ABSTRACT
Multiple formats are available for engineering of monoclonal antibodies (mAbs) by yeast surface display, but they do not all lead to efficient expression of functional molecules. We therefore expressed four anti-tumor necrosis factor and two anti-IpaD mAbs as single-chain variable fragment (scFv), antigen-binding fragment (Fab) or single-chain Fabs and compared their expression levels and antigen-binding efficiency. Although the scFv and scFab formats are widely used in the literature, 2 of 6 antibodies were either not or weakly expressed. In contrast, all 6 antibodies expressed as Fab revealed strong binding and high affinity, comparable to that of the soluble form. We also demonstrated that the variations in expression did not affect Fab functionality and were due to variations in light chain display and not to misfolded dimers. Our results suggest that Fab is the most versatile format for the engineering of mAbs.
Introduction
Over the past 20 years, yeast surface display (YSD) has emerged as a very potent tool for protein engineering. It is a very proficient platform for de novo identification of new binders in naïveCitation 1 or immune libraries.Citation 2 , Citation 3 YSD is also used for the engineering of many antibodies, principally to identify mutants with improved affinityCitation 4 , Citation 5 selectivity, stability and expression.Citation4-Citation7 Affinity-matured antibodies have been developed against multiple antigens such as tumor necrosis factor (TNF),Citation 8 fluorescein,Citation 9 botulinum neurotoxin type A,Citation 10 carcinoembryonic antigen,Citation 11 epidermal growth factor receptorCitation 12 and streptavidin.Citation 13 Engineering of other properties of antibodies such as pH sensitivity or antigen specificity has also been described.Citation 14 , Citation 15
Engineering of pre-existing antibodies using YSD relies on the anchoring of antibody fragments on the surface of yeast cells via fusion with a cell wall protein. Various formats are described in the literature for the functional display of antibody fragments on yeast cells, such as single-chain variable fragment (scFv), antigen-binding fragment (Fab) or single-chain Fab (scFab), although they are known to lead to variable levels of antibody expression.Citation 2 , Citation 4 , Citation 10 , Citation 16 Yeast display of IgG has also been reported with secretion and capture strategies,Citation 17 , Citation 18 but, to our knowledge, not as a conventional fusion with a yeast surface anchor protein. In most cases, antibody fragments are presented using the Aga2p-based display system, which permits either N-terminal or C-terminal fusion,Citation 4 but the lack of comparative studies does not facilitate the choice of the appropriate formats to be used.
ScFv molecules are the smallest functional entity (30 kDa) that includes the complete antigen-binding site of an antibody. This entity comprises the two variables domains VH and VL connected by a flexible linker that prevents their dissociation.
Fab are composed of the light chain (VL + CL) and a heavy chain (VH + CH1) connected by an interchain disulfide bond. Several studies have reported the expression of Fab molecules on the surface of yeast cells using either a bigenic plasmidCitation 13 or two distinct plasmidsCitation 10 , Citation 14 , Citation 19 to allow separate expression of the two chains. Expression levels of Fab molecules are generally considered as lower than those of scFv molecules in E. coli,Citation 20 , Citation 21 at the surface of phagesCitation 22 , Citation 23 and also on the surface of yeast cells.Citation 10
The scFab derived from Fab is obtained through introduction of a polypeptide linker connecting the light chain and the heavy chain. Thus, it combines some advantages of the scFv (single chain and one expression plasmid, level of expression) with the stability and lower tendency to aggregate of the Fab.Citation 2 , Citation 23 The linker prevents the display of unpaired heavy chain antibodies (devoid of light chain), which can sometimes interfere with the selection steps.Citation 2 ScFab molecules carry a flexible linker of variable length (32 to 80 amino acids) and an optional inter-chain disulfide bond.Citation 2 , Citation 24 Deletion of carboxy terminal cysteine residues (ΔC) of the CL and CH1 domains results in a scFabΔC format, which has increased expression levels in E. coli. Citation 23
The scFv is the most popular format for antibody engineering,Citation 4 probably because of its very high level of expression, its small size and the presence of a single peptide chain, which is an advantage for cloning or sequencing procedures. However, Fab and scFab formats have undeniable advantages, especially when it comes to stability or for reformatting to IgG molecules after engineering.Citation 22 To date, there is no clear literature consensus regarding the choice of an antibody fragment format suitable for engineering using yeast cell display. Also, no general study has yet explored the influence of antibody fragment formats on a diversity of antibodies in this particular context.
Here, we report the comparison of 4 antibody fragment formats – scFv, scFab with and without interchain disulfide bond and Fab – for the functional display of 6 antibodies. This study includes a first set of well-known antibodies – adalimumab, infliximab, golimumab and certolizumab – already used in clinical settings. This set of antibodies encompasses humanized, chimeric and fully human antibodies, which share TNF as a common antigen.Citation 25 The second set is composed of two chimeric antibodies targeting the protein IpaD from the needle tip of type III secretion system of Shigella spp obtained from mouse immunization. The objective of this study was to identify the most suitable format for the functional expression of antibody fragments on the surface of yeast cells.
Results
Conception of yeast surface display vectors allowing expression of scFv, scFab and Fab antibody fragments
To determine the influence of formats on the expression and functionality of antibody fragments, a new subset of yeast display plasmids was designed. Four plasmids were constructed for respective expression of scFv, scFab formats and Fab at the surface of S. cerevisiae cells. Those plasmids share a common backbone from previously described pCT-L7.5.1,Citation 26 including inducible promoter Gal 1–10 and Aga2p signal peptide []. Antibody fragments were displayed with a free N-terminus and tethered to the yeast cell wall by C-terminal fusion with Aga2p,Citation 27 in order to avoid possible steric hindrance of the fusion.
Figure 1. Schematic representation of yeast surface display vectors used in this study. Antibody fragment constructs are tethered to the cell wall of S. cerevisiae cells by C-terminal fusion with the Aga2p subunit of the a-agglutinin yeast cell surface anchor protein. Expression is controlled by the tightly regulated galactose-inducible Gal1 promoter. Aga2-ss: native signal sequence of Aga2p for secretion of the constructs. Single-chain antibody fragments (scFv, scFabΔC and sc60Fab) include flexible linkers connecting the light chain and the heavy chain. pNT Fab is a bigenic plasmid including two identical GAL1 promoters for respective expression of light and heavy chains. Cysteine residues responsible for the formation of the inter-chain disulfide bond connecting CK and CH1 are indicated in red. Expression of antibody fragments can be monitored using the HA tag for all constructs. A 6His-Tag sequence can also be used for the monitoring of heavy chain expression when using pNT Fab.
