?Mathematical formulae have been encoded as MathML and are displayed in this HTML version using MathJax in order to improve their display. Uncheck the box to turn MathJax off. This feature requires Javascript. Click on a formula to zoom.
?Mathematical formulae have been encoded as MathML and are displayed in this HTML version using MathJax in order to improve their display. Uncheck the box to turn MathJax off. This feature requires Javascript. Click on a formula to zoom.ABSTRACT
Accurate prediction of the human pharmacokinetics (PK) of a candidate monoclonal antibody from nonclinical data is critical to maximize the success of clinical trials. However, for monoclonal antibodies exhibiting nonlinear clearance due to target-mediated drug disposition, PK predictions are particularly challenging. That challenge is further compounded for molecules lacking cross-reactivity in a nonhuman primate, in which case a surrogate antibody selective for the target in rodent may be required. For these cases, prediction of human PK must account for any interspecies differences in binding kinetics, target expression, target turnover, and potentially epitope. We present here a model-based method for predicting the human PK of MAB92 (also known as BI 655130), a humanized IgG1 monoclonal antibody directed against human IL-36R. Preclinical PK was generated in the mouse with a chimeric rat anti-mouse IgG2a surrogate antibody cross-reactive against mouse IL-36R. Target-specific parameters such as antibody binding affinity (KD), internalization rate of the drug target complex (kint), target degradation rate (kdeg), and target abundance (R0) were integrated into the model. Two different methods of assigning human R0 were evaluated: the first assumed comparable expression between human and mouse and the second used high-resolution mRNA transcriptome data (FANTOM5) as a surrogate for expression. Utilizing the mouse R0 to predict human PK, AUC0-∞ was substantially underpredicted for nonsaturating doses; however, after correcting for differences in RNA transcriptome between species, AUC0-∞ was predicted largely within 1.5-fold of observations in first-in-human studies, demonstrating the validity of the modeling approach. Our results suggest that semi-mechanistic models incorporating RNA transcriptome data and target-specific parameters may improve the predictivity of first-in-human PK.
Introduction
MAB92, also known as BI 655130, is a humanized IgG1 monoclonal antibody engineered for reduced effector function and directed against the human cell-surface receptor, IL1RL2 (IL-36R). Signaling of IL-36R is induced by heterotrimeric binding with its co-receptor, IL-1 receptor accessory protein (IL-1RAP), and one of the three IL-36R cognate agonistic ligands, such as, IL36α, IL-36β, or IL-36γ, resulting in downstream activation of NF-
B and MAPKs and expression of proinflammatory and profibrotic mediators.Citation1–Citation7 An additional ligand, IL-36Ra, competes with the aforementioned ligands, thereby acting as a natural antagonist of IL-36R signaling.Citation1,Citation8 A strong link has been established between IL-36R signaling and skin inflammation as demonstrated by the occurrence of generalized pustular psoriasis in patients with a loss-of-function mutation in IL-36Ra.Citation4,Citation5,Citation8,Citation9 IL-36R agonist ligands are upregulated in psoriatic tissue, and accumulating evidence suggests that the IL-36R signaling pathway plays a role in the pathogenesis of psoriatic and rheumatoid arthritis,Citation10 asthma, chronic obstructive pulmonary disease,Citation11 and inflammatory bowel disease,Citation12–Citation14 making IL-36R signaling an attractive target for therapeutic intervention in the aforementioned and other epithelial-mediated inflammatory diseases. IL-36R is reported to be expressed on dendritic cells, CD4+ T cells, intestinal lymphocytes, and synovial fibroblasts.Citation15 In-house immunohistochemistry (IHC) data for MAB92 in human tissue showed mostly mild-to-moderate staining in a variety of epithelial tissues (bladder, breast, eye, esophagus, lung, pituitary, prostate, salivary gland, skin, thymus, tonsil, ureter, and cervix).
MAB92 shows species-specific binding with high affinity against human IL-36R and > 2000-fold reduced affinity towards IL-36R in mouse, rat, hamster, mini pig, and nonhuman primates (cynomolgus, rhesus, and marmoset). As MAB92 targets a cell-surface receptor, target-mediated drug disposition (TMDD) resulting from internalization and subsequent degradation of the molecule was expected to contribute to overall clearance of the antibody. In order to enable in vivo preclinical studies, we identified a chimeric rat anti-mouse mAb, MAB04 (also known as BI 674304), targeted against mouse IL-36R. MAB04 shares key characteristics with MAB92, including affinity, in vitro functional activity (both within ten-fold), and IL-36R domain-2 epitope binding.Citation16 Intraperitoneal administration of the mouse surrogate antibody, MAB04, in both the imiquimod- and IL36-induced mouse models of skin inflammation resulted in blockade of the swelling response as well as substantial reduction of inflammatory cytokines.Citation16 IHC data were not available for the mouse surrogate antibody against murine IL-36R; therefore, it is unknown if staining patterns and/or intensity were comparable between human and mouse.
