scFv construct as well as directly in the H2 region. The fragment before and containing the H2 was amplified with the pCTFwd Recomb and appropriate PreH2 primers, and the fragment after and also containing the H2 was amplified using the pcTRev Recomb and proper PostH2 primers (Primer sequences in Supplementary Table 1). The pCTCON2 vector was prepared for homologous recombination by digestion with SalI-HF followed by digestion with NheI-HF and BamI-HF (New England Biolabs). The two scFv fragments along with the digested vector were contransformed into chemically competent yeast using the Frozen-EZ Yeast Transformation II Kit (Zymo Research) and resultant clones were sequence verified before experiments.
Modeling
Antibody homology modeling was conducted using Rosetta Antibody through ROSIE using default settings.Citation20-22,Citation40 For all clones, the grafted, relaxed model structure was chosen for further analysis. Structure analysis, alignment and figure generation were conducted using PyMol.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Supplemental_Materials.zip
Download Zip (1 MB)Acknowledgments
R.L.K. was supported by a graduate fellowship from the National Institute of General Medical Sciences Interdepartmental Biotechnology Training Program at the National Institutes of Health [T32 GM008334-25] and the Siebel Scholarship.
References
- Wu H, Pfarr DS, Tang Y, An LL, Patel NK, Watkins JD, Huse WD, Kiener PA, Young JF. Ultra-potent antibodies against respiratory syncytial virus: Effects of binding kinetics and binding valence on viral neutralization. J Mol Biol. 2005;350:126-44. https://doi.org/10.1016/j.jmb.2005.04.049. PMID:15907931.
- Datta-Mannan A, Lu J, Witcher DR, Leung D, Tang Y, Wroblewski VJ. The interplay of non-specific binding, target-mediated clearance and FcRn interactions on the pharmacokinetics of humanized antibodies. MAbs. 2015;7:1084-93. https://doi.org/10.1080/19420862.2015.1075109. PMID:26337808.
- Tiller T, Tsuiji M, Yurasov S, Velinzon K, Nussenzweig MC, Wardemann H. Autoreactivity in human IgG+ memory B cells. Immunity. 2007;26:205-13. https://doi.org/10.1016/j.immuni.2007.01.009. PMID:17306569.
- Ichiyoshi Y, Casali P. Analysis of the structural correlates for antibody polyreactivity by multiple reassortments of chimeric human immunoglobulin heavy and light chain V segments. J Exp Med. 1994;180:885-95. https://doi.org/10.1084/jem.180.3.885. PMID:8064239.
- Aguilera I, Melero J, Nuñez-Roldan A, Sanchez B. Molecular structure of eight human autoreactive monoclonal antibodies. Immunology. 2001;102:273-80. https://doi.org/10.1046/j.1365-2567.2001.01159.x. PMID:11298825.
- West AP, Scharf L, Scheid JF, Klein F, Bjorkman PJ, Nussenzweig MC. Structural Insights on the role of antibodies in HIV-1 vaccine and therapy. Cell. 2014;156:633-48. https://doi.org/10.1016/j.cell.2014.01.052. PMID:24529371.
- Wardemann H, Yurasov S, Schaefer A, Young JW, Meffre E, Nussenzweig MC. Predominant autoantibody production by early human B cell precursors. Science. 2003;301:1374-7. https://doi.org/10.1126/science.1086907. PMID:12920303.
- Mouquet H, Nussenzweig MC. Polyreactive antibodies in adaptive immune responses to viruses. Cell Mol Life Sci. 2012;69:1435-45. https://doi.org/10.1007/s00018-011-0872-6. PMID:22045557.
- Birtalan S, Fisher RD, Sidhu SS. The functional capacity of the natural amino acids for molecular recognition. Mol Biosyst. 2010;6:1186-94. https://doi.org/10.1039/b927393j. PMID:20383388.
- Birtalan S, Zhang Y, Fellouse FA, Shao L, Schaefer G, Sidhu SS. The intrinsic contributions of tyrosine, serine, glycine and arginine to the affinity and specificity of antibodies. J Mol Biol. 2008;377:1518-28. https://doi.org/10.1016/j.jmb.2008.01.093. PMID:18336836.
- Feldhaus MJ, Siegel RW, Opresko LK, Coleman JR, Feldhaus JMW, Yeung YA, Cochran JR, Heinzelman P, Colby D, Swers J, et al. Flow-cytometric isolation of human antibodies from a nonimmune Saccharomyces cerevisiae surface display library. Nat Biotechnol. 2003;21:163-70. https://doi.org/10.1038/nbt785. PMID:12536217.
- Prassler J, Thiel S, Pracht C, Polzer A, Peters S, Bauer M, Nörenberg S, Stark Y, Kölln J, Popp A, et al. HuCAL PLATINUM, a synthetic Fab library optimized for sequence diversity and superior performance in mammalian expression systems. J Mol Biol. 2011;413:261-78. https://doi.org/10.1016/j.jmb.2011.08.012. PMID:21856311.