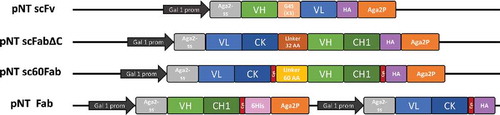
The plasmid pNT scFv encodes a typical scFv composed of VH/VL domains connected by a flexible linker (G4S)3. We also compared two previously described scFab formats, scFabΔCCitation 2 and sc60Fab,Citation 24 by constructing the plasmids pNT scFabΔC and pNT sc60Fab. ScFabs are composed of a VL-CK light chain connected to the VH-CH1 heavy chain by a flexible linker. The difference between the two formats resides in the nature and length of the linker (34 amino acids for scFabΔC versus 60 amino acids for sc60Fab) and the presence of an inter-chain disulfide bond in sc60Fab connecting the two C-terminal cysteine residues of CK and CH1. Display of Fab molecules on yeast without any linker was achieved using the bigenic plasmid pNT Fab where Gal 1–10 promoter and terminator are duplicated to obtain separate expression of the two chains.
Each plasmid includes a hemagglutinin (HA) tag to report expression of the complete antibody fragment. For single chain formats (scFv and scFabs), the HA tag is located between the antibody fragment and the Aga2p subunit. Full Fab expression can be controlled for pNT Fab through the expression of the light chain with the HA tag or by direct labeling with anti-CK antibody. Expression of heavy chain can be revealed by the HIS tag located between the heavy chain and Aga2p.
The 6 selected antibodies are IgG1 molecules with CK light chains. The sequences of TNF-specific antibodies were obtained from the IMGT database.Citation 28 The mAbs IpaD_301 and IpaD_318 were obtained by mouse immunization [data not shown] and engineered to generate chimeric antibodies. These two antibodies derive from the same parental genes IGHV1-14*01/ IGKV1-117*01 from Mus musculus and differ by only 7 and 8 mutations in the heavy chain and the light chain, respectively. Together, 24 plasmids corresponding to the sequences of 6 selected antibodies expressed in the 4 antibody formats were successfully cloned and transformed into yeast. An exhaustive analysis of the 24 constructs was performed in parallel to investigate the influence of the antibody fragment format on antigen binding.
Fab format grants functional expression of the largest number of antibodies
To assess the expression and functionality of the constructs, yeast cells were incubated with the appropriate biotinylated antigen at a concentration far above the expected KD to reach quantitative binding of the antibody fragments to their antigen. When evaluated by fluorescence-activated cell sorting (FACS), cells expressing scFvs of adalimumab, certolizumab, golimumab and IpaD_318 exhibited a high level of fluorescence, indicating quantitative binding of the antigen []. Weaker fluorescence levels were observed for IpaD_301 scFv, whereas no signal was observed for infliximab scFv, indicating antigen binding was weak or absent. Functionality of constructs displaying low or moderate antigen binding was not improved by increased induction times [Supp Data Fig. 3]. Antigen binding was specific, as demonstrated by the lack of cross-reactivity with the non-relevant antigen: anti-TNF antibodies showed no detectable IpaD binding and anti-IpaD antibodies showed no detectable TNF binding [Supp Data Fig. 1].
Figure 2. Antigen binding of the 6 selected antibodies expressed as different antibody fragment formats on the surface of yeast cells. Cells are labeled with biotinylated antigen then with Streptavidin–PE (Target Binding) before FACS analysis. Selected antigen concentrations are far above the expected KD to reach a quantitative binding with the antibody fragments. Respective antigen concentration were 20 nM for anti-TNF antibodies and 50 nM for anti-IpaD antibodies. For each construct, a FACS histogram shows the fluorescence intensity corresponding to antigen binding (x-axis) vs the number of detected events (y-axis). Interval gates (bars) are shown to differentiate cells with detectable antigen binding (mean fluorescence intensity (MFI) score above 103, in green) from cells with no detectable antigen binding (MFI score under 103, in grey).The 103 cutoff has been obtained by measuring MFI of cells with a non-relevant antigen, cf. Supplementary figure 1.
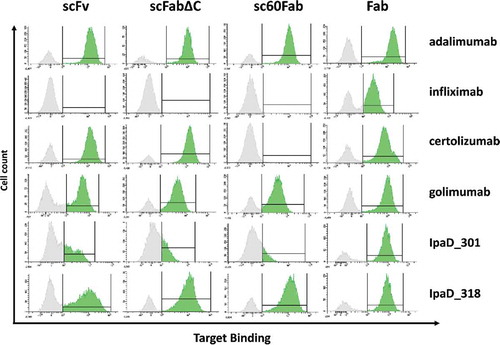
Yeast cells expressing scFab corresponding to adalimumab, golimumab and IpaD_318 displayed quantitative binding to their antigen, both as scFabΔC and sc60Fab formats. Infliximab and IpaD_301 in scFab formats were not or weakly functional, respectively, as attested by the absence of fluorescence for the corresponding cells. Interestingly, certolizumab scFabΔC showed significant binding to TNF, while certolizumab sc60Fab with the 60-amino-acid linker and inter-chain disulfide bond failed to bind TNF. All single-chain constructs were expressed at the surface of cells, as attested by labeling with anti-HA antibodies, with the notable exception of certolizumab sc60Fab [Supp Data Fig. 2]. Except for this specific construct, the absence of binding of the antibody fragments to their antigen could not be explained by lack of expression, but rather by a functional deficiency.
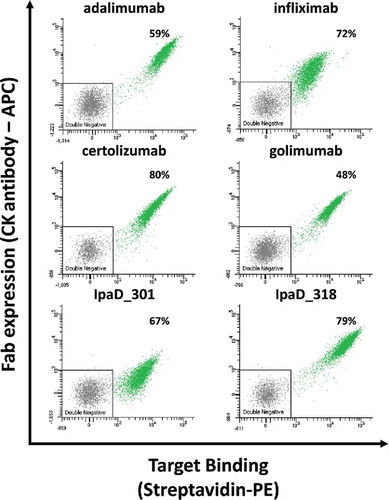
In contrast, all of the 6 tested antibodies bound to their antigen when expressed as Fab at the surface of yeast cells. For each antibody, cells expressing functional Fabs form homogeneous populations with a clear shift in fluorescence levels compared to non-expressing cells. The proportion of cells expressing functional Fabs is relatively high (from 46% to 74%).
