ABSTRACT
Secukinumab, a human monoclonal antibody that selectively neutralizes IL-17A, has consistently shown low anti-drug antibody responses in patients with psoriasis, psoriatic arthritis, and ankylosing spondylitis. Secukinumab has also shown lower in vitro immunogenicity potential compared with other monoclonal antibodies used to treat psoriasis and psoriatic arthritis, and a significantly lower in vitro T cell precursor frequency compared with ixekizumab, which targets the same antigen. Here, secukinumab and ixekizumab were further examined regarding their specific T cell epitopes. Secukinumab- or ixekizumab-specific CD4 T cell lines were generated from 31 healthy, treatment-naïve donors via 28-day co-culture with mature monocyte-derived dendritic cells exposed to either antibody. Consistent with previous data, the frequency of preexisting T cells to secukinumab was significantly lower as compared with ixekizumab. Only two T cell lines from two different donors could be derived for secukinumab, but no specific T cell epitope was identified. In contrast, 32 T cell lines from eight donors were obtained for ixekizumab. For 11 of these T cell lines, the specific T cell epitopes could be identified and confirmed by major histocompatibility complex–associated peptide proteomics as being naturally presented peptides. All identified T cell epitopes cluster in four main regions that are overlapping with the complementarity-determining regions HCDR3, LCDR1, LCDR2 and LCDR3. Interestingly, ixekizumab CDRs contain amino acids that are not found in any of the germline family members. These amino acids may be associated with the higher number of T cell epitopes identified for ixekizumab light chain and may contribute to the increased in vitro immunogenicity potential observed for ixekizumab vs. secukinumab.
Introduction
The pro-inflammatory cytokine interleukin (IL)-17A, produced by the Th17 subset of CD4 T-helper cells, along with other innate and adaptive immune cells, is a cornerstone cytokine in the development of psoriasis (PsO) and psoriatic arthritis (PsA).Citation1–Citation6 The antibody therapeutics secukinumab and ixekizumab, which directly target the IL-17A cytokine, and brodalumab, which target the IL-17A receptor, have been approved for these conditions and various others are in clinical development.Citation7–Citation10 Secukinumab is a human IgG1/kappa antibody generated from human immunoglobulin transgenic mice (Medarex Inc.) immunized with human IL-17A and cloned using classical hybridoma technology. Secukinumab has been demonstrated to be highly efficacious in the treatment of moderate to severe plaque PsO, PsA, and ankylosing spondylitis (AS) with a sustained effect and a favorable safety profile in Phase 3 studies.Citation11–Citation18 Furthermore, even without co-treatment (e.g., methotrexate), the clinical immunogenicity incidence (anti-drug antibody [ADA] response) was consistently below 1% in patients with moderate to severe psoriasis treated with secukinumab for up to 5 years.Citation19–Citation21 Secukinumab was also associated with a low incidence (<1%) of immunogenicity in PsA and AS after 1 year of treatment.Citation22
Ixekizumab is a humanized IgG4 variant/kappa antibody, obtained by immunizing mice with human IL-17A, selecting specific binders using antigen-binding fragment (Fab)-expressing phage display technology, followed by phage display-based humanization and affinity maturation in E. coli.Citation23 Ixekizumab is the second IL-17A monoclonal antibody (mAb) approved for the treatment of moderate to severe plaque PsO and PsA, with clinical trials ongoing in AS.Citation24–Citation27 In a pooled analysis of three Phase 3 studies involving patients with psoriasis, ADA against ixekizumab developed in 9% of patients by Week 12, and those patients with higher titers of ADA experienced a reduction in clinical efficacy.Citation26 According to the product label, approximately 22% of patients treated with ixekizumab develop antibodies during the 60-week treatment period.Citation28
T cell activation assays were previously used to identify potential T cell epitopes/peptides in vitro by major histocompatibility complex (MHC)–associated peptide proteomics (MAPPs): secukinumab demonstrated a low in vitro immunogenic potential that was comparable to, or lower than, five other marketed biotherapeutics (adalimumab, infliximab, rituximab, ustekinumab, and etanercept).Citation29 In an additional in vitro study to assess its immunogenic potential, secukinumab showed both significantly less frequent T cell responses and lower numbers of preexisting T cells than ixekizumab, adalimumab, and infliximab. However, the epitopes that may have activated these T cells remained unknown.Citation30 Therefore, in this study, we aimed to identify the reactive T cell epitopes within secukinumab and ixekizumab. To do so, we mapped the T cell epitopes for each antibody using a two-step procedure using peptides derived from MAPPs analysis and peptides covering the complementarity-determining regions (CDRs).
Results
Assessment of T cell precursor frequencies for secukinumab and ixekizumab
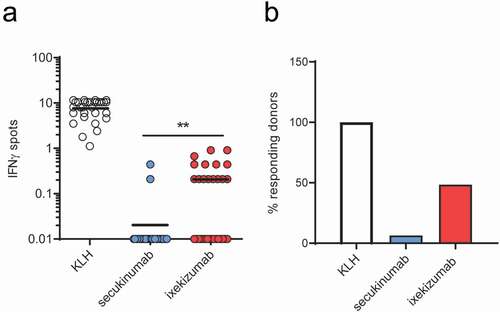
Antibody-specific CD4 T cells were generated as previously describedCitation31 to evaluate the size of the preexisting T cell repertoire specific for secukinumab and ixekizumab. As determined by genotyping, the donors represented the most common Human Leukocyte Antigen – DR isotype (HLA-DR) alleles found in an ethnically mixed European population (Supplementary Table 1). Of the 31 donors, plates from 30 were available to determine the generation of a T cell response to keyhole limpet hemocyanin (KLH [positive control]). All 30 plates demonstrated the ability of the donors to mount an in vitro T cell response to KLH (). Although no positive control KLH data was available for one donor, samples from this donor were subsequently found to react to secukinumab and, therefore, were not excluded from the study. summarizes the number of mAb-specific preexisting T cells per million of CD4 T cells specific to KLH, secukinumab and ixekizumab, and the frequencies of the responding donors. Of the 31 donors, only 2 donors responded to secukinumab with a frequency of 0.21 and 0.44 specific T cells per million of CD4 T cells (mean for all 31 donors: 0.03 specific T cells per million of CD4 T cells). This was significantly lower (p < .01) than response rates to ixekizumab, in which 15 of the 31 donors responded and exhibited a response varying from 0.21–0.91 specific T cells per million of CD4 T cells (mean for all 31 donors: 0.2 specific T cells per million of CD4 T cells).