Although allometric scaling or Dedrick transform of pharmacokinetics (PK) from preclinical species to human is often successful for therapeutic antibodies targeting soluble antigens, prediction of human PK for those targeting cell-associated antigens or otherwise affected by TMDD is significantly more challenging due to potential interspecies differences in target expression or turnover, as well as in binding kinetics.Citation17–Citation19 In these cases, a model-based approach incorporating target-specific parameters may improve the predictivity of human PK.Citation17,Citation20,Citation21 However, additional challenges exist in predicting human PK for molecules lacking cross-reactivity in preclinical species. In these cases, as for MAB92, a surrogate molecule cross-reactive to the target in the preclinical species may be required. As a result, in addition to the aforementioned TMDD challenges, discrepancies in linear PK characteristics, such as neonatal receptor (FcRn) binding and recycling as well as in catabolic susceptibility, may exist between human candidate and surrogate molecule.
The purpose of the experiments outlined herein is to characterize the PK of the anti-mouse IL-36R antibody, MAB04, in mice in support of the first-in-human (FIH) clinical trial. In this retrospective analysis, we incorporated molecule- and species-specific parameters, such as volume of distribution (Vc), intercompartmental transfer rates (k12 and k21), linear elimination (kel), binding affinity (KD), internalization rate of the drug–target complex (kint), target degradation rate (kdeg), and target abundance (R0), into a semi-mechanistic model. Two different methods of assigning target abundance were evaluated: the first assumed comparable expression between human and mouse, and the second utilized FANTOM5 RNA transcriptome data in a subset of matched tissues as a surrogate for expression in each respective species. FANTOM5 is a comprehensive expression dataset that includes ~1000 human and ~400 mouse tissues, primary cells, and cancer cell lines.Citation22 This dataset is based on cap analysis of gene expression (CAGE), a method developed at RIKEN in Japan that characterizes transcription start sites across the entire genome at single-base resolution level.Citation22–Citation26 Since eukaryotic transcription factors are typically activating, the number of transcription factors on a promotor is predictive of breadth of expression.Citation27 Human PK profiles were then simulated based on a semi-mechanistic TMDD model incorporating critical target-specific parameters for both the human candidate and mouse surrogate antibodies with R0 either assumed to be the same as that of mouse or corrected for the differences in RNA transcriptome data between species. For the human model utilizing the model-estimated mouse target abundance, Cmax was well predicted; however, AUC0-∞ was substantially underpredicted. After correcting for relative differences in RNA transcriptome data between species, the model-predicted human AUC0-∞ and Cmax were largely within 1.5-fold that observed for both nonsaturating and saturating doses.
Results
Monkey PK
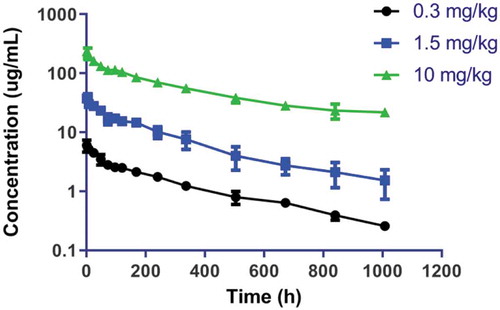
Concentrations versus time curves for MAB92 in cynomolgus monkey following intravenous administration are shown in . Noncompartmental PK analysis showed that clearance was dose linear following the intravenous administration of 0.3, 1.5, and 10 mg/kg of MAB92, as expected due to lack of cross-reactivity of MAB92 against cynomolgus monkey IL-36R (). The clearance, steady-state volume of distribution, and terminal half-life for the three dose groups were similar and were in the range of 0.17–0.22 mL/h/kg, 65.2–83.0 mL/kg, and 284–349 h, respectively. Anti-MAB92 antibodies (ADA) were observed in two of three animals in the 10 mg/kg intravenous dose groups; however, there was no impact of ADA on the PK profile. Monkeys administered 0.3 and 1.5 mg/kg intravenous doses tested negative for ADA.
Table 1. Molecular and functional attributes of MAB04 (mouse-specific mAb) and MAB92 (clinical candidate).
Mouse PK
Noncompartmental analysis for MAB04 PK in female mouse showed dose-dependent clearance with values for clearance (CL/F) of 1.6, 0.47, and 0.13 mL/h/kg for the 0.3, 1.5, and 10 mg/kg doses, respectively, suggestive of TMDD (). At the highest dose evaluated, 10 mg/kg, MAB04 clearance in the mouse was 0.13 mL/h/kg, which is consistent with published values for therapeutic monoclonal antibodies in that species.Citation28 In addition, the mouse clearance was comparable to that observed for a 1.5 mg/kg dose of the non-cross-reactive antihuman monoclonal antibody, MAB92, in the monkey (0.17 ± 0.03 mL/h/kg), suggesting that FcRn recycling and catabolic stability were comparable between molecules and species. No ADA titers were detected in any animals.
Table 2. Non-compartmental analysis parameters for MAB04 in C57BL/6 mouse and MAB92 in cynomolgus monkey following intraperitoneal (clearance only) and intravenous administration, respectively.
Human pharmacokinetic predictions
Modeling approach
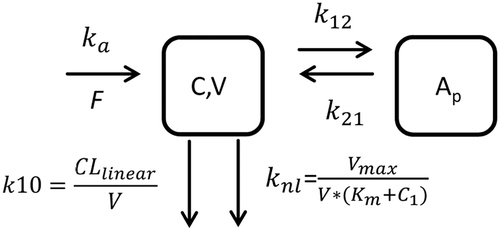
Mouse concentrations versus time data for MAB04 were simultaneously fitted to a 2-compartment (CMT) model () with parallel nonlinear and linear elimination from the central compartment. Vc, k10, Vmax, and Km were derived from the model using mean fit (). Typical linear PK parameter values for mouse (in-house data from a panel of human IgG monoclonal antibodies) were used for absorption rate (ka) and distribution microconstants (k12 and k21). Monoclonal antibody absorption following intraperitoneal dosing in mice has been demonstrated to be rapid and near complete; therefore, bioavailability was assumed to be 100%.Citation29
Table 3. Two-compartment semi-mechanistic model parameters for MAB04 (mouse-specific mAb).