In our set-up, Fab is the only viable format for functional expression of infliximab and IpaD_301 antibody fragments. Besides, for 5 of 6 antibodies tested, the fluorescence of cells expressing functional Fab was higher than the fluorescence of the equivalent cells expressing either scFvs or scFabs. This suggested that Fab molecules were not only active, but also well expressed. For these reasons, we focused on display of Fab molecules in the rest of the study.
Variations of expression in Fab format among antibodies do not affect functionality
We observed that maximal fluorescence levels of Fab-expressing cells vary among antibodies targeting a common antigen. More specifically, the mean fluorescence intensity of cells displaying functional infliximab Fabs was much lower than for adalimumab or golimumab, a phenomenon also observed for IpaD_301 when compared to IpaD_318.
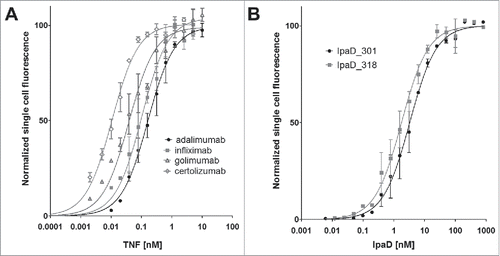
As variations of signal corresponding to antigen binding could be attributed either to differences of expression levels or to functional differences, dissociation constants (KD) were determined for each displayed Fab []. The measured KD values for IpaD_301 and IpaD_318 are 3.2 nM and 1.7 nM, respectively. This result is in accordance with the KD evaluated by OctetRed with measured affinities of 1.1 nM and 0.9 nM (Supplementary Fig. 4). Yeast expressing anti-TNF Fabs show subnanomolar KD, 161 pM for adalimumab, 102 pM for infliximab, 39 pM for golimumab and 11 pM for certolizumab. These KD values are consistent with the reported KD determined for the corresponding IgG molecules using SPR,Citation 25 indicating that Fabs expressed on the surface of yeast cells have a very strong affinity for their antigen.
Figure 3. Affinity of Fab antibody fragments expressed on the surface of yeast cells. The mean fluorescence intensity (MFI), determined by FACS analysis, is plotted against varying concentrations of antigen for every Fab construct, and fit to a monovalent binding model. Measurements were done in triplicate with independent cultures and inductions. (A) Equilibrium binding constants KD of the four anti-TNF Fabs for TNF at 20°C. Curve fitting results in a KD of 114 ± 6 nM for adalimumab Fab, 102 ± 14 pM for infliximab Fab, 39 ± 4 pM for golimumab Fab and 11 ± 1 pM for certolizumab Fab. (B) Equilibrium binding constants KD of the two anti-IpaD Fabs for IpaD at 20°C. KD IpaD_301 Fab = 3.2 ± 0.3 nM, KD IpaD_318 Fab = 1.7 ± 0.2 nM.
Interestingly, higher affinity was not correlated with higher level of binding fluorescence. While having a very good affinity for TNF, infliximab Fab displayed much weaker TNF binding signal compared to adalimumab Fab, suggesting differences in their expression levels. Thus, additional labeling experiments were performed to evaluate different expression reporters. In addition to antigen binding labeling, light chain expression was evaluated with anti-HA or anti-CK antibodies and heavy chain expression was assessed with anti-HIS antibodies [].
Figure 4. Antigen binding and expression levels of heavy and light chains of the 6 selected antibodies expressed as Fab on the surface of yeast cells. Cells were labeled with the appropriate expression reporter and the mean fluorescence intensity of positive cells was determined by FACS analysis. For antigen binding, cells were double-labeled with biotinylated antigen and Streptavidin–PE (target binding) before FACS analysis. (A) Heavy chain expression evaluated with anti-HIS antibody reporter. (B) Light chain expression evaluated with anti-HA antibody reporter (C) Biotinylated antigen binding evaluated with Streptavidin-PE (D) Light chain expression evaluated with anti-CK antibody reporter. Measurements were made in triplicate using cells from independent cultures.
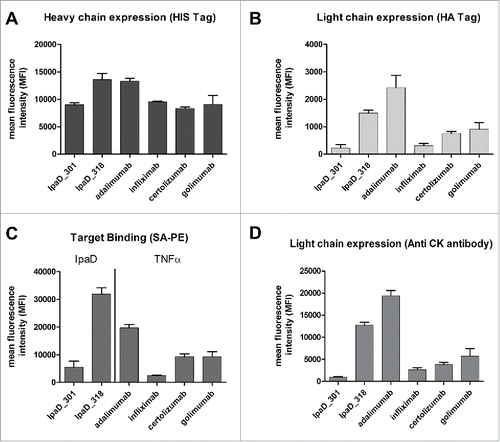
Heavy chain expression showed slight variations between the 6 selected antibodies [a]. Labeling with anti-HIS antibodies showed homogeneous expression of heavy chains, with IpaD_318 and adalimumab being somewhat above average. This observation contrasted with light chain expression, which showed strong differences across the Fab antibodies []. Light chain expression was significantly higher for IpaD_318 and adalimumab Fabs than for IpaD_301 or infliximab, while certolizumab and golimumab had intermediate light chain expression levels. Values observed for both reporters anti-HA or anti-CK were in good agreement, and the relative intensities of constructs were very similar. Anti-CK antibody yielded a very strong fluorescence signal, thus favoring easy discrimination of CK-expressing cells.
Antigen binding signals for IpaD_301 and IpaD_318 correlated well with their relative light chain expression levels [ and ]. Very similarly, the TNF binding signal was proportional to the respective expression levels of adalimumab, infliximab, certolizumab and golimumab light chain of the Fab []. Direct correlation between light chain expression and antigen binding was confirmed by simultaneous labeling with anti-CK antibody conjugated to allophycocyanin (APC) and streptavidin R-phycoerythrin (streptavidin-PE) [].
Figure 5. Representative bivariate flow cytometric analysis of yeast cells expressing Fab antibody fragments expressed on their surface. Each single dot on the plot designates the APC (y-axis) and PE (x-axis) fluorescence intensity values for a single yeast cell. Cells were double-labeled with biotinylated antigen/Streptavidin–PE (target binding) and anti-CK APC labels (Fab expression). Antigen concentrations in labeling experiments were 20 nM for anti-TNF antibodies and 50 nM for anti-IpaD antibodies. Functional subpopulations were gated using an inverted gate of the double negative sub-populations. The percentage number correspond to the mean percentage of cells expressing functional Fabs from three separate inductions.
