ABSTRACT
Site-specific conjugation of small molecules to antibody molecules is a promising strategy for generation of antibody-drug conjugates. In this report, we describe the successful synthesis of a novel bifunctional molecule, 6-(azidomethyl)-2-pyridinecarboxyaldehyde (6-AM-2-PCA), which was used for conjugation of small molecules to peptides and antibodies. We demonstrated that 6-AM-2-PCA selectively reacted with N-terminal amino groups of peptides and antibodies. In addition, the azide group of 6-AM-2-PCA enabled copper-free click chemistry coupling with dibenzocyclooctyne-containing reagents. Bifunctional 6-AM-2-PCA mediated site-specific conjugation without requiring genetic engineering of peptides or antibodies. A key advantage of 6-AM-2-PCA as a conjugation reagent is its ability to modify proteins in a single step under physiological conditions that are sufficiently moderate to retain protein function. Therefore, this new click chemistry-based method could be a useful complement to other conjugation methods.
Introduction
Antibody therapeutics are successfully used for clinical treatment of human diseases, such as cancers, inflammatory diseases, and viral infections.Citation 1 , Citation 2 In particular, antibody-drug conjugates (ADCs) are an important class of highly potent biopharmaceutical drugs for the treatment of cancer.Citation3-Citation6 ADCs are thought to expand and improve the potency and effectiveness of chemotherapies.Citation 7 To date, four FDA-approved ADC drugs have shown promise for cancer therapy, and more than 40 are currently undergoing clinical evaluation.Citation 8 , Citation 9
Typically, ADCs consist of a tumor-specific antibody coupled to cytotoxic drugs through modification and linker technologies. This drug conjugation strategy enables selective targeting and delivery of small cytotoxic molecules to malignant tumor cells.Citation 10 Non-specific coupling of small molecules inevitably generates heterogeneous protein products, resulting in lower bioactivity and potentially protein precipitates or denaturation. Site-specific protein modification is a compelling approach to decrease heterogeneity and improve stability and selectivity.Citation 11 Various site-specific protein conjugation methods have been recently developed, including the use of engineered cysteinesCitation 12 , Citation 13 or lysines,Citation 14 unnatural amino acids,Citation 15 , Citation 16 and enzymatic reactions by glycotransferasesCitation 17 , Citation 18 or transglutaminases.Citation 19 For example, genetic incorporation of unpaired cysteines has become a robust strategy for site-specific modification of the protein. The thiol reaction enables maleimide-bearing linkers to conjugate cysteine residues on a protein.Citation 20 However, this method often leads to a heterogeneous mixture and decrease of protein activity due to the reduction of interchain disulfides in the reaction.Citation 21
The N-terminal α-amine is a crucial site of protein post-translational modification that affects protein activation, conversion, and degradation.Citation 22 The α-amine has gained attention for in vitro protein modification. In addition, N-terminal serines or threonines can be oxidized to aldehydes and react with amines and alkoxyamine groups.Citation23-Citation26 Recent studies showed that the α-amine of proteins can be modified through ketene without interfering with side chains.Citation 27 Nevertheless, the labile property of ketene makes its synthesis difficult. A recent study showed that 2-pyridinecarboxyaldehyde (2-PCA) formed a stable imidazolidinone with N-terminal amines of protein under mild conditions.Citation 28 Although the above-mentioned methods are of great interest, they have several limitations, including difficulties in chemical synthesis, unwanted byproducts, and low reaction efficiency. Therefore, a simple and mild method for coupling a small molecule onto a natural amino acid of peptides or proteins with high selectivity is needed.
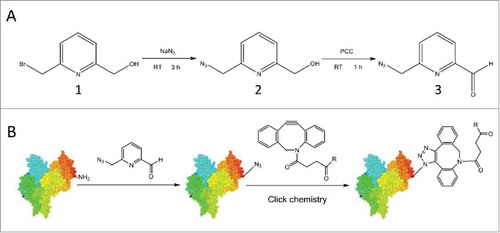
Here, we report a novel azide derivative of 2-PCAs, 6-(azidomethyl)-2-pyridinecarboxyaldehyde (6-AM-2-PCA), which is used for selective N-terminal coupling reactions with peptides or antibodies (). The azido group on 6-AM-2-PCA is compatible with click reactions for a wide range of alkyne-labeled molecules. We demonstrate that this bioconjugation reaction to antibodies is remarkably mild, simple and site-specific for peptide or antibody labeling. Our results are useful for the development of ADC technology and the construction of biomolecular materials.
Figure 1. Synthesis of 6-AM-2-PCA and its application for N-terminal modification of peptides or proteins. (A) 6-AM-2-PCA is synthesized through azidization and subsequent oxidation of 6-(bromomethyl)-2-pyridinemethanol (1) to yield an azido intermediate (2) and ultimately, pyridinecarboxyaldehyde (3). (B) N-terminal modification of a protein with 6-AM-2-PCA results in an azide appended to the protein that orthogonally reacts with a DBCO derivative through the aldehyde-amine reaction.