Figure 2. Two-compartment parallel elimination model utilized for fitting both mouse (0.3, 1.5 and 10 mpk) and Dedrick-transformed monkey (10 mpk) concentration versus time data. ka, absorption rate constant; F, bioavailability; k12, transfer rate constant from central to peripheral compartment; k21, transfer rate constant from peripheral to central compartment; C, mAb concentration in central compartment, V, volume of distribution in central compartment; Ap, amount of mAb in peripheral compartment; Vmax, maximum rate of nonlinear elimination; Km, Michaelis–Menten constant; CLlinear, clearance of the linear elimination pathway; k10, elimination rate constant for the linear elimination pathway; and knl, elimination rate constant for the nonlinear elimination pathway.
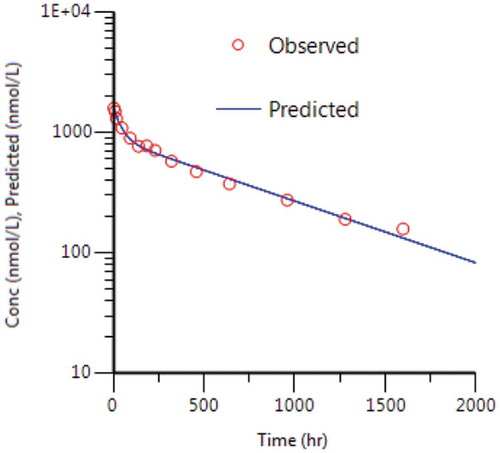
The mouse concentration versus time data were subsequently fitted to a 2-CMT semi-mechanistic model () using central volume (Vc) and linear clearance (kel) derived from the TMDD model fit. Consistent with the parallel elimination model, typical values for human monoclonal antibodies in mouse were utilized for ka, k12, and k21. Average plasma membrane turnover rate of 0.5 h was applied for both kdeg and kint.Citation30 In vitro surface plasmon resonance (SPR) binding affinity data for MAB04 was used for KD, and mouse R0 was estimated by model fitting. The fit and corresponding weighted residual plot for the fit of the mouse data are shown in and , and the model parameters are shown in .
Table 4. Two-compartment semi-mechanistic model parameters for MAB04 (mouse-specific antibody) and MAB92 (clinical candidate).
Figure 3. Schematic of target-mediated drug disposition (TMDD) semi-mechanistic model with rapid-binding approximation. Antibody levels in central and peripheral compartments are denoted by A1 and A2, respectively. Antibody distribution from central to peripheral and peripheral to central compartment is denoted by k12 and k21 respectively. Vc represents the central volume of distribution. The kel, ksyn, kdeg, and kint are first-order rate constants representing antibody clearance, target synthesis, target turnover, and complex degradation rates, respectively, and KD is the equilibrium binding constant.
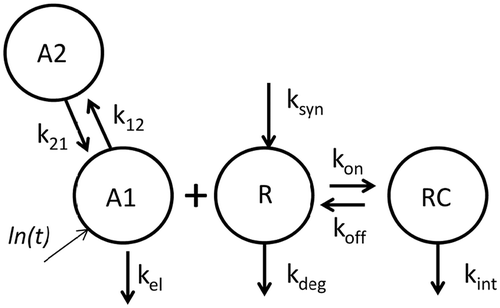
Figure 4. Mean mouse 0.3, 1.5 and 10 mpk i.p. concentration versus time data fitted to a 2-CMT semi-mechanistic TMDD model and corresponding weighted residuals.
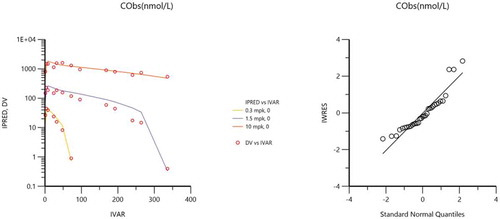
Figure 5. Dedrick transformed mean cynomolgus monkey 10 mg/kg i.v. concentration versus time data fitted to a 2-CMT model. Estimates for the fitted parameters are shown in .
Estimated mouse R0 (0.794 nM) was comparable to the mouse Km (1.18 nM) derived using simultaneous fitting of the mouse data in the 2-CMT TMDD model, suggesting that estimation of baseline of expression in mouse was reasonable. Dedrick transform was then used to scale monkey concentration–time profiles to human using an allometric exponent of 1.0 for volume of distribution (b) and 0.85 for CL (a) according to the following equations.Citation17,Citation31
Dedrick-scaled concentration versus time data were then fitted to the parallel elimination 2-CMT TMDD model previously described in (fit shown in ) to derive the human linear PK parameters (Vc, k10, k12, and k21) shown in . The differential equations for the mouse and human semi-mechanistic TMDD model are shown below and were fitted using rapid-binding approximation such that the free drug, free target, and complex are in rapid equilibrium determined by the equilibrium constant, KD.