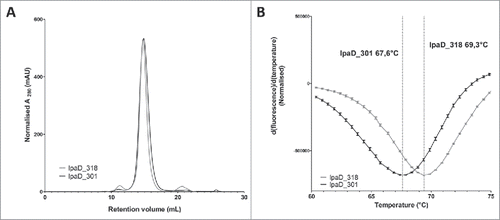
While having high sequence homology and similar affinities, IpaD_301 and IpaD_318 display significant differences in expression and antigen binding in all formats tested. For this reason, we investigated a possible correlation between the biophysical attributes of these two antibodies and their behavior on the surface of yeast cells. Thus, recombinant expression of complete IgG1 IpaD_301 and IpaD_318 was performed by transient transfection of HEK293 cellsCitation 29 followed by protein A purification. Production yield was slightly higher for IpaD_318 than for IpaD_301 (20 and 14 mg per liter, respectively). Size-exclusion chromatography revealed that both antibodies are predominantly produced as full-length IgG with low amounts of aggregates for IpaD_318 and even lower amounts for IpaD_301 []. In contrast, differential scanning fluorimetry (DSF) analysis reveals that IpaD_301 has a lower melting temperature (Tm) than IpaD_318, with Tm values of 67.6°C and 69.3°C, respectively [].
Figure 6. Biophysical characterization of IpaD_301 IgG and IpaD_318 IgG expressed by HEK cells. (A): Size-exclusion chromatograms of the protein A purified IgGs on a Superdex 200 10/300 GL size-exclusion column for IpaD_301 IgG (black) and IpaD_318 IgG (grey). Monomeric IgG species elutes at 15 mL and aggregates or multimeric species elutes between 10 to 12 mL. (B): Tm determination of IgGs by differential scanning fluorimetry. The fluorescence intensities are normalized to peak values. Error bars correspond to experiment triplicates.
Discussion
We carried out a comparative study of expression formats using 6 different antibodies specific for two different antigens, TNF and IpaD. In our hands, Fab appears to be the most versatile expression format for display of functional antibodies on the surface of yeast cells.
ScFv is currently the most widespread format for antibody engineering. It has indisputable advantages, such as its small size, which is particularly convenient for the cloning and sequencing of libraries. Also, numerous studies have used scFvs successfully to engineer clones with improved properties,Citation 4 including clones derived from adalimumab with improved affinity towards TNF.Citation 8 However, some antibodies display reduced affinity and impaired biophysical properties when expressed as scFv as compared to the corresponding Fab molecules.Citation 23 , Citation 30 Indeed, CH1 and CL domains, which are not present in scFv molecules, participate in the greater stability of Fab and scFab.Citation 31 , Citation 32 The choice of an adapted format has also proved to be an important parameter for other approaches such as phage displayCitation 33 , Citation 34 or recombinant expression of antibody fragments.Citation 35
Here, we show that some antibodies do not seem to be active either in the scFv format or in the scFab format when they are expressed at the surface of yeast cells. In our set-up, two antibodies, infliximab and IpaD_301, appeared to lack functionality, as suggested by the total absence of antigen binding in any single-chain format. The presence of CK and CH1 domains, which could have a stabilizing role in the scFab formats, had no effect on the functionality of the corresponding infliximab and IpaD_301 constructs. We cannot exclude that the presence of a linker between the light chain and the heavy chain could also cause steric hindrance that impairs functionality.
We found that adalimumab, golimumab and IpaD_318 were well expressed and perfectly able to bind their antigen as scFv constructs, but also in scFab or Fab formats. Certolizumab was also a very well expressed and functional antibody in these formats, with the notable exception of the sc60Fab format, which exhibited very poor expression and had no detectable antigen binding (Supp. Fig 2). These particular results emphasize that the presence of the two C-terminal cysteine residues in scFab, as well as the nature and length of the linker sequence, can dramatically alter expression of scFab molecules for some antibodies. Moreover, the presence of the interchain disulfide bond can have a major influence on the stability of the antibody fragment, as recently shown for the adalimumab Fab.Citation 36 The length of the linker also proved to be an important parameter for the proper packing of light and heavy chains and possibly the accessibility of the paratope, thus ultimately affecting the function of the scFab fragment.Citation 23 , Citation 24
In this study set-up, Fab was the only format that allowed functional display for each of the 6 tested antibodies. FACS analysis revealed that a very high proportion of induced yeast cells (up to 70%) expressed Fabs and produced a strong fluorescence signal corresponding to antigen binding. In the context of protein engineering or affinity maturation, a strong signal would favor an easy discrimination of high-affinity clones in libraries containing very high numbers of mutants.
Moreover, for anti-TNF molecules, all Fabs expressed on the surface of yeast cells exhibited a high affinity for their antigen, with KD values close to those reported in the literature, strongly suggesting that the KD of a protein-protein binding interaction measured on the surface of yeast cells was essentially equal to that measured using soluble proteins.Citation 37 , Citation 38
Deeper analysis of expression reporters in the Fab format indicated a lower display level of light chains for infliximab and IpaD_301 Fabs. Light chain display correlated perfectly with lower target binding of these two constructs compared to the other anti-TNF Fabs or IpaD_318 Fab, respectively. Coexpression of chaperones such as BiP, use of alternative promoters such as Gal10 for the light chain expression, or introduction of a new secretory leader sequence optimized for antibody secretion such as αMFpp839 might be useful to promote the assembly of light and heavy chains of antibodies during Fab expression. A recent study demonstrated that co-expression of protein disulfide isomerase (PDI) had a positive effect on the functionality of some antibodies expressed as Fab on the surface of yeast cells.Citation 40 In this study, Georgiou and co-workers also showed that pairing of antibody chains could be promoted by fusing the light and heavy chains with dimerization domains such as leucine-zipper proteins. Forced-dimerization of antibody chains restored functionality for 3 of 13 tested anti-HA antibodies, although with a lower proportion of cells displaying functional Fab.
Overall, infliximab and IpaD_301 exhibited relatively low expression in every tested format of antibody fragment. We observed, however, marked differences in the expression profiles of IpaD_301 and IpaD_318 on the surface of yeast cells in spite of their high sequence homology, with only 7 and 8 mutations in the heavy chain and the light chain, respectively. Production of these two antibodies in a full-length IgG format reveal that IpaD_301 has lower production titers in HEK cells and also a lower Tm (67.6°C vs 69.3°C). Interestingly, infliximab is also described as difficult to express in HEK cellsCitation 41 and CHO cells,Citation 42 with relatively low stability over time.Citation 43 A comprehensive study comparing the biophysical properties of clinical-stage antibodies indicated that infliximab has much lower production titers in HEK cells than adalimumab, certolizumab and golimumab (6 mg/L for infliximab vs more than 100 mg/mL for the three other anti-TNF antibodies).Citation 41 Wittrup and co-workers also reported that infliximab had a lower Tm (64.5°C vs at least 70°C for the three other anti-TNF antibodies). These observations are consistent with previously published studies indicating that protein secretion by yeast cells is correlated with thermodynamic stability.Citation 44 , Citation 45 Together, this suggests that antibodies with low thermodynamic stability might have low expression on the surface of yeast cells.