Results
Synthesis of 6-AM-2-PCA
Aldehyde substrates were observed to react with N-terminal residues in 50 mM phosphate buffer at pH 7.5 and 37°C through imidazolidinone formation.Citation 28 We predicted that aldehyde derivatives would be promising reagents for site-specific bioconjugations of peptides or proteins under mild conditions. Azidization of 6-(bromomethyl)-2-pyridinemethanol (1) with NaN3 was performed efficiently. The product was determined to be 6-(azidomethyl)-2-pyridinemethanol (2) by mass spectrometry (MS) (Supplementary Fig. S1A). The calculated m/z ratio for C7H8N4NaO [M+Na]+ was 187.0590, and the experimental m/z ratio was found by MS (electrospray ionization-time-of-flight (ESI-TOF)) to be 187.0589. Mild oxidation of 6-(azidomethyl)-2-pyridinemethanol by pyridinium chlorochromate (PCC) at room temperature also efficiently yielded 6-AM-2-PCA, which was confirmed by MS (Supplementary Fig. S1B). The calculated m/z ratio for C7H6N4NaO [M+Na]+ was 185.0434, and the experimental m/z ratio was found by MS (ESI-TOF) to be 185.0437.
Modification of antibodies with fluorescent dye by click reaction
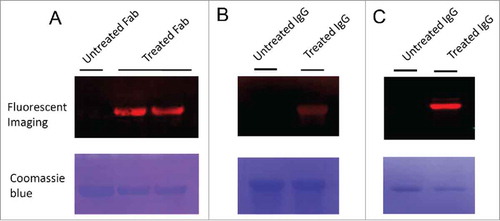
To demonstrate the applicability of 6-AM-2-PCA for protein modification, we examined the linkage by click reaction of a dibenzocyclo-octyne (DBCO) derivative toward antibodies. First, 6-AM-2-PCA–modified anti-human epidermal growth factor receptor (HER) 2 antibodies (both Fab and IgG formats) were fixed on the polyvinylidene difluoride (PVDF) membrane to serve as a matrix. DBCO-coupled Cy5.5 dyes were flowed through the fixed proteins. Fluorescent imaging of antibodies (Fab or IgG) revealed clear bands with the same migration as those of the untreated antibodies, as shown in . In another experiment, naive IgG proteins, directly fixed on the PVDF membrane, were sequentially incubated with 6-AM-2-PCA and DBCO- Cy5.5 dyes. A distinct band was observed after fluorescent imaging, as shown in and . In contrast, antibodies without 6-AM-2-PCA treatment did not show any fluorescent labeling.
Figure 2. Fluorescence imaging of Fab and IgG antibodies modified by 6-AM-2-PCA. (A) Fab antibodies were treated with 6-AM-2-PCA, following by fixing to the membrane and incubation with DBCO-Seta650. Fluorescence signal was detected. (B) IgG antibodies were treated with 6-AM-2-PCA, following by fixing to the membrane and incubation with DBCO-Cy5.5. Fluorescence signal was detected. (C) IgG antibodies were treated with 6-AM-2-PCA, and sequentially labeled with DBCO-Cy5.5. The upper panel is the fluorescent imaging. The lower panel is Coomassie brilliant blue R-250 staining of proteins.
Verification of N-terminal modification by 6-AM-2-PCA
To confirm the site-selectivity of 6-AM-2PCA on one anti-HER2 antibody, a peptide (ADVVMTQSP) derived from the N-terminus of antibody light chains was used. Meanwhile, a peptide with the same sequence was synthesized, but α-amine was blocked by acetylation. As expected (Supplementary Fig. S2A), MS data indicated that the light chain peptide was cross-linked to 6-AM-2-PCA (the peak representing these products was 1113.4752). As further validation of the reaction site specificity, no molecular weight changes were observed in acetylated peptides treated with 6-AM-2-PCA (Supplementary Fig. 2B). Furthermore, a Flag peptide containing an additional C-terminal Cys residue, DYKDDDDKC (Flag-Cys), was also treated with 6-AM-2-PCA and analyzed by MS. Although there are two Lys (K) residues and a Cys (C) in Flag-Cys peptides (MW: 1114.25 Da), MS detection revealed that one 6-AM-2-PCA molecule was coupled to one Flag-Cys peptide (MW: 1282.4418 Da) (Supplementary Fig. 2C). This finding indicates that 6-AM-2-PCA specifically reacted with α-amino residues of peptides, but not to ϵ-amino groups of lysine residues or cysteine. We compared the reaction conditions using different molar ratios of LCN peptide: 6-AM-2PCA. The concentration of 6-AM-2PCA heavily influenced the level of modification (Supplementary Fig. 3). The best conditions were determined to be a 100-fold molar excess of 6-AM-2PCA, resulting in >50% modification. However, we observed that the modification of a DBCO derivative conjugated to azido was relatively low (<50%). The relatively short length of the spacer between azide and pyridine might affect the DBCO conjugation efficiency. Purified modified peptides were analyzed by high-performance liquid chromatography (HPLC) (Supplementary Fig. S4).
We examined the reactivity of 6-AM-2PCA toward proteins using anti-HER2 antibodies. The modification of the anti-HER2 Fab (1 molar equivalent) was conducted in 100 molar equivalents of 6-AM-2PCA in phosphate buffer (PB) at 37°C for 12 h. As shown in , product analysis by LC−MS revealed a peak at 50359 Da, which was assigned to unmodified Fab. On the basis of mass shift, mono- or bis-modified protein was detected in the reaction mixture. The peaks at 50503 Da and 50647 Da are assigned to mono-modified and bis-modified Fab products, respectively, from the completed aldehyde-amine reaction. The peak at 50665 Da represents a hemiaminal product for the aldehyde-amine reaction. Calculations revealed that 79% of Fab proteins were conjugated with 6-AM-2PCA ().