- Tiller T, Schuster I, Deppe D, Siegers K, Strohner R, Herrmann T, Berenguer M, Poujol D, Stehle J, Stark Y, et al. A fully synthetic human Fab antibody library based on fixed VH/VL framework pairings with favorable biophysical properties. MAbs. 2013;5:1-26. https://doi.org/10.4161/mabs.24218. PMID:23254906.
- Knappik A, Ge L, Honegger A, Pack P, Fischer M, Wellnhofer G, Hoess A, Wölle J, Plückthun A, Virnekäs B. Fully synthetic human combinatorial antibody libraries (HuCAL) based on modular consensus frameworks and CDRs randomized with trinucleotides. J Mol Biol. 2000;296:57-86. https://doi.org/10.1006/jmbi.1999.3444. PMID:10656818.
- Wu H, Pfarr D, Johnson S, Brewah Y, Woods R, Patel N, White W, Young J, Kiener P. Development of motavizumab, an ultra-potent antibody for the prevention of respiratory syncytial virus infection in the upper and lower respiratory tract. J Mol Biol. 2007;368:652-65. https://doi.org/10.1016/j.jmb.2007.02.024. PMID:17362988.
- Hotzel I, Theil FP, Bernstein LJ, Prabhu S, Deng R, Quintana L, Lutman J, Sibia R, Chan P, Bumbaca D, et al. A strategy for risk mitigation of antibodies with fast clearance. MAbs. 2012;4:753-60. https://doi.org/10.4161/mabs.22189. PMID:23778268.
- Xu Y, Roach W, Sun T, Jain T, Prinz B, Yu TY, Torrey J, Thomas J, Bobrowicz P, Vásquez M, et al. Addressing polyspecificity of antibodies selected from an in vitro yeast presentation system: A FACS-based, high-throughput selection and analytical tool. Protein Eng Des Sel. 2013;26:663-70. https://doi.org/10.1093/protein/gzt047. PMID:24046438.
- Kelly RL, Sun T, Jain T, Caffry I, Yu Y, Cao Y, Lynaugh H, Brown M, Vásquez M, Wittrup KD, et al. High throughput cross-interaction measures for human IgG1 antibodies correlate with clearance rates in mice. MAbs. 2015;7:770-7. https://doi.org/10.1080/19420862.2015.1043503. PMID:26047159.
- Brezinschek H, Foster S. Analysis of the human VH gene repertoire. Differential effects of selection and somatic hypermutation on human peripheral CD5 (+)/IgM+ and CD5 (−)/IgM+ B cells. J Clin … 1997;99:2488-501. https://doi.org/10.1172/JCI119433. PMID:9153293.
- Sivasubramanian A, Sircar A, Chaudhury S, Gray JJ. Toward high-resolution homology modeling of antibody Fv regions and application to antibody-antigen docking. Proteins. 2009;74:497-514. https://doi.org/10.1002/prot.22309. PMID:19062174.
- Lyskov S, Chou FC, Conchúir SÓ, Der BS, Drew K, Kuroda D, Xu J, Weitzner BD, Renfrew PD, Sripakdeevong P, et al. Serverification of molecular modeling applications: The Rosetta Online Server That Includes Everyone (ROSIE). PLoS One. 2013;8:5-7. https://doi.org/10.1371/journal.pone.0063906. PMID:23717507.
- Marze NA, Lyskov S, Gray JJ. Improved prediction of antibody VL-VH orientation. Protein Eng Des Sel. 2016;29:409-18. https://doi.org/10.1093/protein/gzw013. PMID:27276984.
- Datta-Mannan A, Thangaraju A, Leung D, Tang Y, Witcher DR, Lu J, Wroblewski VJ, Derrick R, Lu J, Wroblewski VJ. Balancing charge in the complementarity-determining regions of humanized mAbs without affecting pI reduces non-specific binding and improves the pharmacokinetics. MAbs. 2015;7:483-93. https://doi.org/10.1080/19420862.2015.1016696. PMID:25695748.
- Boswell CA, Tesar DB, Mukhyala K, Theil FP, Fielder PJ, Khawli L a. Effects of charge on antibody tissue distribution and pharmacokinetics. Bioconjug Chem. 2010;21:2153-63. https://doi.org/10.1021/bc100261d. PMID:21053952.
- Hong G, Chappey O, Niel E, Scherrmann JM. Enhanced cellular uptake and transport of polyclonal immunoglobulin G and Fab after their cationization. J Drug Target. 2000;8:67-77. https://doi.org/10.3109/10611860008996853. PMID:10852339.
- Van Es JH, Raaphorst FM, van Tol MJ, Meyling FH, Logtenberg T. Expression pattern of the most JH-proximal human VH gene segment (VH6) in the B cell and antibody repertoire suggests a role of VH6-encoded IgM antibodies in early ontogeny. J Immunol. 1993;150:161-8. PMID:8417121.