Scaling of baseline expression
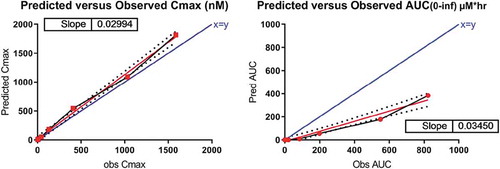
Two different methods of estimating baseline receptor expression in human were evaluated. The first assumed equivalence of IL-36R expression between species and gender such that R0 was expected to be comparable between the preclinical female mice and the FIH male subjects. Therefore, the mouse model-derived R0 was used in the semi-mechanistic model described previously, and human PK profiles corresponding to the doses used in the FIH clinical trial were then simulated. Utilizing this approach, although Cmax was predicted within 1.5-fold that observed in human, the AUC0-∞ was substantially underpredicted ( and , respectively), indicating that IL-36R expression in male human was likely substantially lower than that in female mice.
Figure 7. IL36R is differentially expressed in mouse (female) and human (male). A subset of matched tissues* (non-privileged, gender-specific) were utilized to compare expression in both species.
*heart, lymph node, bone, colon, lung, ovary/uterus/vagina (female only), penis (male only), pancreas, spleen, submandibular gland, tongue and zone of skin.
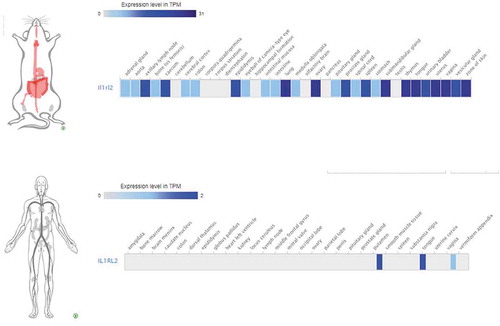
Figure 8. Human PK data against simulation results from the semi-mechanistic model corrected for cross-species differences in RNA expression between mouse and human.
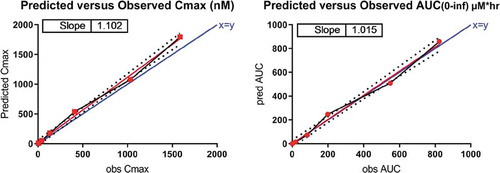
For the second method, integrating mRNA transcriptome (CAGE) data, the model-derived mouse R0 was multiplied by 0.013, the ratio of human to mouse IL-36R transcripts per million (TPM), for a panel of 14 matched, gender-specific, non-privileged tissues (Figure 9). By integrating the RNA transcriptome data, both Cmax and AUC0-∞ were predicted within 1.5-fold (). Within the dose range of 0.030–0.300 mg/kg, exposure of MAB92 increased with increasing dose in a greater than dose-proportional way. However, exposure increased in an approximately dose-proportional manner for the doses of 0.300 mg/kg and higher, suggesting saturation of TMDD within this dose range.
Table 5. Predicted versus observed Cmax and AUC0-∞ for MAB92 in human. Using a semi-mechanistic model incorporating target-specific parameters including a correction for cross-species differences in RNA expression, Cmax and AUC0-∞ for MAB92 in human were predicted largely within 1.5-fold and 1-fold, respectively.
Discussion
Accurate prediction of human PK is critical for effective and efficient design of FIH clinical trials. Although human PK for protein therapeutics targeting soluble or low-expression antigens has been successfully translated from preclinical data using single-species allometric scalingCitation17–Citation19 or Dedrick-transformed monkey PK data,Citation17,Citation31 prediction of PK for TMDD-impacted proteins remains challenging. Some success in overcoming these challenges has been achieved by incorporation of in vitro-derived mechanistic parameters into model-based predictions,Citation32–Citation34 though in these cases assumptions are often made regarding comparable target expression between preclinical species and human. Since the greatest driver of clearance nonlinearity for highly expressed and/or cell-associated targets is typically target density (R0), accurate scaling of this parameter is critical for human predictions. One approach is to experimentally determine target abundance, but converting arbitrary readouts (e.g., light intensity units for flow cytometry) into target concentration does not necessarily translate correctly.Citation35 Similarly, the total number of cells expressing target is typically unknown, so estimation of the number of cells carrying the receptor and the number of receptors per cell may not be valid.Citation35 Liquid chromatography-mass spectrometry (MS)/MS determination of total receptor in preclinical studies is technically feasible, given the identification of a selective, high-affinity, and tissue-penetrant tracer molecule, but translatability to human is still lacking, and assumptions of comparable expression between preclinical species and human may be invalid.