Figure 3. Site-specific attachment of 6-AM-2-PCA to ananti-Her2 antibody analyzed by Q Exactive MS. (A) Fab was reacted with small molecules and analyzed by LC/MS. A peak at 50359 Da (1) was assigned to unmodified Fab. The peaks at 50503 Da (2) and 50647 Da (3) are assigned to mono-modified and bis-modified Fab products, respectively, from the completed aldehyde-amine reaction. The peak at 50665 Da (4) represents a hemiaminal product for the aldehyde-amine reaction. Data are representative of two experimental observations. (B) Two mechanisms of the aldehyde-amine reaction.
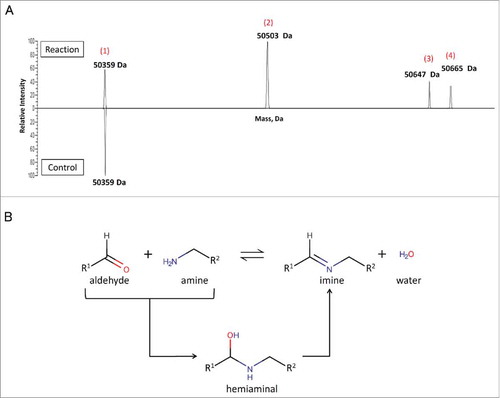
Table 1. Percent values of anti-Her2 Fab modified with 6-AM-2-PCA analyzed by MS.
To identify the conjugation sites, a biotin-PCA-ADC linker (893.39 Da) was prepared by coupling 6-M-2-PCA (1 molar equivalent) and DBCO-(PEG)4-biotin (1 molar equivalent). The modification sites of the linker were confirmed to be the N-terminal residues by LC-MS/MS analysis of trypsin-digested Fab antibodies (Supplementary Fig. 5A). Trypsin peptide mapping analysis resulted in 100% sequence coverage of the heavy chain (Supplementary Fig. 5B). Several conjugated peptides showed the residue sites of biotin-PCA-ADC conjugation in the heavy chain. For example, as shown in Supplementary Fig. S5C, the fragment ion y12 determined the conjugation at E1 of the peptide EVQLVQSGGGVVR. The linker was identified as modified at the N-terminal Glu of the heavy chain. Though we confirmed N-terminal modification of the peptide derived from the N-terminus of the light chain, we failed to identify the N terminus of the light chain in LC-MS/MS analysis. This suggests that 6-M-2-PCA tends to react with the heavy chain.
Bioactivities of antibodies after modification
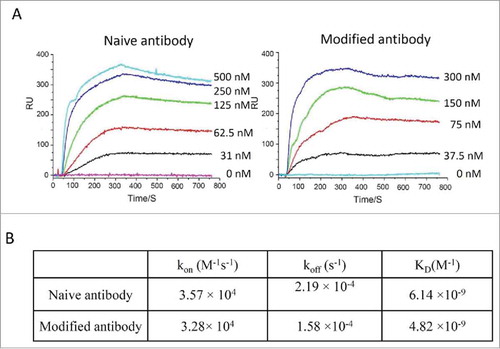
To determine whether modified antibodies retain their function, binding kinetics for IgGs were measured using surface plasmon resonance. Modified anti-HER2 antibody had a similar K D value for HER2 protein compared to unmodified antibody (6 nM vs 4 nM). This confirms that the antigen-binding ability of the antibody was retained through the modification steps. A summary of K D values for anti-HER2 antibodies is shown in .
Figure 4. Binding kinetic analysis of naive and modified anti-Her2 IgG antibodies to Her2 antigen. (A) Sensorgrams of binding kinetics. (B) Summary of binding rate constants and KD values. Experimental details are described in Materials and Methods.
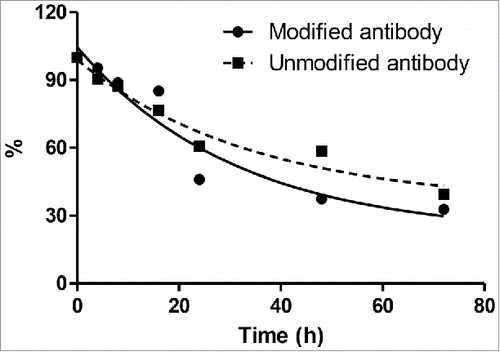
To evaluate the effect of 6-AM-2PCA modification on the stability of anti-HER2 antibodies, modified anti-HER2 antibodies were incubated in mouse plasma at 37oC. The retained amounts of antibodies at different time points were measured by the enzyme-linked immunosorbent assay (ELISA). The values at time points were fit using one-phase decay model. The antibodies were retained with 32–40% after 72 h. In contrast, there was no significant difference in the stability between modified and unmodified for each time point ().