Even as many alternative antibody formats are emerging, most of current therapeutic antibodies on the market are antibodies in the IgG format.Citation 21 Thus, it is essential that the conversion between the screening format – the antibody fragment – and the IgG antibody does not impair the desired properties, and in particular affinity for the antigen. From this perspective, use of Fab has strong advantages. Several reports show that the Fab format tends to retain its binding affinity better than scFv when converted into IgG.Citation 19 Moreover, the presence of a peptide linker in the scFv and scFab formats or introduction of additional domains such as leucine-zipper dimerization proteins might introduce bias in the screening of improved clones because of subtle structural changes. Indeed, some affinity-enhanced molecules engineered in scFv and scFabΔC formats lost affinity after conversion into IgG.Citation 22
In conclusion, in spite of their practical convenience, single-chain antibody fragment formats such as scFv and scFab do not always lead to a functional display of antibodies at the surface of yeast cells. In contrast, we showed that all 6 tested antibodies were functional in the Fab format, with high affinities for their antigen. Using our Fab expression plasmid, high proportions of yeast cells expressed Fab molecules and exhibited strong antigen binding signals in FACS, two important properties when getting started with the affinity maturation process by yeast display in good conditions. These advantages and the lower risk of losing engineered properties upon conversion into IgG suggest that the Fab format is the best alternative for the engineering of antibodies using YSD.
Material and methods
Yeast display vectors construction
Cloning were performed with the SLiCE recombination cloning methodCitation 46 or classic restriction/ligature method using E.coli DH5α strain (Invitrogen). For each anti-TNF antibody, optimized synthetic genes (VH and VL) were ordered from Eurofins Genomics based on the IMGT sequences. All expression plasmids are derived from previously described pCT-L7.5.126 and share the CEN/ARS replication origin, TRP auxotrophy marker, colE1 replication origin, Ampicillin resistance gene and galactose inducible promoter GAL1. Plasmid pCT-L7.5.1 was a gift from Prof. K. Dane Wittrup (Addgene plasmid # 42900).
Plasmids pNT scFv were modified from parental pCT-L7.5.1 plasmid by modifying the aga2p signal sequence with a dipeptide spacerCitation 27 and a NheI restriction site. A stop codon was inserted at the 3′ end of aga2p gene to obtain plasmids allowing the display of scFv with a free N-terminus end and a C-terminal anchoring to the yeast cell wall. Synthetic genes corresponding to each scFv were then cloned using NheI and SpeI restriction sites.
The pNT scFabΔC vector was generated by cloning the coding sequence of the scFabΔC of the previously described pYDscFabCitation 2 between the restriction sites NheI and XhoI of the pNT scFv plasmid. Plasmid pYDscFab was a gift from Prof. Dennis Burton. Heavy chains and light chains of antibodies were cloned between NheI/Pfl23II and NcoI/SalI restrictions sites, respectively.
The pNT sc60Fab vectors were generated from pNT scFab plasmids. The linker and the cysteine residues allowing interchain disulfide bond formation were modified by PCR and homologous recombination between BssHII and NcoI sites. Heavy chains and light chains of antibodies were cloned between NheI-Pfl23II and NcoI-SalI restriction sites, respectively.
The bigenic pNT Fab vector was generated by ligation into pNT scFv between SpeI and XhoI of a synthetic gene (Eurofins Genomics) containing a 6-HIS tag, the Aga2p domain, a Gal1 promoter, an aga2p signal sequence, an HA tag and a transcription terminator. The resulting plasmid has two distinct expression cassettes corresponding to respective expression of light and heavy chains of the Fabs. Heavy chains and light chains of antibodies were then cloned between NheI/Pfl23II and NcoI/SalI restrictions sites, respectively.
Yeast transformation
Preparation of competent yeast cells EBY100 (ATCC® MYA-4941™; a GAL1-AGA1:: URA3 ura3-52 trp1 leu2 Δ1 his3 Δ200 pep4:: HIS2 prb1Δ1.6R can1 GAL) was performed according to Suga et al.Citation 47 Then, 100 µL of EBY100 electro-competent cells were mixed with 1–2 µg plasmid DNA, transferred to a pre-chilled electroporation cuvette (Biorad, 165–2086) and pulsed at 2.5 kV, 25 µF (Biorad Gene Pulser Xcell). Yeast cells were then diluted in 1 mL of sorbitol (1 M), and 200 µL were streaked on SD-CAA agar plates [6.7 g/L yeast nitrogen base without casamino acids, 20 g/L dextrose, 5 g/L casamino acids, 100 mM sodium phosphate pH 6.0].
Growth and expression conditions
Yeast cultures were performed in 24 deep-well round-bottom plates. Pre-cultures were performed by inoculating 3 mL of SD-CAA medium with one colony from selective agar plate and incubated overnight at 30°C, 200 rpm. The saturated pre-culture (typically OD600 of 8–10) was passaged in order to obtain an initial culture OD600 of 0.25-0.50. The culture was grown at 30°C until its OD600 reached 0.5-1.0. Cells were centrifuged and re-suspended in 3 mL of SG-CAA galactose induction medium [6.7 g/L yeast nitrogen base without casamino acids, 20 g/L galactose, 5 g/L casamino acids, 100 mM sodium phosphate, pH 6.0] and induced for 16–36 h at 20°C, 200 rpm.
Yeast labelling and flow cytometry analysis
106 cells were taken from the induced culture and washed with 1 mL PBSF (phosphate-buffered saline (PBS), bovine serum albumin (BSA) 0.1%) buffer. Cells were resuspended in 50 µL of the target protein at the desired concentration and incubated at 20°C, 1000 rpm until equilibrium is reached (depending on target protein concentration). Cells were washed with 1 mL of ice-cold PBSF to avoid dissociation and resuspended in ice-cold PBSF containing the appropriate fluorescent reporters: Streptavidin-PE (Thermo Fisher scientific; catalog number S866; 1:100 dilution), Mouse anti-Human Kappa Light Chain Antibody APC conjugate (Thermo Fisher scientific; catalog number MH10515; 1:100 dilution), 6x-His Epitope Tag antibody Dylight-650 conjugate (Thermo Scientific, catalog number MA1-21315-D650; 1:100 dilution) or HA Tag Monoclonal Antibody APC conjugate (1:50 dilution). Cells were incubated on ice in the dark for 15 minutes and analyzed with BD FACS Aria™ III cytometer.
