ABSTRACT
Transgenic animal platforms for the discovery of human monoclonal antibodies have been developed in mice, rats, rabbits and cows. The immune response to human proteins is limited in these animals by their tolerance to mammalian-conserved epitopes. To expand the range of epitopes that are accessible, we have chosen an animal host that is less phylogenetically related to humans. Specifically, we generated transgenic chickens expressing antibodies from immunoglobulin heavy and light chain loci containing human variable regions and chicken constant regions. From these birds, paired human light and heavy chain variable regions are recovered and cloned as fully human recombinant antibodies. The human antibody-expressing chickens exhibit normal B cell development and raise immune responses to conserved human proteins that are not immunogenic in mice. Fully human monoclonal antibodies can be recovered with sub-nanomolar affinities. Binning data of antibodies to a human protein show epitope coverage similar to wild type chickens, which we previously showed is broader than that produced from rodent immunizations.
Abbreviations
| BDNF | = | brain-derived neurotrophic factor |
| CDR | = | complementarity-determining region |
| BSA | = | bovine serum albumin |
| ELISA | = | enzyme-linked immunosorbent assay |
| FDA | = | Food and Drug Administration |
| GEM | = | Gel Encapsulated Microenvironment |
| HRP | = | horseradish peroxidase |
| IACUC | = | Institutional Animal Care and Use Committee |
| Ig | = | immunoglobulin |
| IgH KO | = | IgH knockout |
| mAb | = | monoclonal antibody |
| PBMC | = | peripheral blood mononuclear cells |
| PBS | = | phosphate-buffered saline |
| PGRN | = | progranulin |
| scFv-Fc | = | single chain variable fragment with fragment crystallizable |
| WT | = | wild type |
Introduction
Monoclonal antibody (mAb) therapy is a crucial segment of the pharmaceutical arsenal, with 57 FDA-approved mAbs, an estimated nine mAbs predicted to be granted their first marketing approvals in 2017,Citation1 and hundreds more in development.Citation2 Therapeutic mAbs should be as similar to native human antibodies as possible to minimize their immunogenicity in patients. Historically, human therapeutic mAbs in the clinical pipeline have been produced from three main technologies: 1) in vitro complementarity-determining region (CDR) grafting of murine antibodies onto human frameworks, 2) in vitro systems such as phage display libraries, and 3) in vivo immune systems of “humanized” mice genetically engineered to express a human immunoglobulin repertoire. To date, the majority of approved human mAbs have been derived from the mouse (wild type (WT) or transgenic) rather than in vitro systems.Citation3,4 Antibodies produced in the intact immune system of an animal have gone through rigorous selection for specific binding to the target, counter-selection to a vast array of endogenous off-target proteins, and high-level expression in plasma cells. The in vitro-derived antibodies do not have the advantage of immune system selection, and therefore require further validation and sequence optimization after initial discovery.
The strategies for expression of human antibodies in transgenic animal platforms have evolved over time,Citation3 and the range of species that have been genetically modified has expanded to include mice, rats, rabbits, and cows.Citation5 As all these transgenic animals are mammals, the accessible epitope space on human protein targets is limited. Immune responses are primarily focused on regions that are evolutionarily divergent between the species, and it is difficult to raise antibodies against epitopes that are shared amongst mammals, i.e., “pan-mammalian” epitopes, in a mammalian host. Nonetheless, species cross-reactive antibodies are highly desirable for drug development because such antibodies facilitate the use of animal models of disease. When a lead therapeutic antibody is not species cross-reactive, it often necessitates a parallel campaign for a “surrogate” antibody.
As an alternative to mammalian species, birds (and in particular, chickens) present an attractive choice because they are phylogenetically distant from humans (having diverged about 300 million years ago,Citation6 compared to 65 million years for rodentsCitation7), produce antibodies of high affinity and specificity, and can recognize unique epitopes not accessible in mice.Citation8 Expanded epitope coverage is an advantage in drug development because it increases the chances of accessing the functionally significant regions of the target, which would likely be conserved. Antibodies with unique characteristics may be identified that provide novel mechanisms of action. Cross-reactive mAbs that recognize the human and mouse orthologs of a targetCitation9 could seamlessly advance from pre-clinical to clinical-stage studies. These benefits are achieved while retaining the general advantage of an in vivo selection process that removes non-specific and poorly expressing clones.
Chickens have a well-developed humoral immune system that is capable of robust immune responses and the production of high-affinity antibodies.Citation8–13 Chickens express serum immunoglobulins with a classical H2L2 structure.Citation14 In developing B cells, the chicken light and heavy chain loci undergo V(D)J rearrangement, leading to expression of the B cell receptor complex on the cell surface and developmental progression of the B cell.Citation14 However, the chicken loci each contain only a single functional V region and a single J region, and a small number of highly-related Ds in the heavy chain locus, so rearrangement produces limited diversity in the somatic repertoire.Citation15,16 After rearrangement, diversity is generated by multiple overlapping rounds of gene conversion from upstream pseudogenes in the heavy and light chain loci that serve to mutate the functional VH and VL genes.Citation15–17 Chickens thus express a single immunoglobulin structural framework consisting of the germline-encoded VH and VL regions, with somatic diversity accumulating mainly in the CDRs.Citation1,18 Gene conversion rarely copies entire pseudogene sequences into the functional V, but rather small sequence stretches, and individual CDRs may be a product of multiple overlapping gene conversion events, and non-templated point mutations. From a drug development standpoint, this focusing of the mutational machinery on the CDR sequences in the chicken could be highly desirable, as a framework could be selected with superior manufacturing, stability, and pharmacodynamic characteristics. Such a strategy of choosing a limited set of frameworks is common practice in library design.Citation2,19
Here we present the OmniChicken, a transgenic chicken carrying humanized immunoglobulin genes that can be used to discover novel, high affinity antibodies, including antibodies against conserved proteins that are not immunogenic in mice. The OmniChicken displays broad epitope coverage and can generate antibodies that are cross-reactive with homologs in mammalian species, such as mice and cynomolgus monkeys, that are relevant to mechanism-of-action and toxicology pre-clinical studies. The human transgenes have been designed to work in the context of the chicken immune system and to take advantage of the restricted frameworks normally found in chickens. The OmniChicken retains the expanded epitope coverage observed in WT chickens,Citation3,8 but in conjunction with human-sequence antibodies.
Results
The complex genetic modifications necessary to make the OmniChicken were produced in cultured germline cells, which were then used to obtain fully transgenic chickens.Citation3,20,21 The immunoglobulin loci of the chicken were modified in a two-step process: 1) targeting of an attP site into the light and heavy chain loci by homologous recombination, then 2) insertion of human sequences using phiC31 integrase (Supplementary Fig. 1). The attP insertion step simultaneously deleted endogenous Ig sequences, producing gene knockouts. Phenotypic analysis of the knockouts confirmed that the correct loci were targeted and that Ig expression was eliminated.Citation5,22,23 In both light and heavy chain knockouts, the endogenous upstream pseudogenes remained intact. In the second step, single functional human VH and VK genes were inserted site-specifically into the attP sites targeted to the Ig loci and were designed to splice to the endogenous chicken constant regions, to ensure proper formation of the B cell receptor complex and engagement of chicken Fc receptors (Supplementary Fig. 1).