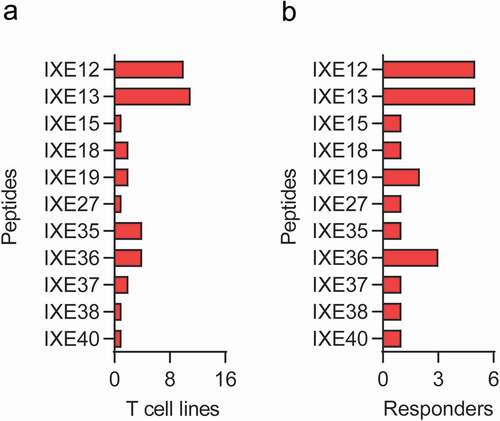
Figure 1. Specific T cell and responding donor frequencies. T cell lines (T cells contained in a single well) were generated by 3 weekly rounds of stimulation with secukinumab, ixekizumab or KLH (positive control) and their specificity was tested by IFN-γ ELISpot. Frequencies of CD4 T cells specific for monoclonal antibodies or KLH (a) and donor response (B, %) for 31 healthy blood donors. A donor was considered a responder if at least one T cell line was detected. The frequency of CD4 T cell precursors was calculated using the Poisson’s distribution. Data was analyzed using Wilcoxon matched-pairs signed rank test, **p < .01. KLH, Keyhole limpet hemocyanin; IFN-γ, interferon gamma
Identification of potential T cell epitopes by MAPPs
Immature monocyte-derived dendritic cells (DC) from healthy human treatment-naïve HLA typed (Supplementary Table 2) blood donors were loaded with secukinumab or ixekizumab in the presence of lipopolysaccharide (LPS). HLA class II-associated peptides were isolated from secukinumab- or ixekizumab-loaded DC by immunoprecipitation with anti-HLA class II beads. Peptides were subsequently analyzed by liquid chromatography-mass spectrometry and identified via database search using the SEQUEST algorithm.Citation29 For both secukinumab and ixekizumab, similar numbers of presented sequence regions, represented as ‘potential’ T cell epitopes, were detected in the variable domains. Secukinumab showed 13 presented sequence regions, 8 in the heavy chain and 5 in the light chain, and ixekizumab showed 12 presented sequence regions, 5 in the heavy chain and 7 in the light chain. Ixekizumab showed the most presented regions in the light chain with a light/heavy chain ratio of 1.4, while secukinumab showed most presented regions in the heavy chain with a light/heavy chain ratio of 0.6. A list of all ixekizumab- and secukinumab- derived peptide sequences identified by MAPPs analysis is provided in Supplementary Table 3.
T cell epitope identification
Two strategies of T cell mapping were followed by utilizing 2 different sets of peptides, as the number of peptides for the complete sets for each antibody was too high to be studied for the individual T cell lines. The standard approach for T cell epitope mapping is a sequence of re-stimulations mediated by autologous antigen presenting cells (APC). Firstly, T cells are stimulated using DC loaded with full length antigen. This is followed by re-stimulation of reactive cell lines with peripheral blood mononuclear cells (PBMC) loaded with pools of overlapping peptides that cover relevant sequence stretches in the antigen; in the case of therapeutic antibodies, the respective variable domains of heavy (VH) and light chains (VL) of the antibodies are usually covered. Then, T cell lines reacting to a peptide pool are re-stimulated with PBMC loaded with individual peptides of positive peptide pools. This approach was applied in the first set of experiments of this study. Here, the peptide pools comprised 20-mer overlapping peptides covering the complete VH and VL sequences of both secukinumab and ixekizumab (Supplementary Tables 4 and 5, respectively; strategy 1 peptides). These peptide sequences were split into 5 individual pools each for both secukinumab and ixekizumab (SEC/IXE Pools 1.1–1.5).
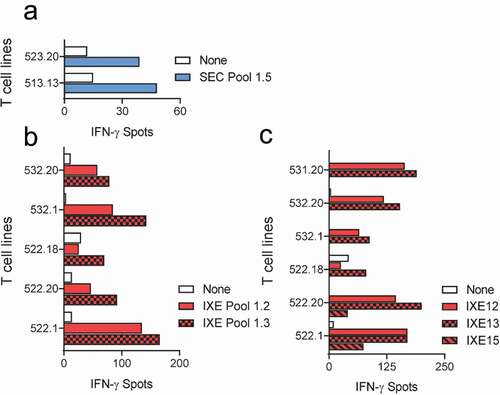
The first set of peptide pools, SEC/IXE Pools 1.1–1.5, were used to map the T cell epitopes of secukinumab and ixekizumab using 16 of the 31 donors (). Only 2 T cell lines raised against secukinumab responded to SEC Pool 1.5 peptides (), but no individual peptide sequence could be identified in the second confirmatory ELISpot. Multiple T cell lines raised against ixekizumab, however, reacted with IXE peptide Pools 1.2 and 1.3, as illustrated in . The second confirmatory ELISpot carried out with the individual peptides from IXE Pools 1.2 and 1.3 revealed that T cell lines reacted with the heavy chain peptides IXE12 and IXE13, which span HCDR3 of ixekizumab, and IXE15 ().