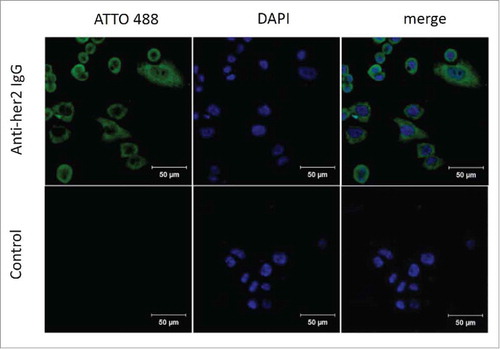
In another assay, the breast cancer cell line SK-BR-3, which strongly expresses HER2 antigens on the surface, was utilized as a model of immunofluorescence detection for modified anti-HER2 antibodies. Fluorescence-coupled anti-HER2 Fabs were mainly distributed at plasma membranes (). These results indicate that 6-AM-2-PCA is bio-compatible and nondestructive towards antibody activity, supporting the use of 6-AM-2-PCA as a feasible candidate for protein modification.
Figure 6. Immunostaining of SK-BR-3 breast cancer cells by 6-AM-2-PCA-conjugated anti-Her2 antibodies. (A) Three panels (left to right) represent fluorescence of SK-BR-3 cells stained with modified anti-Her2 Fab followed by DBCO-ATTO 488, nuclei stained with DPAI, and the mergedimage. (B) Three panels (left to right) represent fluorescence of SK-BR-3 cells stained with DBCO-ATTO 488 alone, nuclei stained with DPAI, and the merged image.
Discussion
Site-specific modification of a protein is often complex, low-yield and not scalable. N-terminal α-amines as sites of protein post-translational modification are particularly useful because they are distant from binding domains. Modifications should not interfere with antibody binding. Pyridoxal-5-phosphate (PLP) is an enzyme cofactor involved in a variety of metabolic transformation reactions, including racemization, elimination, decarboxylation and transamination.Citation 29 , Citation 30 Without enzyme action, PLP mediates the reaction of aldehyde with N-terminal amino groups on a protein to form glyoxyl imine.Citation 31 Similar to PLP, 2-PCAs react as pyridine aldehydes with N-terminal α-amines to form imine, and later generate a stable imidazolidinone that does not undergo hydrolysis.Citation 28
We synthesized a new 2-PCA derivative, 6-AM-2-PCA, that retains the capacity of 2-PCAs to react with N-terminal amino acids of proteins and form stable covalent bonds. In addition, 6-AM-2-PCA is capable of reacting with DBCO derivatives. Site-selectivity of 6-AM-2-PCA for N-terminal α-amines was confirmed by MS analysis. Modification efficiency can be determined by concentrations of free 6-AM-2-PCA. Further experiments demonstrated that 6-AM-2-PCA is able to modify proteins in a single step under physiological conditions that are moderate enough to permit biomolecular activities. MacDonald et al demonstrated that 2-PCA afforded 43% to 95% modification for different proteins.Citation 28 Notably, 33% of uteroglobin, a covalent homodimer, was singly modified at either of two N termini. Our studies demonstrated that 6-AM-2-PCA as a derivative of 2-PCA afforded >79% modification of anti-HER2 Fab with single and double labels. Similar to uteroglobin, we did not obtain the complete formation of double labels per anti-HER2 Fab even at 100-fold excess of reagents. Interestingly, half of bis-modified proteins formed hemiaminal products after the aldehyde-amine reaction. Our further study revealed that 6-AM-2-PCA mainly reacted to the N-terminus of anti-HER2 Fab heavy chain. Steric hindrance from the tertiary structure of the N-terminal region may affect selectivity to either heavy or light chains of the antibody. The 2-PCA method is specific for the native N termini of some antibodies.Citation 28 Therefore, it will be interesting to investigate reaction conditions of 6-AM-2-PCA for different antibodies in future studies.
It should be noted that the length of the linker is important for the click reaction of DBCO. We initially synthesized 6-(azido)-2-PCA that did not exhibit the reactivity to DBCO conjugates. The reaction efficiency was obviously improved by introducing one methyl group between pyridine and azido. In a previous study, we noted that spacer length determined the coupling reaction of one azidolated glycolipid GD2 and DBCO conjugates. Citation 32 Contrary to the short alkyl spacer, the long glycol spacer attenuated steric hindrance and improved the reaction efficiency. Therefore, it is worth investigating the effect of linker length on conjugation in a future study.
Although 6-AM-2-PCA does not exhibit perfect conjugation efficiency, we believe 6-AM-2-PCA has many broad applications. First, 6-AM-2-PCA and its derivatives can be conveniently and efficiently synthesized from commercially available products via two reaction steps. Second, it differs from 6-(piperazin-1-ylmethyl)-2-PCA, which has been reported to require cross-linking to N-hydroxysuccinimide-activated chemicals before it can be conjugated to a protein. This reaction requires organic solvents that could affect the biological functions of antibody molecules. Third, using click chemistry, azide orthogonally reacts with alkyne and DBCO derivatives. We assume that the azido group of 6-AM-2-PCA is compatible with aldehyde chemicals. 6-AM-2-PCA or later derivatives are versatile tools for protein modification, since a series of alkyne and DBCO derivatives can be purchased or synthesized.