The light chain insertion vector (SynVK) contained a functional V kappa gene (rearranged human VK3-15 + JK4), and the heavy chain insertion vector (SynVH) contained a functional VH gene (rearranged and consisting of VH3-23, D1 and JH4 elements). These particular human V region scaffolds were chosen on the basis of prevalence in the population,Citation24 expression levels, manufacturability, and a history of use in marketed therapeutic antibodies.Citation25 Non-coding sequences present in the constructs were from the chicken, so that appropriate expression of the chimeric antibodies in developing and mature B cells is driven by chicken sequences such as promoters, enhancers, and untranslated regions (Supplementary Fig. 1). Arrays of synthetic, human-based pseudogenes were placed upstream of the human functional V regions to provide a pool of sequences for gene conversion (Supplementary Fig. 2). The pseudogenes were designed with very high CDR diversity and limited framework diversity, achieved by matching the pseudogene frameworks to the appropriate V gene family (i.e., VH3 for heavy chain and VK3 for light chain). The sequences of the CDRs in the pseudogenes were derived from naturally occurring, somatic human sequences found in the Expressed Sequence Tags (EST) database at the National Center for Biotechnology Information (NCBI), except for construct SynVH-A7, in which the CDRs are composed entirely of a minimal set of Ser, Trp and Tyr residues. Selectable markers were removed by breeding to a Cre-expressing line of birds.Citation8,26 The SynV transgenic birds were bred with birds containing the knockouts (also with selectable markers removed), leading to the genotype SynVK/IgL KO; SynVH/IgH KO, which we will refer to as the “OmniChicken” genotype. These birds express chimeric antibodies with human V and chicken C for both heavy and light chains.
Lymphocyte populations in OmniChickens
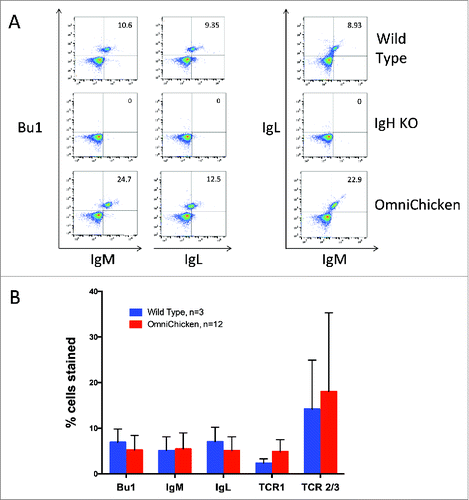
Peripheral blood mononuclear cells (PBMCs) from WT, homozygous heavy-chain knockout (IgH KO), and OmniChickens were labeled with antibodies to chicken IgM and IgL constant regions, the B cell marker Bu-1, and T cell receptors, and analyzed by flow cytometry. In the IgH KO birds, all B lineage cells are missing, and in the OmniChickens, the B cell population was reconstituted to normal levels (). The expression level of surface IgM on circulating cells was normal, and only expressed on B cells (). The percentages of B and T cells in the periphery were normal ().
Figure 1. Flow cytometry analysis shows normal B cell development in the OmniChicken and normal levels of surface IgM. A, Wild-type, IgH knockout (IgH KO), and OmniChicken PBMCs were labeled with antibodies against the B cell marker Bu-1 and either IgL or IgM (left), or with antibodies against IgL and IgM (right). B, Flow cytometry analysis of lymphocytes shows normal levels of B and T cells. Wild-type (N = 3 birds) and OmniChicken (N = 12 birds) PBMCs were labeled with antibodies for Bu-1, IgL, IgM and T cell markers TCR1 and TCR2/3.
Plasma levels of IgM, IgY and IgA
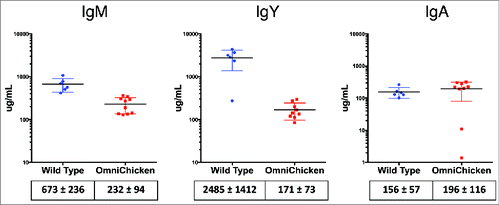
Chickens contain three immunoglobulin isotypes: IgM, IgY and IgA. IgY is functionally analogous to mammalian IgG, although it contains four CH domains and no hinge. As in mammals, IgM is the first expressed isotype in chickens and class switching leads to expression of IgY or IgA. IgA is mainly found in bile and other secretions, but is also found in plasma. Plasma levels of all three isotypes were analyzed by ELISA (). Whereas plasma levels of IgA were normal, levels of plasma IgM and IgY were found to be reduced (3-fold and 15-fold, respectively). The lower relative levels of IgM and IgY were consistent from 4 weeks to 28 weeks (adulthood). In birds expressing a human V-lambda light chain (instead of the V-kappa in the OmniChicken), IgY levels were normal (Supplementary Fig. 3), suggesting that the V-kappa light chain is somehow limiting antibody plasma levels in the OmniChicken.
Figure 2. Plasma levels of IgM and IgY are reduced in OmniChickens but IgA levels are normal. Immunoglobulins were measured by ELISA in wild-type (N = 7) and OmniChickens (N = 9). Ig concentrations were determined by comparison to standards. Mean values (in μg/mL) and standard deviations are shown below the plots.
Immunization and cloning of human mAbs
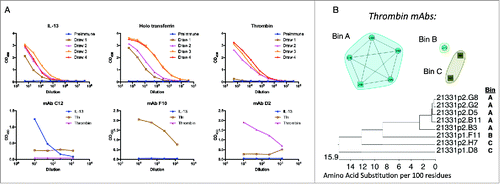
OmniChickens were immunized with a variety of proteins to investigate whether the transgenic birds retained the advantages of the chicken host system, while producing fully human antibodies to a wide range of epitopes on both conserved and non-conserved targets. As a first test of immune competence, OmniChickens were immunized with a cocktail of antigens (bovine thrombin, human transferrin, and murine IL-13). Antigen-specific titer was observed in plasma from birds for all of the antigens (), which increased after subsequent boosts. After reaching an appropriate titer, spleens were obtained and lymphocytes prepared for mAb recovery.
Figure 3. Individual OmniChickens raise specific mAbs to multiple targets. Birds were immunized with a cocktail of mouse IL-13, human transferrin and bovine thrombin, human transferrin, and mouse IL-13. A, Plasma titers to each immunogen from a single bird are shown over multiple draws (top set of graphs). Panels of mAbs to each antigen were identified from that bird, and antigen-specific binding of a single representative mAb for each target is shown (bottom set of graphs). B, epitope binning of eight thrombin antibodies shows three distinct bins. C, phylogenetic tree of the 8 thrombin antibodies, with the epitope bin indicated at right.