Figure 2. T cell lines responsive to secukinumab and ixekizumab Set 1 peptides. T cell lines (T cells contained in a single well) from 16 healthy blood donors were generated and tested for recognition of Set 1 secukinumab (a) and ixekizumab (b) peptides (SEC/IXE Pools 1.1–1.5). Their specificity was tested by IFN-γ ELISpot. T cell lines reacting to ixekizumab peptide pools in (B) were stimulated with individual peptides of the positive pools, and responding donors are demonstrated in (c). IFN-γ, interferon gamma; SEC, secukinumab; IXE, ixekizumab
Besides the large number of peptides that need to be tested for each T cell line, another caveat of this approach is the non-natural nature of these peptides, which do not necessarily represent the peptides that have actually been processed and are presented by professional APC cells such as DC. Therefore, we decided to apply a second, more focused strategy with a reduced number of peptides, covering specifically the CDR regions of the two therapeutic antibodies as well as peptides presented by DC that were identified by MAPPs in the VH and VL regions of secukinumab and ixekizumab (Supplementary Figure 1). The MAPPs peptides were split into 2 pools (SEC/IXE Pools 2.1–2.2) (Supplementary Tables 4 and 5; strategy 2 peptides), while the peptides covering the CDRs of each antibody were combined in one pool only (SEC/IXE pool 2.3) (Supplementary Tables 4 and 5; strategy 3 peptides).
Set 2 peptides pools (SEC/IXE 2.1–2.3) were applied to 15 of the 31 donors (). Two T cell lines reacted with secukinumab peptides from SEC Pool 2.1 (), but again no T cell lines were found to be specific for individual MAPPs peptides in the second confirmatory ELISpot (data not shown). In contrast, seven T cell lines reacted with ixekizumab peptides from IXE Pool 2.2, identified by MAPPs (). In the confirmatory ELISpot using individual peptides from IXE Pool 2.2, T cell lines responded to stimulation with light chain peptides IXE35, IXE36, IXE37, IX38 and IXE40 (,), but not with IXE34 and IXE39 (data not shown). Both peptides, IXE34 and IXE39 are located in framework regions only, while peptides IXE35, IXE36 and IXE37 span the LCDR1, and peptides IXE38 and IXE40 include LCDR2 and LCDR3, respectively. Thus, we found preexisting T cells reacting to peptides from all three CDRs of the light chain variable region of ixekizumab.
Figure 3. T cell lines responsive to secukinumab and ixekizumab set 2 peptides. T cell lines (T cells contained in a single well) from 15 healthy blood donors were generated and tested for recognition of Set 2 secukinumab (a) and ixekizumab (b) peptide pools 2.1–2.3. Their specificity was tested by IFN-γ ELISpot. T cell lines reacting to ixekizumab peptide pools in (B) were retested with individual peptides of the positive pools. (c-d) represents MAPPs peptides while peptides (e-f) cover the CDR regions. IFN-γ, interferon gamma; SEC, secukinumab; IXE, ixekizumab
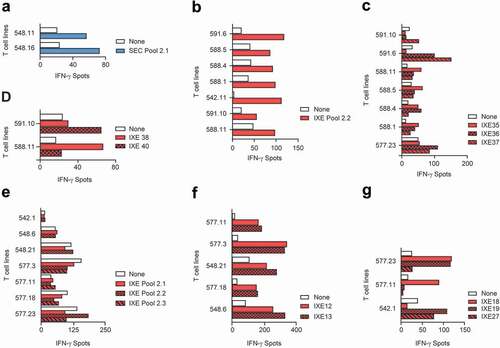
In addition, T cell lines that did not react with any of the pools, but that were positive in the T cell assay used to determine the frequency of antigen-specific T cells (), were further analyzed with individual CDR peptides from IXE Pool 2.3 (Supplementary Table 5; strategy 3 peptides). Five T cell lines were found to be specific for heavy chain peptides IXE12 and IXE13 (). These peptides, which spanned HCDR3 of ixekizumab, were also identified in the first set of experiments. In addition, one T cell line each was specific for both light chain CDR1 peptides IXE18 and IXE19, one for both IXE19 and the LCDR3 peptide IXE27, and one for IXE18 alone (). An overview of T cell lines and donor responses to Set 2 ixekizumab peptides is summarized in .
Discussion
Secukinumab and ixekizumab have revolutionized the treatment of PsO and PsA, but, although they modulate the same cytokine (IL-17A) and have a similar mechanism of action, the clinical immunogenicity of these mAbs differs.Citation30 Data shown here, in combination with our previous study, confirms in almost 50 healthy subjects that the frequency of preexisting T cells to secukinumab is significantly lower compared with ixekizumab.Citation30 Additionally, and consistent with previous findings, secukinumab demonstrated a low immunogenicity potential as indicated by MAPPs analysis and the low number of healthy subjects responding in T cell functional in vitro assays.Citation29 The level of in vitro immunogenicity potential produced by ixekizumab corresponds to proteins that are considered moderately immunogenic (e.g., infliximab, adalimumab, rituximab). The in vitro immunogenicity potential of secukinumab, however, is similar to proteins of low or negligible immunogenicity (e.g., etanercept, trastuzumab).Citation29,Citation32
This is the first study that performed T cell epitope mapping of the human antibody secukinumab and the humanized antibody ixekizumab in order to understand the potential reasons for the observed differences in in vitro immunogenicity potential of the two antibodies. Only 2 T cell lines from 2 different donors could be derived for secukinumab, but no specific T cell epitope could be identified. Previous studies have shown that there can be a mismatch between the number of T cell lines derived in response to an antibody and the number of identified epitopes, especially if the number of preexisting T cells is very low.Citation33 Given the low number of responding donors, the weak T cell responses to secukinumab, and very low number of preexisting T cells, there is a possibility that the assay may be insufficiently sensitive to allow identification of the secukinumab-reactive T cell epitopes. As the ELISpot procedure requires splitting of the T cell line cultures into several separate cultures that are subsequently stimulated with the individual peptides, relatively rare clones may be lost.