In conclusion, we successfully synthesized a novel derivative chemical of 2-PCAs, 6-AM-2-PCA, that specifically links DBCO derivatives to protein N-terminal amino acids under mild reaction conditions. Specific labeling of N-termini using chemicals affords many opportunities to improve native protein function without genetic engineering. 6-AM-2-PCA can be potentially applied as a versatile tool in peptide or protein modification.
Materials and methods
Chemical reagents
6-(bromomethyl)-2-pyridinemethanol (1) was purchased from Tokyo Chemical Industry. Tetrabutylammonium hydrogen sulfate (TBAHS), PCC, diatomite, N,N-dimethylaminopyridine (DMAP), tetrahydrofuran (THF), N,N-dimethylformamide (DMF), and thin layer chromatography (TLC) GF-254 were purchased from Sigma-Aldrich. Methanol, acetonitrile (ACN) and dichloromethane (DCM) were purchased from J&K Scientific. Trifluoroacetic acid (TFA) and sodium azide (NaN3) were purchased from Aladdin. DBCO-ATTO 488 and DBCO-Cy5.5 were purchased from Jena Bioscience. DBCO-PEG4-biotin was purchased from Click Chemistry Tools. Other chemicals were obtained from Sigma-Aldrich.
Synthesis of 6-(azidomethyl)-2-pyridinemethanol (2)
6-(bromomethyl)-2-pyridinemethanol (1) (5.5 mmol) was dissolved in 10 mL THF. A solution of NaN3 (11 mmol) in 10 mL of water was added to the resulting red solution, followed by dropwise addition of TBAHS (to a final concentration of 0.55 mmol). The biphasic mixture was stirred vigorously in the dark at room temperature for 3 h. The completion of the reaction was monitored by TLC GF-254 (DCM and methanol, 10:1, V/V). The layers were then separated and concentrated. The crude product was purified by silica-gel chromatography (hexane/EtOAc) to produce (2) as a solid (820 mg, 91%). 1H nuclear magnetic resonance (NMR) (or 13C NMR) spectra were recorded with a Bruker AV600 or Bruker AV400, at 600 (150) or 400 (100) MHz, respectively. Chemical shifts were referenced either to tetramethylsilane as an internal standard or to the signals resulting from residual solvent. High resolution mass spectra were measured with an ABI Q-star Elite. 1H NMR (13C NMR) spectra were recorded with a Bruker AV600 or Bruker AV400. Resulting spectra were as follows: 1H NMR (400 MHz, dimethyl sulfoxide (DMSO)) δ 7.83 (t, J = 7.7 Hz, 1H), 7.44 (d, J = 7.8 Hz, 1H), 7.28 (d, J = 7.6 Hz, 1H), 5.47 (s, 1H), 4.55 (s, 2H), 4.47 (s, 2H) ppm.
Synthesis of 6-AM-2-PCA (3)
Purified 6-(azidomethyl)-2-pyridinemethanol (2) (5 mmol) was dissolved in 20 mL DCM and cooled to 0 ˚C. After stirring for ten minutes, PCC (5 mmol) was slowly added dropwise. After stirring for 1 h, the reaction mixture was concentrated under reduced pressure. The crude product was purified by silica-gel chromatography (hexane/EtOAc) to result in the final product, in the form of a colorless oil (600 mg, 74%). High resolution mass spectra were measured with an ABI Q-star Elite. 1H NMR (or 13C NMR) spectra were recorded with a Bruker AV600 or Bruker AV400, at 600 (150) or 400 (100) MHz, respectively. Chemical shifts were referenced either to tetramethylsilane as an internal standard or to signals resulting from residual solvent. Resulting spectra were as follows: 1H NMR (400 MHz, CDCl3) δ 10.04 (s, 1H), 7.96 – 7.85 (m, 2H), 7.61 – 7.53 (m, 1H), 4.59 (s, 2H) ppm; 13C NMR (101 MHz, CDCl3) δ 193.08, 156.68, 152.50, 138.11, 126.06, 120.79, 55.07 ppm.
High-performance liquid chromatography
Analytical reversed phase HPLC (RP-HPLC) was performed using an Inertsil ODS-SP column (4.6 × 250 mm, 5 μm, 100 Å) on Shimadzu LC 20 with SPD-20A UV detector at room temperature. The RP-HPLC gradient (A: 0.1% TFA in water, B: MeCN) started with 10% B, and then increased to 100% B over 30 min. Semi-preparative RP-HPLC was performed using the ULTIMAT 3000 Instrument (Thermo Scientific). UV absorbance was measured at 220 and 254 nm using a photodiode array detector. The RP-HPLC gradient (A: 0.1% TFA in water, B: MeCN) started with 10% B, and then increased to 100% B over 30 min.
Antibody expression and purification
Sequences of anti-HER2 monoclonal antibodies were identified using a phage-displayed human antibody library. Fabs of anti-HER2 antibodies were expressed and purified as previously described.Citation 17 Fab sequences in pComb3x vectors were expressed in HB2151. Expression was induced with isopropyl-L-thio-h-D-galactopyranoside. Soluble proteins were released from the periplasm. The clear supernatant was recovered for the purification on Ni-NTA column (QIAGEN). IgGs of anti-HER2 antibodies were expressed in mammalian suspension cells following the previously published protocol.Citation 33 IgGs were expressed in 293 FreeStyle cells. Polyetherimide was used to transfect 293 FreeStyle cells according to manufacturer's instruction (Thermo Fisher). After four days post-transfection, the culture supernatant was harvested. IgGs were purified on protein A resins (GE Healthcare).