A robust hybridoma fusion partner is not available in chickens, so we have developed a single B cell screening method called the GEM assay (for Gel Encapsulated Microenvironment) that can be used to identify antigen-specific antibody-secreting cells from any source, including OmniChickens.Citation9,10 Spleen cells are encapsulated in agarose droplets along with polystyrene beads that have been coated with the target antigen. After incubation to allow secretion of IgY antibody and binding to the antigen on the beads, the IgY is detected with a fluorescent secondary antibody. Bead-positive GEMS are viewed and selected at the microscope, and single cell RT-PCR is used to amplify and clone the variable regions. The V region amplicons from each positive GEM are joined together by a linker sequence to generate a single-chain variable fragment (scFv), and then cloned into a vector containing a human Fc. This expression cassette thus produces a fully human recombinant antibody in the bivalent scFv-Fc format.
GEM screens were performed individually for each of the cocktail antigens and target-specific mAbs were recovered from individual birds for each antigen in the cocktail. These data confirm the ability of the OmniChicken to produce antigen-specific mAbs ().
Array-based surface plasmon resonance imaging (Array SPRi) was used to perform epitope binning experiments on 8 of the thrombin-specific antibodies. These 8 clones clustered into 3 distinct non-overlapping bins (). Sequence analysis () showed that the 5 mAbs in bin A are highly related, whereas the 2 mAbs in bin C are more distantly related, indicating that diverse sequences can converge on the same epitope.
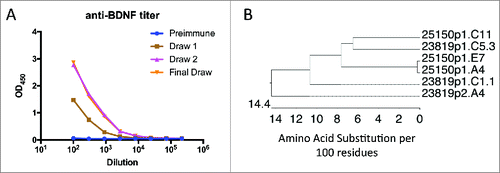
To test more rigorously the ability of OmniChickens to recognize highly-conserved targets, OmniChickens were immunized with human brain-derived neurotrophic factor (BDNF), a protein that is 97% conserved between humans and mice, and 91.5% conserved between humans and chickens. The response to BDNF was variable. Of 9 birds immunized with BDNF protein, 2 had no response, 3 had low titers of under 1:500, but 4 birds had good responses, with titers of 1:4,050 to 1:24,300 (). Spleens were harvested from the immunized birds, and mAbs recovered using the GEM assay. Epitope binning using Array SPRi showed that all six clones mutually blocked one another, and therefore were assigned to the same epitope bin. This is not surprising given the relatively small surface-accessible area of this homodimer (total 28 kDa) that is not buried at the dimer interface.Citation8–13,27 It is striking that the six anti-BDNF mAbs display a high level of sequence diversity, particularly in the heavy chain, despite all converging on the same epitope bin (). This result indicates that it is not possible to state whether antibodies with highly divergent sequences will necessarily bind their target differently; unrelated clones can converge on the same epitope, especially if the available epitope space is restricted.
Figure 4. OmniChickens raise specific mAbs to BDNF, a highly conserved protein. A, Plasma titer to human BDNF in one OmniChicken. B, phylogenetic tree for the 6 mAbs against BDNF.
To further characterize their antibody repertoire, OmniChickens were immunized with human progranulin (PGRN), a multi-domain protein we have used previously to interrogate epitope space covered by WT chickens as compared to WT mice.Citation8,14 Birds were either immunized with human PGRN protein or DNA encoding human PGRN. OmniChickens produced robust titers to PGRN protein (up to 1:36,450), similar to those obtained in WT birds.Citation8,14 A panel of mAbs was produced from both protein and DNA-immunized OmniChickens by screening in GEMs using human PGRN-coated beads.
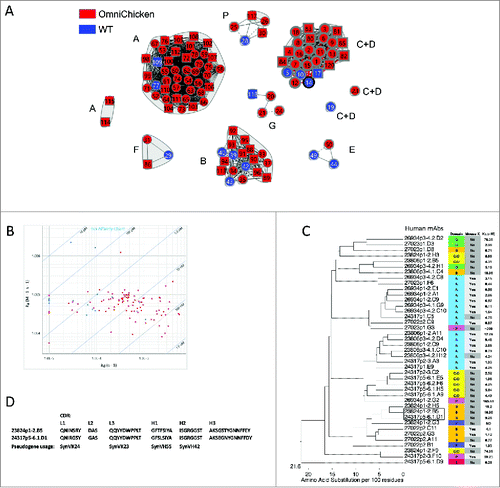
Array SPRi was used to perform epitope binning on a panel of >100 PGRN mAbs from OmniChickens (). All 7 of the granulin domains were recognized by members of the panel, and some members of the panel recognized human/mouse cross-reactive epitopes, thereby mirroring the epitope diversity we reported previously for their WT counterpartsCitation8,15,16 ( and Supplementary Fig. 6). Kinetic analysis showed a range of binding affinities from 0.11 to ∼200 nM (mean value, 25.6 nM), including some in the sub-nanomolar category (), although the highest affinities were found in mAbs from WT birds (the highest being 10 pM). Sequence analysis showed that diverse sequences can converge on the same epitope bin ().
Figure 5. OmniChickens recognize a wide range of epitopes on PGRN, similar to WT birds. A, Network plot for a merged analysis of 90 OmniChicken clones (red) and 16 WT chicken mAb standards (blue), with subdomain assignments indicated. The mAbs were re-numbered to fit into the circles. The WT mAbs were selected fromCitation8 reference standards. B, iso-affinity chart of kinetics of the mAbs in (A). Human mAbs are in red and WT mAbs are in blue. C, phylogenetic tree of 41 selected human mAbs from the group in (A), with the PGRN subdomain, mouse cross-reactivity, and binding affinity for human PGRN of each antibody indicated at right. A box is drawn around a pair of mAbs from different birds that show convergent sequences. D, the CDR sequences from the mAb pair boxed in (C). Where it was possible to determine pseudogene usage in generating the CDR sequences, the pseudogenes are indicated below.