For the prediction of IL-36R TMDD impact and initial exposure in healthy human volunteers, we evaluated two approaches to translating R0 from mouse to human. As both methods required accurate characterization of the TMDD impact in mouse, whole blood microsampling was used to enable a rich sampling scheme from each individual animal. R0 in mouse was first estimated by fitting mouse data to a semi-mechanistic model incorporating mouse linear PK parameters derived from a 2-CMT model fit and binding data derived from MAB04 in vitro studies. For the human semi-mechanistic model, linear PK parameters in human were estimated by a 2-CMT model fit of Dedrick-transformed monkey PK data and binding data from MAB92 in vitro studies were incorporated. In the absence of comparative species expression data, the first approach to translating R0 from mouse to human was to assume equivalent R0 between the two species. This approach is frequently used for translation of TMDD impact from preclinical species to human despite limitations of low predictivity. Although this method resulted in a good prediction of human Cmax, AUC0-∞ was substantially underpredicted at all doses ( and ), demonstrating the limited utility of this approach. The second translation approach was to apply the human to mouse RNA transcriptome ratio for gender- and species-matched organs as a correction factor to the mouse R0. Organs with expected low antibody exposure due to restricted access of large polar molecules (e.g., eye, brain, central nervous system (CNS), and testes) were excluded as exposure to both MAB04 and MAB92 was expected to be limited in those tissues. This method of R0 estimation successfully predicted both Cmax and AUC0-∞ largely within 1.5-fold ().
The plots for the human PK data against simulation results from the semi-mechanistic model corrected for cross-species differences in RNA expression between mouse and human are shown in Figures 10 and 11. A sensitivity analysis within a three-fold range was conducted for the additional target-related parameters (kdeg, kint, and KD). All TMDD-impacted doses were interrogated, with sensitivity defined as greater than 50% change from original AUC. In addition to R0, both kdeg and kint (but not KD) were determined to be sensitive parameters, suggesting that predictivity of human PK might be further improved with actual measured data for target internalization and turnover kinetics (not available for IL-36R). Interestingly, in the case of IL-36R, the assumption of kdeg being equal to kint appeared to be equally valid for both species and may be a useful translation approach when measured values are not available. Although KD was not a sensitive parameter in this model, measured binding affinity (e.g., SPR) may still be useful with the caveat that SPR KD may fail to match cellular or clinical data generated later in the program.Citation15 In cases where TMDD is driven primarily by factors other than R0 (e.g., kint), the FANTOM5-corrected R0 translation approach might not improve preclinical to clinical prediction of PK.
One obvious limitation to utilizing tissue mRNA tissue transcriptome data as a surrogate for expression is the inability to assess inter-species differences in the extent of the shed and/or soluble target. For those cases, quantitation of circulating target in both species and integration of those differences into R0 corrections would likely be required. Similarly, if the target is differentially expressed across organs and between species, an organ-to-body weight ratio correction might be required to improve the predictivity of TMDD impact and therefore PK in human. An additional limitation is the fact that, for some targets, the panel of tissues assessed is not always consistent for both mouse and adult human. For example, in the case of IL-36R, although the target is expected to be expressed in skin, only mouse had TPM values available for that organ. Although TPM was reported for human fetal skin (< 0.5), assumptions cannot be made that the same low expression applies in adult human. Recognition of expression level differences between normal and healthy target tissues is another contributor to translational uncertainty.
In spite of the aforementioned limitations, our results demonstrate the potential utility of a rational, semi-mechanistic approach to predicting human PK using preclinical data generated with a surrogate antibody in combination with CAGE-derived mRNA expression transcriptome data (FANTOM5) and in human in vitro parameters. For molecules like MAB92, which lacks cross-reactivity in nonhuman primate, and requires a rodent or other species cross-reactive surrogate molecule for characterization of TMDD impact and prediction of FIH PK, integration of RNA transcriptome data may enable improved preclinical to clinical translation of PK. An additional benefit is the potential reduction in the use of higher species for preclinical PK characterization since mouse may provide sufficient data to enable human prediction. However, additional studies are required to validate the general applicability of this approach.
Materials and methods
Reagents
MAB04 and MAB92 were produced by the Boehringer Ingelheim Research team (Ridgefield, CT).Citation16 MAB92, also known as BI 655130, is a humanized monoclonal IgG1 antibody produced in Chinese Hamster Ovary (CHO) cells and targeted against human IL-36R. Both L234 and L235 of the heavy chain were mutated to alanine to minimize Fc effector function. MAB04 is a rat/mouse chimeric monoclonal antibody of the IgG2a isotype that is directed against mouse IL-36R. It utilizes a backbone that has two mutations in the Fc region (Asp265Ala and Asn297Ala) to reduce FcγR and complement binding.Citation36 MAB04 is specific to mouse IL-36R and does not cross-react to human, cynomolgus, mini pig, rhesus, marmoset, or rat IL-36R-Fc.
Nonclinical pharmacokinetic studies
All procedures were reviewed and approved by the Institutional Animal Care and Use Committee and conducted in full compliance with the ethical and regulatory principles and local and national licensing regulations.
Mouse studies
In-life studies were conducted in house. MAB04 was administered by intraperitoneal injection to female, C57BL/6 mice at 0.3, 1.5, and 10 mg/kg (N = 3 per dose group) to assess potential TMDD impact and saturability as well as clearance (CL/F) across a dose range intended to cover the human therapeutic dose. Blood samples collected via whole blood microsampling (10 µL) over one and two weeks for the 0.3 mg/kg and higher dose groups, respectively, were diluted in phosphate-buffered saline (PBS) and analyzed using an enzyme-linked immunosorbent assay (ELISA) to determine free concentrations of MAB04.
Nonhuman primate studies
The in-life studies were conducted at Charles River Laboratory (Reno, NV). Nine male drug-naive cynomolgus monkeys (Mauritius origin) weighing 2.3–2.7 kg were each administered a single intravenous dose of MAB92 at 0.3, 1.5, or 10 mg/kg as a 10-min constant rate infusion. Serial blood samples were processed to serum and collected for up to 1008 h (42 d) after dosing. Because MAB92 does not bind to cynomolgus monkey, PK characteristics of that molecule address only the catabolic stability and FcRn recycling properties of the molecule, rather than full PK evaluation including possible TMDD.