Cell-Binding assays for determining the affinity of antibodies for their antigen
Cell binding assays were performed according to Hunter et al.Citation 38 in order to avoid ligand depletion. 104 to 105 cells were incubated in 50–500 µL of the target protein at the desired concentrations and incubated at 20°C, 1000 rpm long enough to reach equilibrium. Cells were analyzed by FACS for their binding fluorescence as described above. Auto-fluorescence was subtracted for each measure and resulting values were normalized with maximum fluorescence. Data are then fitted with a non-linear regression analysis to obtain the KD.
Production and purification of IgG IpaD_301 and IpaD_318
IpaD_301 and IpaD_318 antibodies were produced via transient transfection of HEK293 Freestyle™ cells (HEK 293FS). HEK 293FS were transfected as previously described by Subedi et al. Citation 48 without the use of Valproic acid using the HEK transient transfection plasmids described by Smith et al. Citation 29
The day before transfection, cells were sub-cultured in order to be at a density of 2–3.106 cells/mL the next day. The day of the transfection, cells were centrifuged 5 min at 100 g and resuspended in fresh FreeStyle 293 Expression medium (Thermo-Fisher) to obtain a density of 2.5.106 cells/mL. For a transfection volume of 100 mL, 150 µg of each plasmid (coding for heavy chain and light chain) was added to the cells at a concentration of 0.5 µg/µL. Cells were incubated 5 min at 37°C, 180 rpm, 8% CO2. 1.8 mL of polyethylenimine (PEI) was added at a concentration of 0.5 mg/mL to the cells (PEI:DNA ratio = 3:1). Cells were subsequently incubated 24h at 37°C, 180 rpm, 8% CO2.
A day after transfection, 100 mL of fresh EX-CELL® Serum-Free Medium for HEK 293 (Sigma-Aldrich), supplemented with 6 mM L-Glutamine, was added to the transfected culture. Cells were finally incubated 4 more days at 37°C, 180 rpm, 8% CO2. Five days after transfection, the supernatant was harvested by centrifuging the cells at 1000g for 15 min at 4°C and kept on ice until purification. The supernatant was purified using a HiTrap™ Protein A HP column of 1 mL (GE Healthcare) on an ÄKTA purifier (GE Healthcare) according to the manufacturer's description.
Size-exclusion chromatography
A Superdex-200 10/300 GL (GE Healthcare) column was used on an ÄKTA purifier (GE Healthcare). Runs were monitored with Unicorn™ 5.31 software (GE Healthcare). 500 μL of each sample was applied on the column using a flow rate of 0.7 mL/min and PBS as running buffer.
Fab IpaD_301 and IpaD_318 affinity determination
Fab were prepared from full-length IgG using a Pierce® Fab Micro Preparation Kit. The binding affinity of Fab IpaD_301 and IpaD_318 was evaluated by biolayer interferometry using an Octet RED96 instrument (Pall ForteBio). Biotinylated IpaD was loaded onto streptavidin biosensors (ForteBio) at a concentration of 5 µg/mL for 8 min in PBSF (1x PBS, pH 7.4, 0.1% BSA). Fab IpaD_301 and IpaD_318 were then associated with IpaD using 4 different concentrations (20, 10, 5 and 2.5 nM). Duration of association and dissociation steps were 20 min and 120 min. All steps were performed at 20°C. Data were collected and analyzed using Octet Software version 10.0.3 (Pall ForteBio). Binding kinetics were fitted using a 1:1 Langmuir-binding model.
Thermostability evaluation
DSF was performed as previously described by Niesen et al. Citation 49 using a StepOne Real-Time PCR System from Applied Biosystems. IgG monomers of IpaD_301 and IpaD_318 purified via SEC were mixed with the SYPRO Orange reagent. Final dye concentration was 5x and final IgG concentration was 150 µg/mL. The temperature gradient was run from 20 to 90 °C with 1 min equilibration at each degree centigrade.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Abbreviations
| BSA | = | bovine serum albumin |
| DSF | = | differential scanning fluorimetry |
| Fab | = | antigen-binding fragment |
| MFI | = | mean fluorescence intensity |
| PDI | = | protein disulfide isomerase |
| PEI | = | polyethylenimine |
| scFab | = | single-chain antigen-binding fragment |
| scFv | = | single-chain variable fragment |
| SEC | = | size exclusion chromatography |
| TNF | = | tumor necrosis factor |
| YSD | = | yeast surface display |
2018MABS1709R-s01.pdf
Download PDF (544.6 KB)Acknowledgments
The authors would like to thank Dr Magali Aumont-Nicaise for her assistance with differential scanning fluorimetry experiments and Dr Hervé Volland for anti-HA antibodies.
References
- Chao G, Lau WL, Hackel BJ, Sazinsky SL, Lippow SM, Wittrup KD. Isolating and engineering human antibodies using yeast surface display. Nat Protoc. 2006;1:755–68. doi:10.1038/nprot.2006.94. PMID:17406305.
- Walker LM, Bowley DR, Burton DR. Efficient recovery of high-affinity antibodies from a single-chain Fab yeast display library. J Mol Biol. 2009;389:365–75. doi:10.1016/j.jmb.2009.04.019. PMID:19376130.
- Wang B, Lee CH, Johnson EL, Kluwe CA, Cunningham JC, Tanno H, Crooks RM, Georgiou G. Discovery of high affinity anti-ricin antibodies by B cell receptor sequencing and by yeast display of combinatorial VH:VL libraries from immunized animals. mAbs. 2016;8:1035–44. doi:10.1080/19420862.2016.1190059. PMID:27224530.
- Boder ET, Raeeszadeh-Sarmazdeh M, Price JV. Engineering antibodies by yeast display. Arch Biochem Biophys. 2012;526:99–106. doi:10.1016/j.abb.2012.03.009. PMID:22450168.
- Pepper LR, Cho YK, Boder ET, Shusta EV. A decade of yeast surface display technology: where are we now? Comb Chem High Throughput Screen. 2008;11:127–34. doi:10.2174/138620708783744516. PMID:18336206.
- Angelini A, Chen TF, de Picciotto S, Yang NJ, Tzeng A, Santos MS, Van Deventer JA, Traxlmayr MW, Wittrup KD. Protein engineering and selection using yeast surface display. Methods Mol Biol. 2015;1319:3–36. doi:10.1007/978-1-4939-2748-7_1. PMID:26060067.