Sequence analysis
Sequences of the VK and VH regions were determined for all of the antigen-specific antibodies described above, and the diversity across the panels of antibodies at each position is shown in Supplementary Figures 4 and 5. All of the clones showed accumulation of mutations in both heavy and light chain V regions relative to the original (germline) transgene sequence. Somatic mutations were concentrated in the CDRs, with CDR-H3 showing the most diversity. Gene conversion using sequences donated by the synthetic human pseudogenes was observed in both the VK and VH variable regions, as well as point mutations that cannot be ascribed to gene conversion. All of the human pseudogenes were used in gene conversion; no sequences derived from the chicken pseudogene pool were observed for either light or heavy chain. Sequences of mAbs from different birds were usually quite divergent, although we observed several examples of related sequences from different PGRN-immunized birds that were a result of clear gene conversion leading to similar CDRs being incorporated ( shows one such example). These related antibodies also converged on the same epitope bins. In addition, some residues were commonly found to be introduced by gene conversion into a majority of the antibody sequences. The Arg-Leu-Phe (RLF) motif present in heavy chain framework 3 (IMGT positions 90–92) in the functional V of the SynVH-C construct was gene converted in virtually every antibody sequence to Gln-Met-Asn (QMN) (Supplementary Fig. 4), which can be found in several of the pseudogenes in the SynVH-C array. A Gly residue in CDR-L2 (IMGT position 56) was found to be changed to Asp in about half of the antibody sequences from multiple birds immunized with PGRN. For the antibodies derived from the SynVH-A7 array (Supplementary Fig. 5), in which heavy chain pseudogenes contain only Ser, Trp and Tyr residues in the CDRs, heavy chain gene conversion was more limited than in SynVH-C and consisted of shorter stretches of sequence encoding only one or two amino acids. The longest gene conversion tract was found in CDR-H3 where the residue pair Ser-Tyr was found in a subset of mAbs and was likely derived from pseudogene SynVH17. The majority of the heavy chain diversity in SynVH-A7 mAbs consisted of sequences not found in the pseudogenes.
Some variation in CDR-H3 length was observed, with a range of 9 to 16 amino acids. A substantial proportion (40%) of the sequences contained 13 amino acids in CDR-H3; the germline pre-rearranged CDR-H3 contained 11 amino acids. No cysteine residues were found in any of the CDR-H3 sequences, in contrast to WT chickens, which often contain cysteine in CDR-H3Citation9,15–18 (27/32 clones in a panel of anti-PGRN chicken mAbsCitation8). The only non-canonical cysteine that was found in the human sequences was in one clone (out of ∼150) that had an unpaired cysteine in VH framework 2.
Discussion
Human antibody therapeutics have become an integral part of the pharmaceutical space, and for many indications are a preferred drug format due to their high target-specificity, effector functionality and long serum half-life. During the process of discovery and development, a variety of attributes need to be examined in candidate antibodies, including biophysical properties, immunogenicity, toxicity, efficacy, potency, and pharmacokinetics. Many areas of evaluation require the use of animal models of disease, most commonly in rodents or non-human primates. Therefore, it is advantageous to source antibodies from a phylogenetically distant species that will recognize the human target and its orthologs as foreign and readily elicit antibodies to pan-mammalian epitopes. Chickens have long been used to raise such antibodies against conserved mammalian targets.Citation28–33 Here, we describe an engineered transgenic chicken that retains native antigen recognition, but delivers human sequence antibodies that are suitable for therapeutic use. Replacing the chicken V gene repertoire with a human one allows the transgenic bird to generate human antibodies that retain the benefits of the chicken host, namely broad epitope coverage of the target beyond that available from mammals. The expanded epitope coverage includes a prevalence of species cross-reactive antibodies, which in many cases obviates the need for surrogate antibodies. Targeting novel epitopes can also provide opportunities for therapeutics with different mechanisms of action than previously available and expand the possible routes to drug development. We have shown that the OmniChicken, like WT birds, can recognize highly conserved mammalian proteins such as BDNF, and can be used to provide panels of unique antibodies of high affinity to multiple epitopes on human targets.
The B cells in the OmniChicken are capable of producing a high level of sequence diversity in the human VK and VH genes using both gene conversion and non-templated mutation. In the heavy chain, two different strategies were pursued to produce diversity. In SynVH-C, the pseudogenes contained diverse CDRs and limited diversity in the frameworks, all sourced from naturally occurring human sequences. The power of in vivo selection was evident in that certain sequences, such as the QMN in framework 3, could be obtained from the pseudogene pool and positively selected. In SynVH-A7, the pseudogenes contained a restricted set of residues in the CDRs (only Y, W and S), whereas the CDRs found in the antibody sequences were much more diverse, indicating that additional amino acids were incorporated by somatic hypermutation. Both constructs yielded sequences that could be ascribed to gene conversion, with somatic hypermutation and selection adding another layer of diversity in the CDRs and frameworks. In both the light and heavy chain loci, the chicken pseudogenes are still present upstream of the human transgenes. We saw no evidence of chicken sequence being introduced into the human V regions (references 34 and 35 and Supplementary Figs. 4 and 5). Gene conversion is a homology-based process, and the human V pseudogenes have much higher homology to the human functional V, and they are in closer proximity, which could introduce further bias in their favor.Citation36 Although the SynVK and SynVH transgenes contain fewer pseudogenes than the WT loci (16 vs. 25 for the light chain and 20 vs. about 100 for the heavy chain), not all of the chicken pseudogenes may participate in gene conversion.Citation36 In contrast, all of the human pseudogenes were used in our panels and in previous work in the DT40 model.Citation35 The overall level of sequence diversity in the cohorts of anti-PGRN mAbs derived from OmniChicken and WT birds seemed similar when comparing the sequence dendrograms.Citation8 Most importantly, the repertoire diversity generated by gene conversion or point mutation in OmniChicken and WT birds recognized equivalent epitope space on the PGRN protein.
The OmniChicken expresses chimeric (human variable/chicken constant) immunoglobulins, to ensure the proper engagement of endogenous constant region receptors and to form the B cell receptor complex with Igα/β.Citation37 Once the human V regions are identified by cloning, they can be combined with any Fc region or into any antibody format using standard cloning techniques. Typically, our mAbs are assembled in the scFv-Fc format for initial evaluation. Subsequently, the best candidates are reformatted into full length IgG molecules with equivalent specificity, sensitivity and affinity constants. In addition, they expressed at normal levels in mammalian cell culture.