Peptide modification
Three short peptides were synthesized by Genescript. Peptides were used to test whether 6-AM-2-PCA preferentially reacted to the N-terminal amino groups of antibodies. Synthesized peptides included the N-terminal sequence of antibody light chain (LCN), ADVVMTQSP, and the N-terminal acetylated peptide, Ac-ADVVMTQSP (Ac-LCN), as well as a Flag peptide containing an additional C-terminal Cys, DYKDDDDKC (Flag-Cys). Peptides were dissolved in 50 mM PB, pH 7.5, to a concentration of 10 mM at -20°C. 6-AM-2-PCA was stored as a 400 mM solution in DMSO at -20°C. A 40 µL aliquot of peptides was diluted to 400 µL volume in PB (final concentration 1 mM). Different volumes of 6-AM-2-PCA (final concentration 4, 20, or 100 mM) were added to the peptide solution. The reaction was incubated at 37°C for 12 h. The treated peptides were purified by HPLC. Peptide samples were analyzed using a Nano LC 20AD system (Shimadzu) coupled with an LTQ Orbitrap Velos mass spectrometer (Thermo Scientific).
Antibody modification
The same procedure was used for both anti-HER2 Fab and IgG antibodies. Antibodies were dissolved in PB buffer as a 20 µM solution. A 5 µL aliquot of 6-AM-2-PCA (final concentration 10 mM) was added to 200 µL antibody solution. The reaction was incubated at 37°C for 6 h. The modified antibodies were dialyzed in PB buffer at 4°C to remove residual 6-AM-2-PCA. The final products were concentrated by Amicon Ultra ultrafilters (10 K MWCO, Millipore) at 4°C. Modification of Fab samples were analyzed on an UltiMate™ 3000 RSLCnano system (Thermo Scientific) coupled with a Q-Exactive Quadrupole Orbitrap mass spectrometer (Thermo Scientific). Purity of antibodies was also analyzed using a size-exclusion chromatograph system.
Western blot analysis
The antibodies were analyzed by SDS-PAGE (10% for Fab and 8% for IgG) without the denaturing agent β-mercaptoethanol and transferred to a PVDF membrane. Membranes were blocked with 5% (w/v) bovine serum albumin (BSA) for 1 h and incubated with DBCO-Cy5.5 at room temperature for 3 h. The membranes were washed with phosphate-buffered saline (PBS) 5 times. Fluorescence on the PVDF membranes was scanned using the ODYSSEY CLx infrared imaging system (Li-COR).
Binding kinetic analysis
Kinetics and affinities between antibodies and antigens were analyzed using a PlexArray HT instrument (Plexera). Briefly, HER2 antigens were directly immobilized onto the Dextran sensor chip. The reference surface had immobilized BSA. For analysis of interaction kinetics, varying concentrations of antibodies were injected at a flow rate of 15 μL/s using assay buffer. The data were collected with the Plexera SPRi system. All of the kinetic experiments were performed at 25°C.
In vitro plasma stability
Mouse plasma was prepared according to the published protocol.Citation 34 The in vitro stability of antibodies was studied in mouse plasma. The analysis was performed by incubating modified or unmodified antibodies in diluted plasma at 37oC. The aliquots were collected at 0, 4, 8, 16, 24, 48 and 72 h. Samples were quantified in the ELISA assay. The stability data were fit using one-phase decay model of Graphpad Prism software.
Immunostaining on tumor cells
SK-BR-3 breast cancer cells were cultured on chamber slides and fixed with 4% paraformaldehyde at room temperature for 20 min and washed with PBS for 1 time. The fixed cells were blocked with 5% BSA in PBS at room temperature for 1 h. Cells were incubated with 6-AM-2-PCA-conjugated anti-HER2 Fabs in the dark at room temperature for 2 h followed by staining with DBCO-ATTO 488 (1:100 dilution). Next, cells were stained with DAPI at room temperature for 10 min and washed with PBS 3 times. The cell slides were sealed with 50% glycerol and observed under an OLYMPUS-LX71 microscope.