- Cuisinier AM, Guigou V, Boubli L, Fougereau M, Tonnelle C. Preferential expression of VH5 and VH6 immunoglobulin genes in early human B-cell ontogeny. Scand J Immunol. 1989;30:493-7. https://doi.org/10.1111/j.1365-3083.1989.tb02455.x. PMID:2814367.
- Berman JE, Nickerson KG, Pollock RR, Barth JE, Schuurman RK, Knowles DM, Chess L, Alt FW. VH gene usage in humans: Biased usage of the VH6 gene in immature B lymphoid cells. Eur J Immunol. 1991;21:1311-4. https://doi.org/10.1002/eji.1830210532. PMID:1903708.
- Tiumentseva M, Morozova V, Zakabunin A, Korobko D, Malkova N, Filipenko M, Tikunova N. Use of the VH6-1 gene segment to code for anti-interleukin-18 autoantibodies in multiple sclerosis. Immunogenetics. 2016;68:237-46. https://doi.org/10.1007/s00251-015-0895-5. PMID:26743536.
- Tyumentseva MA, Morozova VV, Lebedev LR, Babkin IV, Tikunova NV. Presence of aberrant VH6 domains in anti-interferon-γ autoantibodies in multiple sclerosis. Hum Antibodies. 2013;22:31-49. doi:10.3233/HAB-130269. PMID:24284307.
- Logtenberg T, Young FM, van Es J, Gmelig-Meyling FH, Berman JE, Alt FW. Frequency of VH-gene utilization in human EBV-transformed B-cell lines: The most JH-proximal VH segment encodes autoantibodies. J Autoimmun. 1989;2 Suppl:203-13. https://doi.org/10.1016/0896-8411(89)90132-7. PMID:2550019.
- van Dijk-Hard I, Feld S, Holmberg D, Lundkvist I. Increased utilization of the VH6 gene family in patients with autoimmune idiopathic thrombocytopenic purpura. J Autoimmun. 1999;12:57-63. https://doi.org/10.1006/jaut.1998.0257. PMID:10028023.
- Chukwuocha RU, Hsiao ET, Shaw P, Witztum JL, Chen PP. Isolation, characterization and sequence analysis of five IgG monoclonal anti-beta 2 glycoprotein-1 and anti-prothrombin antigen-binding fragments generated by phage display. J Immunol. 1999;163:4604-11. PMID:10510405.
- Logtenberg T, Young FM, Van Es JH, Gmelig-Meyling FH, Alt FW. Autoantibodies encoded by the most Jh-proximal human immunoglobulin heavy chain variable region gene. J Exp Med. 1989;170:1347-55. https://doi.org/10.1084/jem.170.4.1347. PMID:2507728.
- Kallewaard NL, Corti D, Collins PJ, Neu U, McAuliffe JM, Benjamin E, Wachter-Rosati L, Palmer-Hill FJ, Yuan AQ, Walker PA, et al. Structure and function analysis of an antibody recognizing all influenza a subtypes. Cell. 2016;166:596-608. https://doi.org/10.1016/j.cell.2016.05.073. PMID:27453466.
- Andris JS, Brodeur BR, Capra JD. Molecular characterization of human antibodies to bacterial antigens: Utilization of the less frequently expressed VH2 and VH6 heavy chain variable region gene families. Mol Immunol. 1993;30:1601-16. https://doi.org/10.1016/0161-5890(93)90452-H. PMID:8247031.
- VanDeventer JA, Wittrup KD. Yeast surface display for antibody isolation: Library construction, library screening, and affinity maturation [Internet]. In: Ossipow V, Fischer N, editors. Monoclonal Antibodies. Totowa, NJ: Humana Press. 2014. p. 151-81.
- Chao G, Lau WL, Hackel BJ, Sazinsky SL, Lippow SM, Wittrup KD. Isolating and engineering human antibodies using yeast surface display. Nat Protoc. 2006;1:755-68. https://doi.org/10.1038/nprot.2006.94. PMID:17406305.
- Brochet X, Lefranc MP, Giudicelli V. IMGT/V-QUEST: The highly customized and integrated system for IG and TR standardized V-J and V-D-J sequence analysis. Nucleic Acids Res. 2008;36:503-8. https://doi.org/10.1093/nar/gkn316. PMID:18503082.
- Sircar A, Kim ET, Gray JJ. RosettaAntibody: Antibody variable region homology modeling server. Nucleic Acids Res. 2009;37:W474-9. https://doi.org/10.1093/nar/gkp387. PMID:19458157.
- Lefranc MP, Pommié C, Kaas Q, Duprat E, Bosc N, Guiraudou D, Jean C, Ruiz M, Da Piédade I, Rouard M, et al. IMGT unique numbering for immunoglobulin and T cell receptor constant domains and Ig superfamily C-like domains. Dev Comp Immunol. 2005;29:185-203. https://doi.org/10.1016/j.dci.2004.07.003. PMID:15572068.