Levels of plasma Ig were normal or near normal for IgM and IgA isotypes, but lower for IgY. Reduced Ig levels have also been observed in many of the humanized rodent platforms.Citation5 Circulating IgY is produced by terminally-differentiated plasma cells, which contain a greatly expanded secretory apparatus for high-level secretion of antibody. It is possible that either the number of plasma cells is reduced, or that IgY expression per cell is lower than normal. In chickens, markers to specifically assess the number of plasma cells are lacking, but the numbers of splenocytes from immunized OmniChickens were comparable to those from immunized WT birds (2.2e9 splenocytes averaged over 25 OmniChickens compared to 2.5e9 splenocytes averaged over 28 WT birds). Reduced expression of the human kappa chimeric light chain in IgY-switched plasma cells seems to be the main cause of the IgY reduction, as a human lambda chimeric light chain resulted in normal IgY levels. The reduced level of kappa light chain that is available for secretion may restrict the level of IgY, but that level is sufficient for IgM and IgA, which are normally found at lower levels than IgY. The wild type chicken light chain V gene is more homologous to human lambda (65% amino acid identity to VL3-19 used here) than to human kappa (45% identity to human VK3-15 used here). The human lambda V region may thus be a better match with the chicken CL to fold properly into a functional light chain than the kappa V region. Kappa was initially chosen since most therapeutic antibodies contain a kappa light chain,Citation25 and it was fused to the chicken CL region to ensure pairing with the chicken heavy chain CH1 domains. The lambda birds were more recently obtained and are under evaluation. Despite the reduction in bulk IgY levels, when compared to WT birds immunized with the same antigen, the OmniChickens were able to produce similar antigen-specific titers and antibodies with similar epitope coverage.Citation8
In conclusion, we demonstrated that the active chicken V gene repertoire can be fully replaced by sequences derived from human V genes. The transgenes are expressed in the context of chicken C regions, and are diversified in developing B cells by the native chicken process that includes both gene conversion and somatic hypermutation. The resulting transgenic chickens are phenotypically normal in B cell development and deliver affinity-matured human-sequence antibodies, yet share the immune recognition characteristics of WT chickens, including responsiveness to mammalian-conserved targets and a propensity to generate pan-mammalian cross-reactive antibodies. Interestingly, the “chicken-like” immune recognition is preserved in the OmniChicken despite it being genetically constrained to generate its repertoire based upon a restricted set of V gene frameworks. By focusing on selected frameworks chosen for developability, it is expected that the OmniChicken will yield high quality therapeutic leads at an improved frequency relative to other antibody discovery approaches.
Materials and methods
Constructs used to make OmniChickens
Production of the IgL and IgH knockouts was previously described.Citation22,23
The human kappa construct, SynVK-CK, was identical to the SynVK-C construct previously describedCitation35 except for a modified Kozak consensus sequence in the functional V. The functional V region consists of a rearranged human VK3-15/JK4 gene and was obtained from screening a small library of VK3 family sequences from human PBMC RNA. For the VK pseudogenes, a diverse set of CDRs was selected from the naturally-occurring somatic human sequences found in the NCBI EST database, which was queried with the VK3-15 germline gene. Individual pseudogenes may consist of CDRs from different ESTs, and some framework diversity was also included in some of the pseudogenes. Framework 4 sequence was not included in the pseudogenes. For the human lambda construct, the functional V was swapped for a rearranged VL1-44 gene and VL-based pseudogenes. For the human heavy chain, the human functional VH was obtained from screening a small library of VH3 family sequences from human PBMC RNA. The selected V region is based on germline VH3-23 with a few somatic changes and is a rearranged V region consisting of V, D and JH4 elements. For the VH pseudogenes, two versions were made, and OmniChickens contained either one or the other. In construct SynVH-C (used in the BDNF and PGRN campaigns), a diverse set of CDRs was selected from the NCBI EST database, which was queried with the VH3-23 germline gene (Supplementary Fig. 2). Pseudogenes may have CDRs from different ESTs, some framework 1–3 diversity was included, but no framework 4 sequence was present in the pseudogenes. In construct SynVH-A7 (used in the cocktail immunization), the CDRs were composed of S, Y and W residues, with no changes in the frameworks relative to the functional VH (Supplementary Fig. 2). The pseudogenes were synthesized (Genewiz) with flanking 50 base pair spacers derived from the chicken heavy chain pseudogene locus and cloned into an array, all in the opposite orientation to the functional V. The IgH promoter region was amplified from chicken genomic DNA. The functional V and a short segment of the chicken J-Cμ intron were synthesized. These parts were assembled with a β-actin promoter and attB site for integration into the attP-neo present in the IgH KO.Citation22
Germ cell culture
Germ cell derivation and culture were performed as previously described.Citation21,38,39 Re-derived Knockout cell lines IgL KO 229–92Citation23 and IgH KO 472–138Citation40 were transfected with the SynVK and SynVH constructs, respectively, using phiC31 integrase as described.Citation34
Breeding
Animal experiments were done in accordance to Institutional Animal Care and Use Committee (IACUC)-approved protocols and under supervision of the IACUC. To obtain OmniChickens with the genotype SynVK/IgL KO; SynVH/IgH KO, crosses between SynVK/+; IgH KO/+ and IgL KO/+; SynVH/+ birds were performed. Progeny were genotyped by PCR using DNA obtained from comb biopsy. Males and females were kept for analysis.
Flow cytometry
PBMCs were isolated using Histopaque (Sigma,10771), labeled on ice for 1 h with the following reagents from Southern Biotech: ms anti-Bu1 (8395–01), ms anti-ch IgM (8310–01), ms anti-ch IgL (8340–01), ms anti-ch ΤCR-γδ (8230–01), ms anti-TCRαβ/Vβ1 (8240–01) or ms anti-TCRαβ/Vβ2 (8250–01). All antibodies were diluted in 1% bovine serum albumin (BSA)-phosphate-buffered saline (PBS). Following incubation in primary antibody, samples were washed and then incubated with AF-647 anti-ms IgG (Thermo Fisher, 115-605-205). For double-labeling experiments, direct conjugates (Southern Biotech) were used. Data was collected using an Attune Acoustic Focusing Cytometer (Thermo Fisher) and analyzed using FloJo.
Elisa
ELISA plates were coated with ms anti-ch IgA (Southern Biotech, 2330–01), ms anti-ch IgM (Southern Biotech, 8310–01)) or rb anti-ch IgY (Sigma, C2288) at 2 ug/mL in PBS, washed in PBS/0.5% Tween-20 (PBST), and blocked for 1 h in PBST/3% nonfat dry milk. The blocking buffer was aspirated and serial dilutions of plasma in blocking buffer added for 1 h at RT. Plates were washed, incubated with goat anti-ch IgA-HRP (Bethyl, A30-103P), anti-ch IgM-HRP (Bethyl, A30-102P) or anti-ch IgY-HRP (Sigma, A9046) for 1 h at RT, washed, developed with TMB (Thermo Fisher) for 10 min, and the reaction stopped with 1 N HCl. Plates were read at 450 nm on a BioTek microplate reader. Purified IgM (Rockland, 003–0107) and IgY (GenScript, A01010) were used to quantitate plasma Ig titers. IgA was quantified against IgA purified and quantified from chicken plasma (Kaspers Lab, University of Munich).
To assess antigen-specific titer, ELISA plates were coated with 2 ug/mL antigen in PBS. Plates were blocked as described above, incubated with plasma at the indicated dilutions for 1 h, washed, incubated with rb anti-ch IgY-HRP (Sigma, A9046) for 1 h washed, and developed as above.
Protein and DNA immunizations
Immunogens were: recombinant human PGRN (Sino Biologicals, 10826-H08H), human BDNF (Sino Biologicals, 50240-MNAS), bovine thrombin (Sigma, T4648), murine IL-13 (Peprotech, 210–13) and human Holo-transferrin (R&D Systems, 2914-HT). Human PGRN was also cloned with a C-terminal 6X-His tag and expressed in Expi293 cells (see below).