Abbreviations
| 2-PCA | = | 2-pyridinecarboxyaldehyde; |
| 6-AM-2-PCA | = | 6-(azidomethyl)-2-pyridinecarboxyaldehyde; |
| ACN | = | acetonitrile; |
| ADC | = | antibody-drug conjugate; |
| BSA | = | bovine serum albumin; |
| DBCO | = | dibenzocyclooctyne; |
| DCN | = | dichloromethane; |
| DMAP | = | N,N-dimethylaminopyridine; |
| DMF | = | N,N-dimethylformamide; |
| DMSO | = | dimethyl sulfoxide; |
| ELISA | = | the enzyme-linked immunosorbent assay; |
| ESI-TOF | = | electrospray ionization-time-of-flight; |
| HER | = | human epidermal growth factor receptor; |
| HPLC | = | high-performance liquid chromatography; |
| MS | = | mass spectroscopy; |
| NaN3 | = | sodium azide; |
| NMR | = | nuclear magnetic resonance; |
| PB | = | phosphate buffer; |
| PBS | = | phosphate-buffered saline; |
| PCC | = | pyridinium chlorochromate; |
| PLP | = | pyridoxal-5-phosphate; |
| PVDF | = | polyvinylidene difluoride; |
| RP-HPLC | = | reversed phase HPLC; |
| TBAHS | = | tetrabutylammonium hydrogen sulfate; |
| TFA | = | trifluoroacetic acid; |
| THF | = | tetrahydrofuran; |
| TLC | = | thin layer chromatography. |
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
2017MABS1593R1-s02.pdf
Download PDF (655.6 KB)Acknowledgments
This work was supported by the Science and Technology Development Fund of Macau (FDCT/131/2016/A3), Start-up Research Grand (SRG2016-00082-FHS) and the intramural research program of Faculty of Health Sciences, University of Macau, Shenzhen's Sanming Project (SZSM201612071), the Guangzhou Science and Technology Program (201807010004), the Guangdong Science and Technology Program (2016A050502034), Natural Science Foundation of Guangdong (Grand No. 2015A030313741), and National Natural Science Fund of China (31440041, 81373321). We thank Dr. Yan Ren of BGI-Shenzhen for the MS analysis of peptides.
Additional information
Funding
References
- Leavy O. Therapeutic antibodies: past, present and future. Nat Rev Immunol. 2010;10:297. doi:10.1038/nri2763. PMID:20422787.
- Adler MJ, Dimitrov DS. Therapeutic antibodies against cancer. Hematol Oncol Clin North AMa. 2012;26:447–81. vii. doi:10.1016/j.hoc.2012.02.013. PMID:22520975.
- Bakhtiar R. Antibody drug conjugates. Biotechnol lett. 2016;38:1655–64. doi:10.1007/s10529-016-2160-x. PMID:27334710. DOI.
- Borcoman E, Le Tourneau C. Antibody drug conjugates: the future of chemotherapy? Curr Opin Oncol. 2016;28:429–36. doi:10.1097/CCO.0000000000000310. PMID:27366963.
- Diamantis N, Banerji U. Antibody-drug conjugates–an emerging class of cancer treatment. Br J Cancer. 2016;114:362–7. doi:10.1038/bjc.2015.435. PMID:26742008.
- Thomas A, Teicher BA, Hassan R. Antibody-drug conjugates for cancer therapy. Lancet Oncol. 2016;17:e254–62. doi:10.1016/S1470-2045(16)30030-4. PMID:27299281.
- Boutureira O, Bernardes GJ. Advances in chemical protein modification. Chem Rev. 2015;115:2174–95. doi:10.1021/cr500399p. PMID:25700113.
- Chudasama V, Maruani A, Caddick S. Recent advances in the construction of antibody-drug conjugates. Nat Chem. 2016;8:114–9. doi:10.1038/nchem.2467. PMID:26892562.
- Hoffmann RM, Coumbe BGT, Josephs DH, Mele S, Ilieva KM, Cheung A, Tutt AN, Spicer JF, Thurston DE, Crescioli S, et al. Antibody structure and engineering considerations for the design and function of Antibody Drug Conjugates (ADCs). Oncoimmunology. 2018;7:e1395127. DOI:10.1080/2162402X.2017.1395127. PMID:29375935.
- Peters C, Brown S. Antibody-drug conjugates as novel anti-cancer chemotherapeutics. Biosci Rep. 2015;35:e00225. doi:10.1042/BSR20150089. PMID:26182432.
- Panowski S, Bhakta S, Raab H, Polakis P, Junutula JR. Site-specific antibody drug conjugates for cancer therapy. mAbs. 2014;6:34–45. PMID:24423619.
- Junutula JR, Raab H, Clark S, Bhakta S, Leipold DD, Weir S, Chen Y, Simpson M, Tsai SP, Dennis MS, et al. Site-specific conjugation of a cytotoxic drug to an antibody improves the therapeutic index. Nat Biotechnol. 2008;26:925–32. doi:10.1038/nbt.1480. PMID:18641636.
- Casi G, Huguenin-Dezot N, Zuberbuhler K, Scheuermann J, Neri D. Site-specific traceless coupling of potent cytotoxic drugs to recombinant antibodies for pharmacodelivery. J Am Chem Soc. 2012;134:5887–92. doi:10.1021/ja211589m. PMID:22394212.
- Basle E, Joubert N, Pucheault M. Protein chemical modification on endogenous amino acids. Chem Biol. 2010;17:213–27. doi:10.1016/j.chembiol.2010.02.008. PMID:20338513.
- Axup JY, Bajjuri KM, Ritland M, Hutchins BM, Kim CH, Kazane SA, Halder R, Forsyth JS, Santidrian AF, Stafin K, et al. Synthesis of site-specific antibody-drug conjugates using unnatural amino acids. Proc Natl Acad Sci U S A. 2012;109:16101–6. doi:10.1073/pnas.1211023109. PMID:22988081.
- Zimmerman ES, Heibeck TH, Gill A, Li X, Murray CJ, Madlansacay MR, Tran C, Uter NT, Yin G, Rivers PJ, et al. Production of site-specific antibody-drug conjugates using optimized non-natural amino acids in a cell-free expression system. Bioconjug Chem. 2014;25:351–61. doi:10.1021/bc400490z. PMID:24437342.