In contrast, 27 T cell lines from 15 different donors could be derived from ixekizumab, and for 19 of these T cell lines, the specific T cell epitope could be identified. Most T cell epitopes were identified by both 20mer CDR peptides (IXE Pool 2.3) or naturally presented sequences identified via MAPPs analysis (IXE Pools 2.1 and 2.2). Interestingly, the T cell epitope in LCDR2 could only be identified using a peptide length variant identified via MAPPs assay (IXE38), but not using the corresponding CDR peptide IXE22 with a slightly downstream shifted frame. In contrast, the epitope in HCDR3 was only identified using the CDR peptide IXE12, but not using the MAPPs assay-derived length variant IXE32. These discrepancies may be explained by the shift in peptide frames between MAPPs and CDR peptides, enabled by the open HLA class II peptide binding groove, thereby resulting in a change of T cell recognition,Citation34 as well as by differences in the HLA class II genes that were expressed by the different donor sets used for the MAPPs and T cell assay experiments. Another explanation may be limitations of assay sensitivity due to the low T cell precursor frequency observed. Overall, the approach of MAPPs-assisted T cell epitope mapping appeared to be superior as compared to the more “traditional” peptide-scanning approach as it allowed identification of more functional T cell epitopes. This is in particular interesting since these functional T cell epitopes identified by MAPPs represent antigenic regions that were presented by APC after intracellular processing of the full-length protein and may thus present “authentic” T cell epitopes. Further, those peptide sequences that were found by MAPPs but didn’t comprise a T cell epitope may be of interest for future research in order to understand better and eventually be able to predict which sequence characteristics do make functional T cell epitopes.
Interestingly, the T cell epitopes found in ixekizumab cluster in four main regions that overlap with HCDR3, LCDR1, LCDR2 and LCDR3; thus, the CDR appeared to be the main driver of the in vitro T cell responses (). This is in line with the hypothesis that immunogenicity of human or humanized antibodies resides in the CDR regions.Citation36 Therefore, it is not a surprise that, depending on their sequence, some human or humanized antibodies show very little immunogenicity in the clinic while others show increased immunogenicity, which also affects safety and efficacy. Although this appears obvious, it is not trivial to obtain or engineer antibodies with low or absent T cell epitope content in these regions without negatively impacting affinity or the structural integrity of the therapeutic antibody. Ixekizumab LCDRs contain amino acids that are derived from the mouse parent sequence or have been introduced during antibody engineering, which are atypical at these positions considering the amino acid sequence of germline family members ().
Figure 5. T cell epitopes identified in ixekizumab and secukinumab. Secukinumab (a) heavy and light chain were aligned to closest germline IGHV3-7*01|IGHJ2*01 and IGKV3-20*01|IGKJ5*01, respectively. Ixekizumab (b) heavy and light chain were aligned to closest germline IGHV1-69*01|IGHJ1*01 and IGKV2D-29*02|IGKJ2*01, respectively. For ixekizumab (B), deviations are labeled according to their origin.Citation23 Deviations in the amino acid sequence are highlighted along with the number of germline family members. CDR and MAPPs derived peptides are mapped graphically below the amino acid sequence and colored according to their reactivity in the T cell assay. The definition of framework and CDR is as previously described.Citation35 CDR, complementarity determining region; MAPPs, major histocompatibility complex–associated peptide proteomics
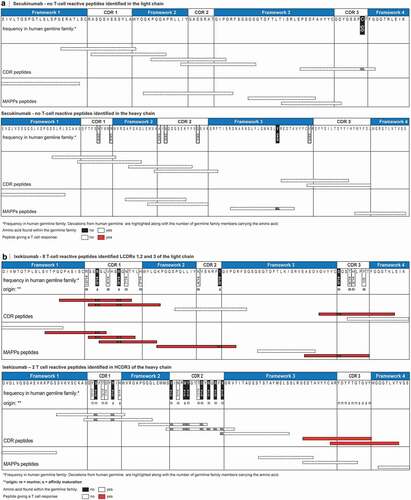
The reactive T cell epitopes in the heavy and light chain CDRs of ixekizumab contain amino acid residues originating from parental clone 2321 mouse antibody or were introduced by mutagenesis to improve affinity to the target or remove a potential N-glycosylation site.Citation23 Many of these amino acid residues are either absent or rare in human germline genes based on our analysis (summarized in ) and the analysis by Liu et al.Citation23 Therefore, it is possible that both the transfer of binding-critical residues of mouse origin during the process of humanization, affinity maturation and/or the engineering process of ixekizumab may have generated peptide sequences that are more prone to being presented to, and recognized by T cells, leading to an increased potential of in vitro immunogenicity. Aligning the amino acid sequence of ixekizumab heavy chain to the closest human germline sequence IGHV1-69*01|IGHJ1*01Citation37 identified 15 deviations, with 9 of them not being present in any germline of the same family.Citation37 All of these deviations are residues in HCDR1 and HCDR2. Although multiple CDR peptides were designed to cover this region of the heavy chain, none of them led to a T cell response. Interestingly, MAPPs didn’t reveal any peptide sequence in that region, an indication that this area of ixekizumab may not be presented by APC at all and thus, may not contribute to the in vitro immunogenicity of ixekizumab, despite the presence of these 9 atypical amino acid residues.
Similarly, 12 deviations to the closest germline IGKV2D-29*02|IGKJ2*01Citation37 were found in the light chain of ixekizumab. Four of them cannot be found in any germline of the same family.Citation37 Surprisingly, all of the found T cell reactive peptides contain at least one of those four residues, indicating that these residues may be the drivers of the increased in vitro immunogenicity potential observed for ixekizumab.
In contrast, secukinumab contains only very few amino acids that are atypical at these positions in the human germline and are likely the result of the natural process of somatic hypermutation in the B cells (). However, most of these amino acids in secukinumab deviating from the closest germline are present in several germline sequences of the same family. There are only two mutations that are atypical, namely a valine in the heavy chain framework 3 and a cysteine in LCDR3. While no peptide was found by MAPPs covering LCDR3, a peptide including the valine in heavy chain framework 3 could be identified (SEC34). However, no T cell line was reactive to this peptide. In fact, the close relationship of the secukinumab CDRs to the germline sequences and the nature of the changes may be an explanation as to why we have not seen frequent and strong in vitro T cell responses and were not able to identify T cell epitopes for secukinumab.