- Gera N, Hussain M, Rao BM. Protein selection using yeast surface display. Methods. 2013;60:15–26. doi:10.1016/j.ymeth.2012.03.014. PMID:22465794.
- Rajpal A, Beyaz N, Haber L, Cappuccilli G, Yee H, Bhatt RR, Takeuchi T, Lerner RA, Crea R. A general method for greatly improving the affinity of antibodies by using combinatorial libraries. Proc Natl Acad Sci U S A. 2005;102:8466–71. doi:10.1073/pnas.0503543102. PMID:15939870.
- Boder ET, Midelfort KS, Wittrup KD. Directed evolution of antibody fragments with monovalent femtomolar antigen-binding affinity. Proc Natl Acad Sci U S A. 2000;97:10701–5. doi:10.1073/pnas.170297297. PMID:10984501.
- Lou J, Geren I, Garcia-Rodriguez C, Forsyth CM, Wen W, Knopp K, Brown J, Smith T, Smith LA, Marks JD. Affinity maturation of human botulinum neurotoxin antibodies by light chain shuffling via yeast mating. Protein Eng Des Sel. 2010;23:311–9. doi:10.1093/protein/gzq001. PMID:20156888.
- Graff CP, Chester K, Begent R, Wittrup KD. Directed evolution of an anti-carcinoembryonic antigen scFv with a 4-day monovalent dissociation half-time at 37 degrees C. Protein Eng Des Sel. 2004;17:293–304. doi:10.1093/protein/gzh038. PMID:15115853.
- Zhou Y, Drummond DC, Zou H, Hayes ME, Adams GP, Kirpotin DB, Marks JD. Impact of single-chain Fv antibody fragment affinity on nanoparticle targeting of epidermal growth factor receptor-expressing tumor cells. J Mol Biol. 2007;371:934–47. doi:10.1016/j.jmb.2007.05.011. PMID:17602702.
- van den Beucken T, Pieters H, Steukers M, van der Vaart M, Ladner RC, Hoogenboom HR, Hufton SE. Affinity maturation of Fab antibody fragments by fluorescent-activated cell sorting of yeast-displayed libraries. FEBS Lett. 2003;546:288–94. doi:10.1016/S0014-5793(03)00602-1. PMID:12832056.
- Schroter C, Gunther R, Rhiel L, Becker S, Toleikis L, Doerner A, Becker J, Schönemann A, Nasu D, Neuteboom B, et al. A generic approach to engineer antibody pH-switches using combinatorial histidine scanning libraries and yeast display. mAbs. 2015;7:138–51. doi:10.4161/19420862.2014.985993. PMID:25523975.
- Garcia-Rodriguez C, Levy R, Arndt JW, Forsyth CM, Razai A, Lou J, Geren I, Stevens RC, Marks JD. Molecular evolution of antibody cross-reactivity for two subtypes of type A botulinum neurotoxin. Nat Biotechnol. 2007;25:107–16. doi:10.1038/nbt1269. PMID:17173035.
- Rosowski S, Becker S, Toleikis L, Valldorf B, Grzeschik J, Demir D, Willenbücher I, Gaa R, Kolmar H, Zielonka S, et al. A novel one-step approach for the construction of yeast surface display Fab antibody libraries. Microb Cell Fact. 2018;17:3. doi:10.1186/s12934-017-0853-z. PMID:29316915.
- Rhiel L, Krah S, Gunther R, Becker S, Kolmar H, Hock B. REAL-Select: full-length antibody display and library screening by surface capture on yeast cells. PLoS One. 2014;9:e114887. doi:10.1371/journal.pone.0114887. PMID:25501029.
- Rakestraw JA, Aird D, Aha PM, Baynes BM, Lipovsek D. Secretion-and-capture cell-surface display for selection of target-binding proteins. Protein Eng Des Sel. 2011;24:525–30. doi:10.1093/protein/gzr008. PMID:21402751.
- Weaver-Feldhaus JM, Lou JL, Coleman JR, Siegel RW, Marks JD, Feldhaus MJ. Yeast mating for combinatorial Fab library generation and surface display. FEBS Lett. 2004;564:24–34. doi:10.1016/S0014-5793(04)00309-6. PMID:15094038.
- Gaciarz A, Veijola J, Uchida Y, Saaranen MJ, Wang C, Horkko S, Ruddock LW. Systematic screening of soluble expression of antibody fragments in the cytoplasm of E. coli. Microb Cell Fact. 2016;15:22. doi:10.1186/s12934-016-0419-5. PMID:26809624.
- Holliger P, Hudson PJ. Engineered antibody fragments and the rise of single domains. Nat Biotechnol. 2005;23:1126–36. doi:10.1038/nbt1142. PMID:16151406.
- Steinwand M, Droste P, Frenzel A, Hust M, Dubel S, Schirrmann T. The influence of antibody fragment format on phage display based affinity maturation of IgG. mAbs. 2014;6:204–18. doi:10.4161/mabs.27227. PMID:24262918.
- Hust M, Jostock T, Menzel C, Voedisch B, Mohr A, Brenneis M, Kirsch MI, Meier D, Dübel S. Single chain Fab (scFab) fragment. BMC Biotechnol. 2007;7:14. doi:10.1186/1472-6750-7-14. PMID:17346344.
- Koerber JT, Hornsby MJ, Wells JA. An improved single-chain Fab platform for efficient display and recombinant expression. J Mol Biol. 2015;427:576–86. doi:10.1016/j.jmb.2014.11.017. PMID:25481745.
- van Schie KA, Ooijevaar-de Heer P, Dijk L, Kruithof S, Wolbink G, Rispens T. Therapeutic TNF inhibitors can differentially Stabilize Trimeric TNF by inhibiting monomer exchange. Scientific Reports. 2016;6:32747. doi:10.1038/srep32747. PMID:27605058.
- Hackel BJ, Kapila A, Wittrup KD. Picomolar affinity fibronectin domains engineered utilizing loop length diversity, recursive mutagenesis, and loop shuffling. J Mol Biol. 2008;381:1238–52. doi:10.1016/j.jmb.2008.06.051. PMID:18602401.
- Wang Z, Mathias A, Stavrou S, Neville DM, Jr. A new yeast display vector permitting free scFv amino termini can augment ligand binding affinities. Protein Eng Des Sel. 2005;18:337–43. doi:10.1093/protein/gzi036. PMID:15976011.