Clinical pharmacokinetic study
Human PK data from a Phase 1, single rising dose study of MAB92 (NCT02525679) were used to validate the PK predictions based on the models. The FIH dose selection was based on a MABEL approach using an in vitro IC10 as the target with predicted exposure based on TMDD saturation (linear) PK. The PK data were obtained from healthy male volunteers, and the available data from the 0.03, 0.05, 0.1, 0.3, 1, 3, 6, and 10 mg/kg intravenous doses were used to verify human PK predictions. Noncompartmental analyses were utilized to compare the observed versus predicted Cmax and AUC0-∞. All studies were conducted in accordance with the ethical standards of the Declaration of Helsinki.
Bioanalytical assay
MAB04 quantitation in mouse
MAB04 concentration in the mouse whole blood samples was determined using a protein-capture ELISA. Briefly, recombinant mouse IL-1 Rrp2/IL-1 R6 (R&D Systems, Minneapolis, MN) was immobilized onto Nunc MaxiSorp 96-well plates (ThermoFisher, Waltham, MA). The plates were washed and then blocked with 5% bovine serum albumin in PBS (w/v). Matrix reference standards, quality control, and test samples (10 µL whole blood diluted into 90 µL PBS) were transferred to the blocked plates. The plates were washed again, and horseradish peroxidase (HRP)-conjugated secondary antibody (Southern Biotech, cat#1080–05) was added. The plates were washed, and the BioFx (SurModics) substrate TMBW was added. The plates were allowed to develop at room temperature, and then, the BioFx liquid stop solution (0.2 M H2SO4) was added before the plates were analyzed on a SpectraMax (Molecular Devices) M5 Plate Reader at OD 450 nM. Drug concentrations were derived from the four-parameter fitting model from Softmax Pro software (Molecular Devices). Equivalent MAB04 concentration in mouse serum was derived from hematocrit factor (0.45)-corrected whole blood concentration.
MAB92 quantitation in cynomolgus monkeys
Serum concentrations of humanized anti-IL36 monoclonal antibody MAB92 in cynomolgus monkey serum were measured using a drug-specific capture ELISA method. Microtiter plates (Nunc Maxisorp) were coated overnight with hsIL1RL2 (ECD) provided by the Boehringer Ingelheim reagent group. Plates were washed with wash buffer (1X PBS with 0.05% Tween 20, Sigma) and blocked with blocking buffer (5% milk in 1X PBS, Sigma) and then incubated shaking for 1 h at room temperature. Plates were washed again, and calibration standards, quality controls, and monkey serum samples diluted in blocking buffer were added to the plate and incubated shaking for 1 h at room temperature. Plates were washed, and HRP-conjugated goat antihuman IgG, adsorbed against monkey (Southern Biotech, cat#2041–05), detection antibody was added at 1:10,000 dilution and incubated shaking for 1 h at room temperature. Following three plate washes, bound HRP-conjugate was detected using tetramethylbenzidine (TMB) substrate. The reaction was stopped with 1 M H2SO4, and the absorbance was measured using a SpectraMax M3 microplate reader at 450 and 650 nm dual wavelength. Softmax Pro software (v5.4) was used for calibration standard curve fitting using a four-parameter logistics model and back calculation of all unknown sample concentrations.
MAB92 quantitation in human
The concentration of MAB92 in human plasma (K2EDTA) was measured using a validated ELISA method utilizing a 96-well microtiter format. The plate was coated with an anti-MAB92 monoclonal antibody (5C8). Test material (blanks, calibration standards, QCs, and study samples) was diluted at the minimum required dilution of 1:20 (5% matrix), added to the 5C8 coated and blocked microtiter plate, and then incubated for 60 ± 5 min on a plate shaker at room temperature. After washing the microtiter plate, anti-MAB92 monoclonal antibody (biotinylated 8H11) was added and incubated on the plate for an additional 60 ± 5 min. The plate was washed again, streptavidin-HRP was added, and then the mixture was incubated on the plate for an additional 30 ± 5 min. The plate was washed again, TMB was added to the wells to generate a chromophore, and the development of color was stopped by the addition of a stopping solution. The absorbance at 450 and 650 nm was measured, and the MAB92 concentrations were calculated using a four-parameter standard calibration curve.
FANTOM5 RNA transcriptome (mouse and human)
Mouse and human heat maps of FANTOM5 RNA transcriptome data (0.5 TPM cutoff) were extracted from the European Bioinformatics Institute (EMBL-EBI) using EBI Search, a scalable text search engine providing access to the biological data resources hosted at the EMBL-EBI.Citation37 Transcriptome data were then exported to excel, and the total TPM for 14 gender-matched organs was recorded for each species. Organs with expected low antibody exposure due to restricted access of large polar molecules (e.g., eye, brain, CNS, and testes) were excluded as contributions to TMDD in those organs were expected to be low. The ratio of human to mouse TPM for IL-36R was 0.013. TPM in remaining tissues was below the quantitation limit in both species.