While the results presented here are in line with reports of the clinical incidence of ADA, more research is required to confirm an association between in vitro immunogenicity potential and the emergence of clinical immunogenicity events. However, our study offers potential learnings for antibody development and antibody engineering, including: (1) appropriate steps should be taken to avoid ‘rare’ or ‘atypical’ mutations, and (2) MAPPs-assisted T cell epitope mapping should be applied during drug development at the design and engineering stage, to reduce risk of ADA.
Materials and methods
Proteins and peptides
KLH was purchased from Thermo Fisher Scientific (Brebières, France). Secukinumab (150 mg/mL) and ixekizumab (90 mg/mL) were obtained from the campus pharmacy (Campus Apotheke, Basel, Switzerland) and stored according to the instructions provided. Peptides were purchased from Pepscan (Lelystad, The Netherlands). The sequences of the mAbs were retrieved from the international ImMunoGeneTics information system (IMGT) and sequence alignment was performed using the IMGT/DomainGapAlign tool.Citation38 Perfect 9mer content was calculated based on the Human String Content (HSC) method.Citation39
Characterization of antibody-specific CD4 T cell lines
An overview of the T cell assay procedure and epitope mapping strategy is presented in Supplementary Figure 1. Secukinumab and ixekizumab were tested in an in vitro T cell assay and the frequency of specific T cells present in the blood of the donors was evaluated for each antibody.
PBMC were obtained from blood cells collected at the Etablissement Francais du Sang (EFS, Rungis, France), as buffy-coat preparations from 31 HLA-DRB1c-typed (Supplementary Table 2), anonymous, healthy donors who gave informed consent in accordance with EFS guidelines. All the samples were genotyped for HLA-DR using the Gold SSP DRB typing kit (One lambda) after DNA extraction from PBMC with NucleoSpin Blood QuickPure Kit (Macherey-Nagel). Antibody-specific CD4 T cell lines were generated as described previously.Citation31 DC were produced from plastic-adherent cells of PBMC, while CD4 T cells were isolated from PBMC by using magnetic microbeads (Miltenyi Biotech, Paris, France). The quality of the DC was assessed by labeling of markers HLA-DR, HLA-A,B,C, CD14, DC-SIGN, CD86 and CD80. DC were separately loaded overnight at 37°C with KLH (Sigma; 0.25 µM), used as a positive control or with the therapeutic antibodies (1 µM) and matured with LPS (1 µg/mL). CD4 T cells (200,000/well) were stimulated by protein-loaded DC (20,000/well) and cultured for 28 days. The number of antigen specific T cells was identified by interferon (IFN)-γ ELISpot assay using an AID ELISpot Reader System. CD4 T cell lines were considered as specific when a spot count was 2-fold higher in the presence of the protein or the peptide than in their absence, with a minimal difference of 25 spots. The frequency of preexisting CD4 T cells specific for secukinumab, ixekizumab or KLH was calculated by considering that the T cell distribution follows the Poisson distribution, as previously described.Citation30
Based on previous experiments performed with immunogenic and non-immunogenic proteins,Citation31,Citation32 the levels of immunogenicity potential are discriminated as described previously and summarized (high >1; moderate 0.1–1; low <0.1).Citation29,Citation32
MAPPS assay
Naturally presented HLA class II–associated peptides were identified via the MAPPs assay from 38 healthy donors’ monocyte-derived DC exposed to either of the therapeutic antibodies secukinumab or ixekizumab. Peptides generated in the endolysosomal cellular compartment typically occur in multiple length variants sharing the same HLA class II binding core. Based on the HLA haplotypes of the donors in the test set (Supplementary Table 3), the same sequence region may be presented by several donors. Each presented sequence region may potentially, but not necessarily, be recognized as T cell epitopes.
CD14-positive mononuclear cells were purified from PBMC collected from consented healthy donors (Blood Donation Center Bern, Bern, Switzerland) and differentiated into immature DC.Citation29 Immature DC were loaded with either secukinumab or ixekizumab, matured by LPS (1 µg/mL, Sigma) and incubated for 24 hours at 37°C and 5% CO2. HLA class II molecules were immunoprecipitated with anti-HLA-DR/DP/DQ Mab IVA12-conjugated beads and peptides were eluted from HLA class II molecules by adding 0.1% trifluoroacetic acid (Fluka, Buchs, Switzerland) at 37°C. Lyophilized peptides were re-suspended in hydrophilic buffer containing 5% acetonitrile and 1.1% formic acid. Peptide composition was analyzed by liquid chromatography (nano capillary system, Dionex Corporation, Sunnyvale, California, USA) on a self-packed fused-silica C18 reversed-phase nano-high-performance liquid chromatography column connected to a mass spectrometer (Q-Exactive, Thermo, California, USA) via electrospray ionization (LC-ESI-MS/MS). Peptides were identified via a database search approach using the SEQUEST algorithm as detailed previously.Citation29
Epitope mapping two-step ELISpot assay
In parallel to the identification of the number of specific T cells, the epitopes of the derived CD4 T cell lines were evaluated in two rounds of ELISpot: in the first round ELISpot assay, CD4 T cell lines specific for either secukinumab or ixekizumab (200,000/well) were stimulated with PBMC that were loaded with peptide pools for either mAb. In the second round ELISpot assay, individual peptides from the active pools were used in the stimulation assay.
In the first set of experiments with 16 donors, the peptide pools comprised 20-mer overlapping peptides covering the complete VH and VL sequences of both secukinumab and ixekizumab. These peptide sequences were split into 5 individual pools each for both secukinumab and ixekizumab, SEC/IXE Pools 1.1–1.5 (Supplementary Tables 4 and 5, respectively).