- Lefranc MP, Giudicelli V, Duroux P, Jabado-Michaloud J, Folch G, Aouinti S, Carillon E, Duvergey H, Houles A, Paysan-Lafosse T, et al. IMGT(R), the international ImMunoGeneTics information system(R) 25 years on. Nucleic Acids Res. 2015;43:D413–22. doi:10.1093/nar/gku1056. PMID:25378316.
- Smith K, Garman L, Wrammert J, Zheng NY, Capra JD, Ahmed R, Wilson PC. Rapid generation of fully human monoclonal antibodies specific to a vaccinating antigen. Nat Protoc. 2009;4:372–84. doi:10.1038/nprot.2009.3. PMID:19247287.
- Worn A, Pluckthun A. Stability engineering of antibody single-chain Fv fragments. J Mol Biol. 2001;305:989–1010. doi:10.1006/jmbi.2000.4265. PMID:11162109.
- Rothlisberger D, Honegger A, Pluckthun A. Domain interactions in the Fab fragment: a comparative evaluation of the single-chain Fv and Fab format engineered with variable domains of different stability. J Mol Biol. 2005;347:773–89. doi:10.1016/j.jmb.2005.01.053. PMID:15769469.
- Quintero-Hernandez V, Juarez-Gonzalez VR, Ortiz-Leon M, Sanchez R, Possani LD, Becerril B. The change of the scFv into the Fab format improves the stability and in vivo toxin neutralization capacity of recombinant antibodies. Mol Immunol. 2007;44:1307–15. doi:10.1016/j.molimm.2006.05.009. PMID:16814388.
- Chan CE, Chan AH, Lim AP, Hanson BJ. Comparison of the efficiency of antibody selection from semi-synthetic scFv and non-immune Fab phage display libraries against protein targets for rapid development of diagnostic immunoassays. J Immunol Methods. 2011;373:79–88. doi:10.1016/j.jim.2011.08.005. PMID:21856306.
- Hust M, Jostock T, Menzel C, Voedisch B, Mohr A, Brenneis M, Kirsch MI, Meier D, Dübel S. Single chain Fab (scFab) fragment. BMC Biotechnol. 2007;7:14. doi:10.1186/1472-6750-7-14. PMID:17346344.
- Rao MF, Li YJ, Dong JX, Wu WJ, Xu ZL, Sun YM, et al. Production and characterization of a single-chain Fab fragment for the detection of O,O-diethyl organophosphorus pesticides. Anal Methods-Uk. 2016;8:3140–7. doi:10.1039/C6AY00224B.
- Nakamura H, Oda-Ueda N, Ueda T, Ohkuri T. A novel engineered interchain disulfide bond in the constant region enhances the thermostability of adalimumab Fab. Biochem Biophys Res Commun. 2018;495:7–11. doi:10.1016/j.bbrc.2017.10.140. PMID:29097200.
- Cherf GM, Cochran JR. Applications of yeast surface display for protein engineering. Methods Mol Biol. 2015;1319:155–75. doi:10.1007/978-1-4939-2748-7_8. PMID:26060074.
- Hunter SA, Cochran JR. Cell-binding assays for determining the affinity of protein-protein interactions: Technologies and considerations. Methods Enzymol. 2016;580:21–44. doi:10.1016/bs.mie.2016.05.002. PMID:27586327.
- Rakestraw JA, Sazinsky SL, Piatesi A, Antipov E, Wittrup KD. Directed evolution of a secretory leader for the improved expression of heterologous proteins and full-length antibodies in Saccharomyces cerevisiae. Biotechnol Bioeng. 2009;103:1192–201. doi:10.1002/bit.22338. PMID:19459139.
- Wang B, DeKosky BJ, Timm MR, Lee J, Normandin E, Misasi J, Kong R, McDaniel JR, Delidakis G, Leigh KE, et al. Functional interrogation and mining of natively paired human VH:VL antibody repertoires. Nat Biotechnol. 2018;36(2):152–15. doi:10.1038/nbt.4052. PMID:29309060.
- Jain T, Sun T, Durand S, Hall A, Houston NR, Nett JH, Sharkey B, Bobrowicz B, Caffry I, Yu Y, et al. Biophysical properties of the clinical-stage antibody landscape. Proc Natl Acad Sci U S A. 2017;114:944–9. doi:10.1073/pnas.1616408114. PMID:28096333.
- Le Fourn V, Girod PA, Buceta M, Regamey A, Mermod N. CHO cell engineering to prevent polypeptide aggregation and improve therapeutic protein secretion. Metab Eng. 2014;21:91–102. doi:10.1016/j.ymben.2012.12.003. PMID:23380542.
- Lerch TF, Sharpe P, Mayclin SJ, Edwards TE, Lee E, Conlon HD, Polleck S, Rouse JC, Luo Y, Zou Q. Infliximab crystal structures reveal insights into self-association. mAbs. 2017;9:874–83. doi:10.1080/19420862.2017.1320463. PMID:28421849.
- Kowalski JM, Parekh RN, Mao J, Wittrup KD. Protein folding stability can determine the efficiency of escape from endoplasmic reticulum quality control. J Biol Chem. 1998;273:19453–8. doi:10.1074/jbc.273.31.19453. PMID:9677365.
- Kowalski JM, Parekh RN, Wittrup KD. Secretion efficiency in Saccharomyces cerevisiae of bovine pancreatic trypsin inhibitor mutants lacking disulfide bonds is correlated with thermodynamic stability. Biochemistry. 1998;37:1264–73. doi:10.1021/bi9722397. PMID:9477952.
- Zhang Y, Werling U, Edelmann W. SLiCE: a novel bacterial cell extract-based DNA cloning method. Nucleic Acids Res. 2012;40:e55. doi:10.1093/nar/gkr1288. PMID:22241772.
- Suga M, Isobe M, Hatakeyama T. Cryopreservation of competent intact yeast cells for efficient electroporation. Yeast. 2000;16:889–96. doi:10.1002/1097-0061(200007)16:10%3c889::AID-YEA582%3e3.0.CO;2-R. PMID:10870100.
- Subedi GP, Johnson RW, Moniz HA, Moremen KW, Barb A. High yield expression of recombinant human proteins with the transient transfection of HEK293 cells in suspension. J Vis Exp. 2015;106:e53568. PMID:26779721.
- Niesen FH, Berglund H, Vedadi M. The use of differential scanning fluorimetry to detect ligand interactions that promote protein stability. Nat Protoc. 2007;2:2212–21. doi:10.1038/nprot.2007.321. PMID:17853878.