Abbreviations
| IL-36R | = | interleukin-1 receptor-like 2 |
| IL-1RAcP | = | IL-1 receptor accessory protein |
| mAb | = | monoclonal antibody |
| PK | = | pharmacokinetics |
| Cmax | = | maximum concentration |
| AUC0-∞ | = | area under the curve from time 0 to infinity |
| t1/2 | = | terminal half-life |
| CL | = | clearance |
| V | = | volume of distribution |
| FIH | = | First-in-Human |
| TMDD | = | target-mediated drug disposition |
| ITE | = | Indirect target engagement |
| FANTOM | = | Functional Annotation of the Human Genome |
| CAGE | = | Cap Analysis of Gene Expression. |
Acknowledgments
The authors acknowledge the following individuals for their contributions in reviewing the manuscript or providing technical expertise: Chia-Hung Tsai, Frank Li, Steven Cafiero, Sally Ye, and Jon Hill.
References
- Towne JE, Renshaw BR, Douangpanya J, Lipsky BP, Shen M, Gabel CA, Sims JE. Interleukin-36 (il-36) ligands require processing for full agonist (il-36alpha, il-36beta, and il-36gamma) or antagonist (il-36ra) activity. J Biol Chem. 2011;286:42594–602.
- Towne JE, Sims JE. Il-36 in psoriasis. Curr Opin Pharmacol. 2012;12:486–90.
- Cowen EW, Goldbach-Mansky R. Dira, ditra, and new insights into pathways of skin inflammation: what’s in a name? Arch Dermatol. 2012;148:381–84.
- Marrakchi S, Guigue P, Renshaw BR, Puel A, Pei XY, Fraitag S, Zribi J, Bal E, Cluzeau C, Chrabieh M, et al. Interleukin-36-receptor antagonist deficiency and generalized pustular psoriasis. N Engl J Med. 2011;365(7):620–28.
- Blumberg H, Dinh H, Dean C Jr., Trueblood ES, Bailey K, Shows D, Bhagavathula N, Aslam MN, Varani J, Towne JE, et al. Il-1rl2 and its ligands contribute to the cytokine network in psoriasis. J Immunol. 2010;185(7):4354–62.
- Sims JE, Nicklin MJ, Bazan JF, Barton JL, Busfield SJ, Ford JE, Kastelein RA, Kumar S, Lin H, Mulero JJ, et al. A new nomenclature for il-1-family genes. Trends Immunol. 2001;22(10):536–37.
- Saha SS, Singh D, Raymond EL, Ganesan R, Caviness G, Grimaldi C, Woska JR Jr., Mennerich D, Brown SE, Mbow ML, et al. Signal transduction and intracellular trafficking by the interleukin 36 receptor. J Biol Chem. 2015;290(39):23997–4006.
- Afonina IS, Muller C, Martin SJ, Beyaert R. Proteolytic processing of interleukin-1 family cytokines: variations on a common theme. Immunity. 2015;42:991–1004.
- Onoufriadis A, Simpson MA, Pink AE, Di Meglio P, Smith CH, Pullabhatla V, Knight J, Spain SL, Nestle FO, Burden AD, et al. Mutations in il36rn/il1f5 are associated with the severe episodic inflammatory skin disease known as generalized pustular psoriasis. Am J Hum Genet. 2011;89(3):432–37.
- Frey S, Derer A, Messbacher ME, Baeten DL, Bugatti S, Montecucco C, Schett G, Hueber AJ. The novel cytokine interleukin-36alpha is expressed in psoriatic and rheumatoid arthritis synovium. Ann Rheum Dis. 2013;72:1569–74.
- Chen H, Wang Y, Bai C, Wang X. Alterations of plasma inflammatory biomarkers in the healthy and chronic obstructive pulmonary disease patients with or without acute exacerbation. J Proteomics. 2012;75:2835–43.
- Medina-Contreras O, Harusato A, Nishio H, Flannigan KL, Ngo V, Leoni G, Neumann PA, Geem D, Lili LN, Ramadas RA, et al. Cutting edge: il-36 receptor promotes resolution of intestinal damage. J Immunol. 2016;196(1):34–38.
- Scheibe K, Backert I, Wirtz S, Hueber A, Schett G, Vieth M, Probst HC, Bopp T, Neurath MF, Neufert C. Il-36r signalling activates intestinal epithelial cells and fibroblasts and promotes mucosal healing in vivo. Gut. 2017;66:823–38.
- Russell SE, Horan RM, Stefanska AM, Carey A, Leon G, Aguilera M, Statovci D, Moran T, Fallon PG, Shanahan F, et al. Il-36alpha expression is elevated in ulcerative colitis and promotes colonic inflammation. Mucosal Immunol. 2016;9(5):1193–204.
- Wang M, Kussrow AK, Ocana MF, Chabot JR, Lepsy CS, Bornhop DJ, O’Hara DM. Physiologically relevant binding affinity quantification of monoclonal antibody pf-00547659 to mucosal addressin cell adhesion molecule for in vitro in vivo correlation. Br J Pharmacol. 2017;174:70–81.
- Ganesan R, Raymond EL, Mennerich D, Woska JR Jr., Caviness G, Grimaldi C, Ahlberg J, Perez R, Roberts S, Yang D, et al. Generation and functional characterization of anti-human and anti-mouse il-36r antagonist monoclonal antibodies. MAbs. 2017;9(7):1143–54.