- Zhu Z, Ramakrishnan B, Li J, Wang Y, Feng Y, Prabakaran P, Colantonio S, Dyba MA, Qasba PK, Dimitrov DS. Site-specific antibody-drug conjugation through an engineered glycotransferase and a chemically reactive sugar. MAbs. 2014;6:1190–200. doi:10.4161/mabs.29889. PMID:25517304.
- Zhou Q, Stefano JE, Manning C, Kyazike J, Chen B, Gianolio DA, Park A, Busch M, Bird J, Zheng X, et al. Site-specific antibody-drug conjugation through glycoengineering. Bioconjug Chem. 2014;25:510–20. doi:10.1021/bc400505q. PMID:24533768.
- Jeger S, Zimmermann K, Blanc A, Grunberg J, Honer M, Hunziker P, Struthers H, Schibli R. Site-specific and stoichiometric modification of antibodies by bacterial transglutaminase. Angew Chem. 2010;49:9995–7. doi:10.1002/anie.201004243. PMID:21110357.
- Shen BQ, Xu K, Liu L, Raab H, Bhakta S, Kenrick M, Parsons-Reponte KL, Tien J, Yu SF, Mai E, et al. Conjugation site modulates the in vivo stability and therapeutic activity of antibody-drug conjugates. Nat Biotechnol. 2012;30:184–9. doi:10.1038/nbt.2108. PMID:22267010.
- Stephanopoulos N, Francis MB. Choosing an effective protein bioconjugation strategy. Nat Chem Biol. 2011;7:876–84. doi:10.1038/nchembio.720. PMID:22086289.
- Varland S, Osberg C, Arnesen T. N-terminal modifications of cellular proteins: The enzymes involved, their substrate specificities and biological effects. Proteomics. 2015;15:2385–401. doi:10.1002/pmic.201400619. PMID:25914051.
- Chelius D, Shaler TA. Capture of peptides with N-terminal serine and threonine: a sequence-specific chemical method for Peptide mixture simplification. Bioconjug Chem. 2003;14:205–11. doi:10.1021/bc025605u. PMID:12526710.
- Witus LS, Francis M. Site-Specific Protein Bioconjugation via a Pyridoxal 5′-Phosphate-Mediated N-Terminal Transamination Reaction. Curr Protoc Chem Biol. 2010;2:125–34. doi:10.1002/9780470559277.ch100018. PMID:23836553.
- Witus LS, Netirojjanakul C, Palla KS, Muehl EM, Weng CH, Iavarone AT, Francis MB. Site-specific protein transamination using N-methylpyridinium-4-carboxaldehyde. J Am Chem Soc. 2013;135:17223–9. doi:10.1021/ja408868a. PMID:24191658.
- Obermeyer AC, Jarman JB, Francis MB. N-terminal modification of proteins with o-aminophenols. J Am Chem Soc. 2014;136:9572–9. doi:10.1021/ja500728c. PMID:24963951.
- Chan AO, Ho CM, Chong HC, Leung YC, Huang JS, Wong MK, Che CM. Modification of N-terminal alpha-amino groups of peptides and proteins using ketenes. J Am Chem Soc. 2012;134:2589–98. doi:10.1021/ja208009r. PMID:22288779.
- MacDonald JI, Munch HK, Moore T, Francis MB. One-step site-specific modification of native proteins with 2-pyridinecarboxyaldehydes. Nat Chem Biol. 2015;11:326–31. doi:10.1038/nchembio.1792. PMID:25822913.
- Eliot AC, Kirsch JF. Pyridoxal phosphate enzymes: mechanistic, structural, and evolutionary considerations. Annu Rev Biochem. 2004;73:383–415. doi:10.1146/annurev.biochem.73.011303.074021. PMID:15189147.
- Toney MD. Reaction specificity in pyridoxal phosphate enzymes. Arch Biochem Biophys. 2005;433:279–87. doi:10.1016/j.abb.2013.10.002. PMID:24121043.
- Gilmore JM, Scheck RA, Esser-Kahn AP, Joshi NS, Francis MB. N-terminal protein modification through a biomimetic transamination reaction. Angewandte Chemie. 2006;45:5307–11. doi:10.1002/anie.200600368. PMID:16847857.
- Zhao Q, Ahmed M, Guo HF, Cheung IY, Cheung NK. Alteration of Electrostatic Surface Potential Enhances Affinity and Tumor Killing Properties of Anti-ganglioside GD2 Monoclonal Antibody hu3F8. J Biol Chem. 2015;290:13017–27. doi:10.1074/jbc.M115.650903. PMID:25851904.
- Zhao Q, Feng Y, Zhu Z, Dimitrov DS. Human monoclonal antibody fragments binding to insulin-like growth factors I and II with picomolar affinity. Mol Cancer Ther. 2011;10:1677–85. doi:10.1158/1535-7163.MCT-11-0281. PMID:21750218.
- Argmann CA, Auwerx J. Collection of blood and plasma from the mouse. Curr Protoc Mol Biol. 2006;Chapter 29:Unit 29A 3. doi:10.1002/0471142727.mb29a03s75. PMID:18265383.