In the second set of experiments with 15 additional donors, PBMC were loaded with three peptide pools, 2 of the pools comprising peptides from the MAPPs analysis (SEC/IXE Pools 2.1 and 2.2) and the third pool (SEC/IXE Pool 2.3) comprising peptides covering the CDR region of the two antibodies. In parallel, the individual peptides of the CDR pool were tested for T cell lines that did not react with any of the pools, but that were positive in the T cell assay used to determine the frequency of antigen-specific T cells. (Supplementary Tables 4 and 5 and Supplementary Figure 1).
Sequence alignment
Amino acid sequences of secukinumab and ixekizumab were aligned to germline sequences published on imgt.org.Citation37 In the case of ixekizumab, IGHV1-69*01|IGHJ1*01 and IGKV2D-29*02|IGKJ2*01 were found as the germline sequences with the highest sequence identity for the heavy and light chain, respectively. In the case of secukinumab, IGHV3-7*01|IGHJ2*01 and IGKV3-20*01|IGKJ5*01 were found as closest germline sequences for heavy and light chain, respectively. To assess whether amino acids deviating from the closest germline are found in other germline family members the following sequences were considered from IMGT database:Citation37
For ixekizumab heavy chain: IGHV1-2; IGHV1-3; IGHV1-8; IGHV1-18; IGHV1-24; IGHV1-38-4; IGHV1-45; IGHV1-46; IGHV1-58; IGHV1-68; IGHV1-69; IGHV1-69-2; IGHV1-69D; IGHV1-NL1.
For ixekizumab light chain: IGKV2-4; IGKV2-18; IGKV2-24; IGKV2-28; IGKV2-29; IGKV2-30; IGKV2-40; IGKV2D-24; IGKV2D-26; IGKV2D-28; IGKV2D-29; IGKV2D-30; IGKV2D-40; IGKV2D-18.
For secukinumab heavy chain: IGHV3-7; IGHV3-9; IGHV3-11; IGHV3-13; IGHV3-15; IGHV3-16; IGHV3-19; IGHV3-20; IGHV3-21; IGHV3-22; IGHV3-23; IGHV3-23D; IGHV3-25; IGHV3-29; IGHV3-30; IGHV3-30-2; IGHV3-30-3; IGHV3-30-5; IGHV3-30-22; IGHV3-30-33; IGHV3-30-42; IGHV3-30-52; IGHV3-32; IGHV3-33; IGHV3-33-2; IGHV3-35; IGHV3-38; IGHV3-38-3; IGHV3-43; IGHV3-47; IGHV3-48; IGHV3-49; IGHV3-52; IGHV3-53; IGHV3-54; IGHV3-62; IGHV3-63; IGHV3-64; IGHV3-66; IGHV3-69-1; IGHV3-71; IGHV3-72; IGHV3-73; IGHV3-74; IGHV3-NL1.
For secukinumab light chain: IGKV3-7; IGKV3-11; IGKV3-15; IGKV3-20; IGKV3D-7; IGKV3D-11; IGKV3D-15; IGKV3D-20; IGKV4-1; IGKV5-2; IGKV6-21; IGKV6D-21; IGKV6D-41; IGKV7-3.
Disclosure of Potential Conflicts of Interest
S Gottlieb, T Huber, A Karle, F Kolbinger, are employees of, and have stocks and/or stock options in Novartis. S Spindeldreher is a former employee and has stocks and/or stock options in Novartis. B Maillere, E Correia, and M Tenon have received research grants from Novartis.
Data availability
Novartis is committed to sharing with qualified external researchers, access to patient-level data and supporting clinical documents from eligible studies. These requests are reviewed and approved by an independent review panel on the basis of scientific merit. All data provided is anonymized to respect the privacy of patients who have participated in the trial in line with applicable laws and regulations.
Supplemental Material
Download MS Word (166.8 KB)Acknowledgments
The authors thank Trudy McGarry, PhD and Jackie L. Johnson, PhD of Novartis Ireland Ltd for providing medical writing support/editorial support, which was funded by Novartis Pharma GmbH, Nürnberg, Germany in accordance with Good Publication Practice (GPP3) guidelines (http://www.ismpp.org/gpp3). The authors thank Stephan Koepke for conducting the LC-MS analysis for the MAPPs assay.
Supplementary material
Supplemental data for this article can be accessed on the publisher’s website.
Additional information
Funding
References
- Lin AM, Rubin CJ, Khandpur R, Wang JY, Riblett M, Yalavarthi S, Villanueva EC, Shah P, Kaplan MJ, Bruce AT. Mast cells and neutrophils release IL-17 through extracellular trap formation in psoriasis. J Immunol. 2011;187(1):490–10. doi:https://doi.org/10.4049/jimmunol.1100123.
- Lowes MA, Kikuchi T, Fuentes-Duculan J, Cardinale I, Zaba LC, Haider AS, Bowman EP, Krueger JG. Psoriasis vulgaris lesions contain discrete populations of Th1 and Th17 T cells. J Invest Dermatol. 2008;128(5):1207–11. doi:https://doi.org/10.1038/sj.jid.5701213.
- Martin DA, Towne JE, Kricorian G, Klekotka P, Gudjonsson JE, Krueger JG, Russell CB. The emerging role of IL-17 in the pathogenesis of psoriasis: preclinical and clinical findings. J Invest Dermatol. 2013;133(1):17–26. doi:https://doi.org/10.1038/jid.2012.194.
- Res PC, Piskin G, de Boer OJ, van der Loos CM, Teeling P, Bos JD, Teunissen MB. Overrepresentation of IL-17a and IL-22 producing CD8 T cells in lesional skin suggests their involvement in the pathogenesis of psoriasis. PLoS One. 2010;5:e14108.
- Johansen C, Usher PA, Kjellerup RB, Lundsgaard D, Iversen L, Kragballe K. Characterization of the interleukin-17 isoforms and receptors in lesional psoriatic skin. Br J Dermatol. 2009;160(2):319–24. doi:https://doi.org/10.1111/bjd.2009.160.issue-2.