- Deng R, Iyer S, Theil FP, Mortensen DL, Fielder PJ, Prabhu S. Projecting human pharmacokinetics of therapeutic antibodies from nonclinical data: what have we learned? MAbs. 2011;3:61–66.
- Ling J, Zhou H, Jiao Q, Davis HM. Interspecies scaling of therapeutic monoclonal antibodies: initial look. J Clin Pharmacol. 2009;49:1382–402.
- Oitate M, Nakayama S, Ito T, Kurihara A, Okudaira N, Izumi T. Prediction of human plasma concentration-time profiles of monoclonal antibodies from monkey data by a species-invariant time method. Drug Metab Pharmacokinet. 2012;27:354–59.
- Mager DE, Jusko WJ. General pharmacokinetic model for drugs exhibiting target-mediated drug disposition. J Pharmacokinet Pharmacodyn. 2001;28:507–32.
- Mager DE, Neuteboom B, Efthymiopoulos C, Munafo A, Jusko WJ. Receptor-mediated pharmacokinetics and pharmacodynamics of interferon-beta1a in monkeys. J Pharmacol Exp Ther. 2003;306:262–70.
- Consortium F, Forrest AR, Kawaji H, Rehli M, Baillie JK, de Hoon MJ, Haberle VLassmann T, Kulakovskiy IV, Lizio M, et al. A promoter-level mammalian expression atlas. Nature. 2014;507(7493):462–70.
- Kanamori-Katayama M, Itoh M, Kawaji H, Lassmann T, Katayama S, Kojima M, Bertin N, Kaiho A, Ninomiya N, Daub CO, et al. Unamplified cap analysis of gene expression on a single-molecule sequencer. Genome Res. 2011;21(7):1150–59.
- Noguchi S, Arakawa T, Fukuda S, Furuno M, Hasegawa A, Hori F, Ishikawa-Kato S, Kaida K, Kaiho A, Kanamori-Katayama M, et al. Fantom5 cage profiles of human and mouse samples. Sci Data. 2017;4:170112.
- Lizio M, Harshbarger J, Abugessaisa I, Noguchi S, Kondo A, Severin J, Mungall C, Arenillas D, Mathelier A, Medvedeva YA, et al. Update of the fantom web resource: high resolution transcriptome of diverse cell types in mammals. Nucleic Acids Res. 2017;45(D1):D737–D743.
- Kodzius R, Kojima M, Nishiyori H, Nakamura M, Fukuda S, Tagami M, Sasaki D, Imamura K, Kai C, Harbers M, et al. Cage: cap analysis of gene expression. Nat Methods. 2006;3(3):211–22.
- Hurst LD, Sachenkova O, Daub C, Forrest AR, Huminiecki L, Consortium F. A simple metric of promoter architecture robustly predicts expression breadth of human genes suggesting that most transcription factors are positive regulators. Genome Biol. 2014;15:413.
- Betts A, Keunecke A, van Steeg TJ, van der Graaf PH, Avery LB, Jones H, Berkhout J. Linear pharmacokinetic parameters for monoclonal antibodies are similar within a species and across different pharmacological targets: a comparison between human, cynomolgus monkey and hfcrn tg32 transgenic mouse using a population-modeling approach. MAbs. 2018;10:751–64.
- Shah DK, Veith J, Bernacki RJ, Balthasar JP. Evaluation of combined bevacizumab and intraperitoneal carboplatin or paclitaxel therapy in a mouse model of ovarian cancer. Cancer Chemother Pharmacol. 2011;68:951–58.
- Mellman I, Fuchs R, Helenius A. Acidification of the endocytic and exocytic pathways. Annu Rev Biochem. 1986;55:663–700.
- Dedrick RL. Animal scale-up. J Pharmacokinet Biopharm. 1973;1:435–61.
- Luu KT, Bergqvist S, Chen E, Hu-Lowe D, Kraynov E. A model-based approach to predicting the human pharmacokinetics of a monoclonal antibody exhibiting target-mediated drug disposition. J Pharmacol Exp Ther. 2012;341:702–08.
- Singh AP, Krzyzanski W, Martin SW, Weber G, Betts A, Ahmad A, Abraham A, Zutshi A, Lin J, Singh P. Quantitative prediction of human pharmacokinetics for mabs exhibiting target-mediated disposition. Aaps J. 2015;17:389–99.
- Parng C, Singh P, Pittman DD, Wright K, Leary B, Patel-Hett S, Rakhe S, Stejskal J, Peraza M, Dufield D, et al. Translational pharmacokinetic/pharmacodynamic characterization and target-mediated drug disposition modeling of an anti-tissue factor pathway inhibitor antibody, pf-06741086. J Pharm Sci. 2018:1995–2004.
- Ng CM, Joshi A, Dedrick RL, Garovoy MR, Bauer RJ. Pharmacokinetic-pharmacodynamic-efficacy analysis of efalizumab in patients with moderate to severe psoriasis. Pharm Res. 2005;22:1088–100.
- Michaelsen TE, Sandlie I, Bratlie DB, Sandin RH, Ihle O. Structural difference in the complement activation site of human igg1 and igg3. Scand J Immunol. 2009;70:553–64.
- Park YM, Squizzato S, Buso N, Gur T, Lopez R. The ebi search engine: ebi search as a service-making biological data accessible for all. Nucleic Acids Res. 2017;45:W545–W549.