200 ug of protein was emulsified in 250 uL of Freund's Complete Adjuvant (Thermo Fisher) and injected into the breast tissue. Fourteen days later, chickens were boosted with 200 ug protein and 250 uL of Freund's Incomplete Adjuvant (Thermo Fisher). Animals were subsequently boosted every other week, with blood drawn on the off-week to assess antigen-specific titer. Four days before euthanization and spleen harvesting, animals received a final intravenous (IV) boost with no adjuvant.
DNA immunizations were performed using the Helios Gene Gun (BioRad). Animals received DNA immunizations of an expression vector containing 6X-His tagged full length human PGRN alone, with the vector pCMVi-LSrCOMPTT (Arizona State University), or an expression vector containing full length chicken granulocyte/monocyte stimulating factor. pCMVi-LSrCOMPTT contains the sequences of various adjuvants shown to enhance the response to DNA immunization, including tetanus toxoid p30 and N-terminal tetanus toxin epitopes.Citation41 50 ug of PGRN DNA, alone or with up to 50 ug of adjuvant DNA, was precipitated onto 25 mg gold according to the manufacturer's instructions. Birds received two shots in each breast at 550 psi, and were kept on the same boost/blood draw schedule as described above. Four days before euthanization, and spleen harvesting, animals received a final IV boost of protein with no adjuvant.
Single B cell GEM screening
We employed a single lymphocyte screening method, the Gel-Encapsulated Microenvironment (GEM) assay (US Patents 8030095 and 84151738) to identify antigen-specific mAbs from immunized chickens. Five μm aldehyde-latex beads (Thermo Fisher) were coated with either purified antigen, or with streptavidin followed by biotinylated antigen, overnight, blocked with 3% milk-PBS, and tested by labeling with plasma from immunized animals. GEMs were preparedCitation10 containing a single secreting B cell and antigen-coated beads and incubated for 3 h at 37°C in RPMI/10% fetal calf serum containing 2 ug/mL Alexa Fluor 594 anti-ch IgY.
Single cell PCR to amplify VH and VK regions and clone as scFv-Fc
Cells secreting antigen-specific mAbs were captured and their VH and VK regions were amplified by a two-step, semi-nested strategy as described.Citation10 Primer sequences are given in Supplementary Table 1. The variable regions were assembled with human Fc by overlap extension PCR.
Expression of scFv-Fc antibodies
Recombinant scFv-Fc was expressed in Expi293 cells as described.Citation10 Recombinant PGRN was expressed using the same transfection method in 30 mL with 30 ug of DNA.
Biosensor analysis
Interaction analysis studies were performed by Array SPRi at 25°C in a running buffer of PBS + 0.05% Tween20 + 1 g/L BSA (unless stated otherwise) using a MX96 SPR imager (IBIS, Netherlands) equipped with Xantec CMD 200M sensor chips. Epitope binning was performed as described previouslyCitation42 with the following modifications. All HuMabs were used directly from crude supernatant and typically diluted 10-fold into 10 mM sodium acetate pH 4.5 + 0.01% Tween-20 (coupling buffer) to amine-couple them onto the chip. The BDNF HuMabs were analyzed using a “premix” assay format in a running buffer of 10 mM Hepes pH7.4, 150 mM ammonium sulfate, 0.05% Tween-20, and 1 g/L BSA.Citation43 Briefly, this involved premixing 43 nM binding sites of BDNF with titrating levels of each HuMab (representing final dilutions of 1/15, 1/30, 1/60, and 1/120 of the supernatant) and injecting these equilibrated mixtures over the coupled HuMab array. Binding responses were compared to those for BDNF alone to determine whether the premixed HuMabs blocked the coupled HuMabs, which were regenerated with a cocktail of 4:1 v/v Pierce Elution buffer (pH2.8):5 M NaCl. The bovine thrombin and PGRN HuMabs were analyzed using a “classical sandwich” assay format in a running buffer of PBS pH7.4, 0.05% Tween-20, and 1 g/L BSA and the arrayed HuMabs were regenerated with 75 mM phosphoric acid. By merging the HuMabs with a panel of purified mAb standards from the WT chicken,Citation8 we inferred the subdomain (or “granulin”) specificitiesCitation44 of the PGRN HuMabs by their cross-blockade of the standards. As a complementary approach, the subdomain assignments of the human-specific PGRN HuMabs were determined using a chimeric swap epitope mapping strategy by injecting the HuMabs as analytes over spots coupled with human PGRN, mouse PGRN, five previously described human/mouse chimeras,Citation8 and a 19-kDa fragment of human PGRN expressing only the D+E subdomains. Epitope binning and mapping of the PGRN HuMabs were performed in the same experiment on the same chip onto which the mAbs and antigen variants were coupled. Binning and mapping data were processed (calibrated, locally-referenced, aligned, and zeroed) in IBIS's SPRINT software and analyzed in the Epitope Binning Tool v 2.0 (Carterra, Inc., Salt Lake City, UT) to generate network blocking plots.
Affinities of the PGRN HuMabs and WT standards were determined using a capture-based kinetic analysis on an amine-coupled “lawn” of the capture reagent (goat anti-human Fc polyclonal, Southern Biotech, 2047–01). Coupling was performed in the MX96 SPR imager using coupling buffer (as above, pH 4.5) as the running buffer. Three solutions were injected for 7 min, one after another as follows: 1) the surface was activated with a freshly prepared mixture of 40 mM EDC + 10 mM sulfo-NHS (representing a 10-fold dilution of their stock concentrations), 2) the capture reagent was coupled, and 3) excess reactive esters on the surface were blocked with 1 M ethanolamine.HCl pH 8.5, resulting in a final coupling level of about 12,000 response units (or 1,200 millidegrees). The anti-human-Fc-coated chip was then inserted into a continuous flow microspotter (CFM) (Carterra, Inc) and the HuMabs (diluted 3- to10-fold in PBS + 0.05% Tween-20 + 1 g/L BSA were captured for 15 min using a 48-channel printhead. The purified WT mAb standards were diluted to 2–5 µg/mL and printed in parallel. Two consecutive prints were used to array 96 mAbs per chip. The printed chip was then redocked into the MX96 SPR imager and primed with PBS + 0.05% Tween-20 + 1 g/L BSA as running buffer. PGRN was prepared as a 3-fold dilution series at nominal concentrations of 0, 0, 7, 22, 67, and 200 nM and injected in increasing concentrations for 3 min each, allowing a 15-min dissociation phase, without any regeneration. Data were processed in SPRINT (as above) with a double-referencing stepCitation45 and the double-referenced data were analyzed globally in Carterra's kinetic tool “KIT” using a simple Langmuir model with mass transport, allowing each spot its own Rmax value. The affinity or equilibrium dissociation constant (KD) for PGRN interacting with each captured mAb, was determined by the ratio of the kinetic rate constants, KD = kd/ka.
Suppl_Mat_1386825.zip
Download Zip (543.6 KB)Acknowledgment
We would like to thank Prof. Bernd Kaspers for generously providing purified chicken IgA standard.