- Ortega C, Fernandez AS, Carrillo JM, Romero P, Molina IJ, Moreno JC, Santamaria M. IL-17-producing CD8+ T lymphocytes from psoriasis skin plaques are cytotoxic effector cells that secrete Th17-related cytokines. J Leukoc Biol. 2009;86(2):435–43. doi:https://doi.org/10.1189/JLB.0109046.
- Garcia-Montoya L, Marzo-Ortega H. The role of secukinumab in the treatment of psoriatic arthritis and ankylosing spondylitis. Ther Adv Musculoskelet Dis. 2018;10(9):169–80. doi:https://doi.org/10.1177/1759720X18787766.
- Wendling D, Verhoeven F, Prati C. Anti-IL-17 monoclonal antibodies for the treatment of ankylosing spondylitis. Expert Opin Biol Ther. 2019;19(1):55–64. doi:https://doi.org/10.1080/14712598.2019.1554053.
- Sekhon S, Jeon C, Nakamura M, Yan D, Afifi L, Bhutani T, Levin E. Clinical utility of ixekizumab in the treatment of moderate-to-severe plaque psoriasis. Psoriasis (Auckl). 2017;7:65–72. doi:https://doi.org/10.2147/PTT.S129792.
- Foulkes AC, Warren RB. Brodalumab in psoriasis: evidence to date and clinical potential. Drugs Context. 2019;8:212570.
- Blauvelt A, Prinz JC, Gottlieb AB, Kingo K, Sofen H, Ruer-Mulard M, Singh V, Pathan R, Papavassilis C, Cooper S, et al. Secukinumab administration by pre-filled syringe: efficacy, safety and usability results from a randomized controlled trial in psoriasis (feature). Br J Dermatol. 2015;172(2):484–93. doi:https://doi.org/10.1111/bjd.2015.172.issue-2.
- Langley RG, Elewski BE, Lebwohl M, Reich K, Griffiths CE, Papp K, Puig L, Nakagawa H, Spelman L, Sigurgeirsson B, et al. Secukinumab in plaque psoriasis–results of two phase 3 trials. N Engl J Med. 2014;371(4):326–38. doi:https://doi.org/10.1056/NEJMoa1314258.
- Paul C, JP L, Tedremets L, Kreutzer K, Jazayeri S, Adams S, Guindon C, You R, Papavassilis C; group Js. Efficacy, safety and usability of secukinumab administration by autoinjector/pen in psoriasis: A randomized, controlled trial (juncture). J Eur Acad Dermatol Venereol. 2015;29(6):1082–90. doi:https://doi.org/10.1007/s13555-018-0220-y.
- Mease PJ, McInnes IB, Kirkham B, Kavanaugh A, Rahman P, van der Heijde D, Landewe R, Nash P, Pricop L, Yuan J, et al. Secukinumab inhibition of interleukin-17a in patients with psoriatic arthritis. N Engl J Med. 2015;373(14):1329–39. doi:https://doi.org/10.1056/NEJMoa1412679.
- Mrowietz U, Leonardi CL, Girolomoni G, Toth D, Morita A, Balki SA, Szepietowski JC, Regnault P, Thurston H, Papavassilis C, et al. Secukinumab retreatment-as-needed versus fixed-interval maintenance regimen for moderate to severe plaque psoriasis: A randomized, double-blind, noninferiority trial (sculpture). J Am Acad Dermatol. 2015;73(1):27–36 e21. doi:https://doi.org/10.1016/j.jaad.2015.04.011.
- Rich P, Sigurgeirsson B, Thaci D, Ortonne JP, Paul C, Schopf RE, Morita A, Roseau K, Harfst E, Guettner A, et al. Secukinumab induction and maintenance therapy in moderate-to-severe plaque psoriasis: A randomized, double-blind, placebo-controlled, phase II regimen-finding study. Br J Dermatol. 2013;168(2):402–11. doi:https://doi.org/10.1111/bjd.12070.
- Balato A, Scala E, Balato N, Caiazzo G, Di Caprio R, Monfrecola G, Raimondo A, Lembo S, Ayala F. Biologics that inhibit the Th17 pathway and related cytokines to treat inflammatory disorders. Expert Opin Biol Ther. 2017;17(11):1363–74. doi:https://doi.org/10.1080/14712598.2017.1363884.
- McInnes IB, Mease PJ, Kirkham B, Kavanaugh A, Ritchlin CT, Rahman P, van der Heijde D, Landewe R, Conaghan PG, Gottlieb AB, et al. Secukinumab, a human anti-interleukin-17a monoclonal antibody, in patients with psoriatic arthritis (future 2): A randomised, double-blind, placebo-controlled, phase 3 trial. Lancet (London, England). 2015;386(9999):1137–46. doi:https://doi.org/10.1016/S0140-6736(15)61134-5.
- Strand V, Balsa A, Al-Saleh J, Barile-Fabris L, Horiuchi T, Takeuchi T, Lula S, Hawes C, Kola B, Marshall L. Immunogenicity of biologics in chronic inflammatory diseases: A systematic review. BioDrugs. 2017;31(4):299–316. doi:https://doi.org/10.1007/s40259-017-0231-8.
- Reich K, Blauvelt A, Armstrong A, Langley RG, Fox T, Huang J, Papavassilis C, Liang E, Lloyd P, Bruin G. Secukinumab, a fully human anti-interleukin-17a monoclonal antibody, exhibits minimal immunogenicity in patients with moderate-to-severe plaque psoriasis. Br J Dermatol. 2017;176(3):752–58. doi:https://doi.org/10.1111/bjd.14965.
- Reich K, Blauvelt A, Armstrong A, Langley RG, de Vera A, Kolbinger F, Spindeldreher S, Ren M, Bruin G. Secukinumab, a fully human anti-interleukin-17a monoclonal antibody, exhibits low immunogenicity in psoriasis patients treated up to 5 years. J Eur Acad Dermatol Venereol. 2019. doi:https://doi.org/10.1111/jdv.15637.
- Deodhar A, Gladman DD, McInnes IB, Spindeldreher S, Martin R, Pricop L, Porter B, Safi J Jr., Shete A, Bruin G. Secukinumab immunogenicity over 52 weeks in patients with psoriatic arthritis and ankylosing spondylitis. J Rheumatol. 2019. doi:https://doi.org/10.3899/jrheum.190116.
- Liu L, Lu J, Allan BW, Tang Y, Tetreault J, Chow CK, Barmettler B, Nelson J, Bina H, Huang L, et al. Generation and characterization of ixekizumab, a humanized monoclonal antibody that neutralizes interleukin-17a. J Inflamm Res. 2016;9:39–50. doi:https://doi.org/10.2147/JIR.S100940.
- van der Heijde D, Cheng-Chung Wei J, Dougados M, Mease P, Deodhar A, Maksymowych WP, Van den Bosch F, Sieper J, Tomita T, Landewe R, et al. Ixekizumab, an interleukin-17a antagonist in the treatment of ankylosing spondylitis or radiographic axial spondyloarthritis in patients previously untreated with biological disease-modifying anti-rheumatic drugs (coast-v): 16 week results of a phase 3 randomised, double-blind, active-controlled and placebo-controlled trial. Lancet (London, England). 2018;392(10163):2441–51. doi:https://doi.org/10.1016/S0140-6736(18)31946-9.
- Genovese MC, Combe B, Kremer JM, Tsai TF, Behrens F, Adams DH, Lee C, Kerr L, Nash P. Safety and efficacy of ixekizumab in patients with psa and previous inadequate response to tnf inhibitors: week 52 results from spirit-p2. Rheumatology (Oxford). 2018;57(11):2001–11. doi:https://doi.org/10.1093/rheumatology/key182.
- Gordon KB, Blauvelt A, Papp KA, Langley RG, Luger T, Ohtsuki M, Reich K, Amato D, Ball SG, Braun DK, et al. Phase 3 trials of ixekizumab in moderate-to-severe plaque psoriasis. N Engl J Med. 2016;375(4):345–56. doi:https://doi.org/10.1056/NEJMoa1512711.
- Leonardi C, Maari C, Philipp S, Goldblum O, Zhang L, Burkhardt N, Ball S, Mallbris L, Gonzalez P, Fernandez-Penas P, et al. Maintenance of skin clearance with ixekizumab treatment of psoriasis: three-year results from the uncover-3 study. J Am Acad Dermatol. 2018;79(5):824–830 e822. doi:https://doi.org/10.1016/j.jaad.2018.05.032.
- Eli lilly and co. 2016 Taltz (ixekizumab) [package insert]. [accessed 06th aug 2019] https://www.Accessdata.Fda.Gov/drugsatfda_docs/label/2016/125521s000lbl.Pdf.
- Karle A, Spindeldreher S, Kolbinger F. Secukinumab, a novel anti-IL-17A antibody, shows low immunogenicity potential in human in vitro assays comparable to other marketed biotherapeutics with low clinical immunogenicity. MABS. 2016;8(3):536–50. doi:https://doi.org/10.1080/19420862.2015.1136761.
- Spindeldreher S, Maillere B, Correia E, Tenon M, Karle A, Jarvis P, Kolbinger F. Secukinumab demonstrates significantly lower immunogenicity potential compared to ixekizumab. Dermatol Ther (Heidelb). 2018;8:57–68.
- Delluc S, Ravot G, Maillere B. Quantitative analysis of the CD4 T-cell repertoire specific to therapeutic antibodies in healthy donors. Faseb J. 2011;25(6):2040–48. doi:https://doi.org/10.1096/fj.10-173872.
- Delluc S, Ravot G, Maillere B. Quantification of the preexisting CD4 T-cell repertoire specific for human erythropoietin reveals its immunogenicity potential. Blood. 2010;116(22):4542–45. doi:https://doi.org/10.1182/blood-2010-04-280875.
- Hamze M, Meunier S, Karle A, Gdoura A, Goudet A, Szely N, Pallardy M, Carbonnel F, Spindeldreher S, Mariette X, et al. Characterization of CD4 T cell epitopes of infliximab and rituximab identified from healthy donors. Front Immunol. 2017;8:500. doi:https://doi.org/10.3389/fimmu.2017.00500.
- Mohan JF, Petzold SJ, Unanue ER. Register shifting of an insulin peptide-MHC complex allows diabetogenic T cells to escape thymic deletion. J Exp Med. 2011;208(12):2375–83. doi:https://doi.org/10.1084/jem.20111502.
- Honegger A, Pluckthun A. Yet another numbering scheme for immunoglobulin variable domains: an automatic modeling and analysis tool. J Mol Biol. 2001;309(3):657–70. doi:https://doi.org/10.1006/jmbi.2001.4662.
- Harding FA, Stickler MM, Razo J, DuBridge RB. The immunogenicity of humanized and fully human antibodies: residual immunogenicity resides in the CDR regions. MABS. 2010;2(3):256–65. doi:https://doi.org/10.4161/mabs.2.3.11641.
- Lefranc MP. IMGT, the international immunogenetics database. Nucleic Acids Res. 2001;29(1):207–09. doi:https://doi.org/10.1093/nar/29.1.207.
- Lefranc MP, Giudicelli V, Duroux P, Jabado-Michaloud J, Folch G, Aouinti S, Carillon E, Duvergey H, Houles A, Paysan-Lafosse T, et al. IMGT(r), the international immunogenetics information system(r) 25 years on. Nucleic Acids Res. 2015;43(Database issue):D413–422. doi:https://doi.org/10.1093/nar/gku1056.
- Lazar GA, Desjarlais JR, Jacinto J, Karki S, Hammond PW. A molecular immunology approach to antibody humanization and functional optimization. Mol Immunol. 2007;44(8):1986–98. doi:https://doi.org/10.1016/j.molimm.2006.09.029.