References
- Reichert JM. Antibodies to watch in 2017. MAbs. 2017;9:167–81. doi:10.1080/19420862.2016.1269580. PMID:27960628
- Ecker DM, Jones SD, Levine HL. The therapeutic monoclonal antibody market. MAbs. 2015;7:9–14. doi:10.4161/19420862.2015.989042. PMID:25529996
- Green LL. Transgenic mouse strains as platforms for the successful discovery and development of human therapeutic monoclonal antibodies. Curr Drug Discov Technol. 2014;11:74–84. doi:10.2174/15701638113109990038. PMID:23978036
- Frenzel A, Schirrmann T, Hust M. Phage display-derived human antibodies in clinical development and therapy. MAbs. 2016;8:1177–94. doi:10.1080/19420862.2016.1212149. PMID:27416017
- Bruggemann M, Osborn MJ, Ma B, Hayre J, Avis S, Lundstrom B, Buelow R. Human antibody production in transgenic animals. Arch Immunol Ther Exp (Warsz). 2015;63:101–8. doi:10.1007/s00005-014-0322-x. PMID:25467949
- Dodgson JB, Delany ME, Cheng HH. Poultry genome sequences: progress and outstanding challenges. Cytogenet Genome Res. 2011;134:19–26. doi:10.1159/000324413. PMID:21335957
- O'Leary MA, Bloch JI, Flynn JJ, Gaudin TJ, Giallombardo A, Giannini NP, Goldberg SL, Kraatz BP, Luo Z-X, Meng J, et al. The placental mammal ancestor and the post-K-Pg radiation of placentals. Science. 2013;339:662–7. doi:10.1126/science.1229237. PMID:23393258
- Abdiche YN, Harriman R, Deng X, Yeung YA, Miles A, Morishige W, Boustany L, Zhu L, Izquierdo SM, Harriman W. Assessing kinetic and epitopic diversity across orthogonal monoclonal antibody generation platforms. MAbs. 2016;8:264–77. doi:10.1080/19420862.2015.1118596. PMID:26652308
- Könitzer JD, Pramanick S, Pan Q, Augustin R, Bandholtz S, Harriman W, Izquierdo S. Generation of a highly diverse panel of antagonistic chicken monoclonal antibodies against the GIP receptor. MAbs. 2017;9:536–49. doi:10.1080/19420862.2016.1276683. PMID:28055305
- Mettler-Izquierdo S, Varela S, Park M, Collarini EJ, Lu D, Pramanick S, Rucker J, Lopalco L, Etches R, Harriman W. High-efficiency antibody discovery achieved with multiplexed microscopy. Microscopy (Oxf). 2016;65:341–52. doi:10.1093/jmicro/dfw014. PMID:27107009
- Finlay WJJ, Shaw I, Reilly JP, Kane M. Generation of high-affinity chicken single-chain Fv antibody fragments for measurement of the Pseudonitzschia pungens toxin domoic acid. Appl Environ Microbiol. 2006;72:3343–9. doi:10.1128/AEM.72.5.3343-3349.2006. PMID:16672476
- Shih HH, Tu C, Cao W, Klein A, Ramsey R, Fennell BJ, Lambert M, Ní Shúilleabháin D, Autin B, Kouranova E, et al. An ultra-specific avian antibody to phosphorylated tau protein reveals a unique mechanism for phosphoepitope recognition. J Biol Chem. 2012;287:44425–34. doi:10.1074/jbc.M112.415935. PMID:23148212
- Bowes T, Hanley SA, Liew A, Eglon M, Mashayekhi K, O'Kennedy R, Barry F, Taylor WR, O'Brien T, Griffin MD, et al. Developing Cell-Specific Antibodies to Endothelial Progenitor Cells Using Avian Immune Phage Display Technology. J Biomolecular Screening. 2011;16:744–54. doi:10.1177/1087057111407067. PMID:21593485
- Ratcliffe MJH. Antibodies, immunoglobulin genes and the bursa of Fabricius in chicken B cell development. Dev Comp Immunol. 2006;30:101–18. doi:10.1016/j.dci.2005.06.018. PMID:16139886
- Reynaud C-A, Anquez V, Grimal H, Weill J-C. A hyperconversion mechanism generates the chicken light chain preimmune repertoire. Cell. 1987;48:379–88. doi:10.1016/0092-8674(87)90189-9. PMID:3100050
- Reynaud C, Dahan A, Anquez V, Weill J. Somatic hyperconversion diversifies the single Vh gene of the chicken with a high incidence in the D region. Cell. 1989;59:171–83. doi:10.1016/0092-8674(89)90879-9. PMID:2507167
- Carlson LM, McCormack WT, Postema CE, Humphries EH, Thompson CB. Templated insertions in the rearranged chicken IgL V gene segment arise by intrachromosomal gene conversion. Genes Dev. 1990;4:536–47.
- Wu L, Oficjalska K, Lambert M, Fennell BJ, Darmanin-Sheehan A, Ní Shúilleabháin D, Autin B, Cummins E, Tchistiakova L, Bloom L, et al. Fundamental characteristics of the immunoglobulin VH repertoire of chickens in comparison with those of humans, mice, and camelids. J Immunol. 2012;188:322–33. doi:10.4049/jimmunol.1102466. PMID:22131336
- Mahon CM, Lambert MA, Glanville J, Wade JM, Fennell BJ, Krebs MR, Armellino D, Yang S, Liu X, O'Sullivan CM, et al. Comprehensive interrogation of a minimalist synthetic CDR-H3 library and its ability to generate antibodies with therapeutic potential. J Mol Biol. 2013;425:1712–30. doi:10.1016/j.jmb.2013.02.015. PMID:23429058
- van de Lavoir M-C, Diamond JH, Leighton PA, Mather-Love C, Heyer BS, Bradshaw R, Kerchner A, Hooi LT, Gessaro TM, Swanberg SE, et al. Germline transmission of genetically modified primordial germ cells. Nature. 2006;441:766–9. doi:10.1038/nature04831. PMID:16760981
- Collarini E, Leighton P, Pedersen D, Harriman B, Jacob R, Mettler-Izquierdo S, Yi H, van de Lavoir M-C, Etches RJ. Inserting random and site-specific changes into the genome of chickens. Poult Sci. 2014. PMID:25332140
- Schusser B, Collarini EJ, Yi H, Izquierdo SM, Fesler J, Pedersen D, Klasing KC, Kaspers B, Harriman WD, van de Lavoir MC, et al. Immunoglobulin knockout chickens via efficient homologous recombination in primordial germ cells. Proc Natl Acad Sci USA 2013; 110:20170–5. doi:10.1073/pnas.1317106110. PMID:24282302
- Schusser B, Collarini EJ, Pedersen D, Yi H, Ching K, Izquierdo S, Thoma T, Lettmann S, Kaspers B, Etches RJ, et al. Expression of heavy chain-only antibodies can support B-cell development in light chain knockout chickens. Eur J Immunol. 2016;46(9):2137–48 doi:10.1002/eji.201546171. PMID:27392810
- Tiller T, Schuster I, Deppe D, Siegers K, Strohner R, Herrmann T, Berenguer M, Poujol D, Stehle J, Stark Y, et al. A fully synthetic human Fab antibody library based on fixed VH/VL framework pairings with favorable biophysical properties. MAbs. 2013;5:445–70. doi:10.4161/mabs.24218. PMID:23571156
- Reichert JM. Marketed therapeutic antibodies compendium. MAbs. 2012;4:413–5. doi:10.4161/mabs.19931. PMID:22531442
- Leighton PA, Pedersen D, Ching K, Collarini EJ, Izquierdo S, Jacob R, van de Lavoir M-C. Generation of chickens expressing Cre recombinase. Transgenic Res. 2016;25(5):609–16. doi:10.1007/s11248-016-9952-6. PMID:27034267
- Miller S, Lesk AM, Janin J, Chothia C. The accessible surface area and stability of oligomeric proteins. Nature. 1987;328:834–6. doi:10.1038/328834a0. PMID:3627230
- Carroll S, Stollar B. Antibodies to calf thymus RNA polymerase II from egg yolks of immunized hens. J Biol Chem. 1983;258:24–6. PMID:6336747
- Burger D, Ramus M, Schapira M. Antibodies to human plasma kallikrein from egg yolks of an immunized hen: preparation and characterization. Thromb Res. 1985;40:283–8. doi:10.1016/0049-3848(85)90340-8. PMID:3852552
- Stuart C, Pietrzyk R, Furlanetto R, Green A. High affinity antibody from hen's eggs directed against the human insulin receptor and the human IGF-I receptor. Anal Biochem. 1988;173:142–50. doi:10.1016/0003-2697(88)90171-6. PMID:2973262
- Gassmann M, Thommes P, Weiser T, Hubscher U. Efficient production of chicken egg yolk antibodies against a conserved mammalian protein. FASEB J. 1990;4:2528–32. PMID:1970792
- Lemamy G, Roger P, Mani J, Robert M, Rochefort H, Brouillet J. High-affinity antibodies from hen's-egg yolks against human mannose-6-phosphate/insulin-like growth-factor-II receptor (M6P/IGFII-R): characterization and potential use in clinical cancer studies. Int J Cancer. 1999;80:896–902. doi:10.1002/(SICI)1097-0215(19990315)80:6%3c896::AID-IJC16%3e3.0.CO;2-J. PMID:10074924
- Camenisch G, Tini M, Chilov D, Kvietikova I, Srinivas V, Caro J, Spielmann P, Wenger R, M G. General applicability of chicken egg yolk antibodies: the performance of IgY immunoglobulins raised against the hypoxia-inducible factor 1alpha. FASEB J. 1999;13:81–8. PMID:9872932
- Schusser B, Yi H, Collarini EJ, Izquierdo SM, Harriman WD, Etches RJ, Leighton PA. Harnessing Gene Conversion in Chicken B Cells to Create a Human Antibody Sequence Repertoire. PLoS One. 2013;8:e80108.
- Leighton PA, Schusser B, Yi H, Glanville J, Harriman W. A Diverse Repertoire of Human Immunoglobulin Variable Genes in a Chicken B Cell Line is Generated by Both Gene Conversion and Somatic Hypermutation. Front Immun. 2015;6:126. doi:10.3389/fimmu.2015.00126.
- McCormack WT, Thompson CB. Chicken IgL variable region gene conversions display pseudogene donor preference and 5′ to 3′ polarity. Genes Dev. 1990;4:548–58. doi:10.1101/gad.4.4.548. PMID:2113879
- Ma B, Osborn MJ, Avis S, Ouisse L-H, Ménoret S, Anegon I, Buelow R, Bruggemann M. Human antibody expression in transgenic rats: comparison of chimeric IgH loci with human VH, D and JH but bearing different rat C-gene regions. J Immunol Methods. 2013;400–401:78–86. doi:10.1016/j.jim.2013.10.007. PMID:24184135
- Song Y, Duraisamy S, Ali J, Kizhakkayil J, Jacob VD, Mohammed MA, Eltigani MA, Amisetty S, Shukla MK, Etches RJ, et al. Characteristics of long-term cultures of avian primordial germ cells and gonocytes. Biol Reprod. 2014;90:15. doi:10.1095/biolreprod.113.113381. PMID:24337317
- van de Lavoir M-C, Collarini EJ, Leighton PA, Fesler J, Lu DR, Harriman WD, Thiyagasundaram TS, Etches RJ. Interspecific germline transmission of cultured primordial germ cells. PLoS One. 2012;7:e35664. doi:10.1371/journal.pone.0035664. PMID:22629301
- Dimitrov L, Pedersen D, Ching KH, Yi H, Collarini EJ, Izquierdo S, van de Lavoir M-C, Leighton PA. Germline gene editing in chickens by efficient CRISPR-mediated homologous recombination in primordial germ cells. PLoS One. 2016;11:e0154303. doi:10.1371/journal.pone.0154303. PMID:27099923
- Hansen DT, Robida MD, Craciunescu FM, Loskutov AV, Dörner K, Rodenberry J-C, Wang X, Olson TL, Patel H, Fromme P, et al. Polyclonal Antibody Production for Membrane Proteins via Genetic Immunization. Sci Rep. 2016;6:21925. doi:10.1038/srep21925. PMID:26908053
- Abdiche YN, Miles A, Eckman J, Foletti D, Van Blarcom TJ, Yeung YA, Pons J, Rajpal A. High-throughput epitope binning assays on label-free array-based biosensors can yield exquisite epitope discrimination that facilitates the selection of monoclonal antibodies with functional activity. PLoS One. 2014;9:e92451. doi:10.1371/journal.pone.0092451. PMID:24651868
- Teng HK, Teng KK, Lee R, Wright S, Tevar S, Almeida RD, Kermani P, Torkin R, Chen Z-Y, Lee FS, et al. ProBDNF induces neuronal apoptosis via activation of a receptor complex of p75NTR and sortilin. J Neurosci. 2005;25:5455–63. doi:10.1523/JNEUROSCI.5123-04.2005. PMID:15930396
- Bhandari V, Palfree RG, Bateman A. Isolation and sequence of the granulin precursor cDNA from human bone marrow reveals tandem cysteine-rich granulin domains. Proc Natl Acad Sci USA. 1992;89:1715–9. doi:10.1073/pnas.89.5.1715. PMID:1542665
- Myszka DG. Improving biosensor analysis. J Mol Recognit. 1999;12:279–84. doi:10.1002/(SICI)1099-1352(199909/10)12:5%3c279::AID-JMR473%3e3.0.CO;2-3. PMID:10556875
