ABSTRACT
LINGO-1 is a membrane protein of the central nervous system (CNS) that suppresses myelination of axons. Preclinical studies have revealed that blockade of LINGO-1 function leads to CNS repair in demyelinating animal models. The anti-LINGO-1 antibody Li81 (opicinumab), which blocks LINGO-1 function and shows robust remyelinating activity in animal models, is currently being investigated in a Phase 2 clinical trial as a potential treatment for individuals with relapsing forms of multiple sclerosis (AFFINITY: clinical trial.gov number NCT03222973). Li81 has the unusual feature that it contains two LINGO-1 binding sites: a classical site utilizing its complementarity-determining regions and a cryptic secondary site involving Li81 light chain framework residues that recruits a second LINGO-1 molecule only after engagement of the primary binding site. Concurrent binding at both sites leads to formation of a 2:2 complex of LINGO-1 with the Li81 antigen-binding fragment, and higher order complexes with intact Li81 antibody. To elucidate the role of the secondary binding site, we designed a series of Li81 variant constructs that eliminate it while retaining the classic site contacts. These Li81 mutants retained the high affinity binding to LINGO-1, but lost the antibody-induced oligodendrocyte progenitor cell (OPC) differentiation activity and myelination activity in OPC- dorsal root ganglion neuron cocultures seen with Li81. The mutations also attenuate antibody-induced internalization of LINGO-1 on cultured cortical neurons, OPCs, and cells over-expressing LINGO-1. Together these studies reveal that engagement at both LINGO-1 binding sites of Li81 is critical for robust functional activity of the antibody.
Introduction
LINGO-1 (leucine-rich repeat and Ig containing Nogo receptor interacting protein-1), also known as LERN1 and LRRN6A, is selectively expressed by oligodendrocytes and neurons in the central nervous system (CNS).Citation1–Citation4 LINGO-1 expression regulates the timing of CNS myelination during development and LINGO-1 upregulation in neurological disorders suggests a deleterious role for the endogenous protein.Citation1,Citation2,Citation5,Citation6 Blocking LINGO-1 function leads to robust remyelination in chemical- and immune-induced demyelination animal models.Citation7–Citation10 The biological consequences of blocking LINGO-1 function have been substantiated using small interfering ribonucleic acid (siRNA), soluble versions of the LINGO-1 extracellular domain, anti-LINGO-1 antibodies, and LINGO-1-null mice.Citation1,Citation6–Citation8,Citation10−Citation14 LINGO-1 is a 581 amino acid transmembrane protein. The extracellular domain of LINGO-1 is heavily glycosylated and contains 12 leucine rich repeat (LRR) motifs with N- and C-terminal caps, an immunoglobulin (Ig) domain, and a stalk region attached to a transmembrane region and a short distal cytoplasmic tail in the full length protein.Citation1,Citation15 The Ig domain of LINGO-1 plays an important role in its biological function. Structure-activity relationship studies suggest that the Ig domain alone is sufficient for its activity.Citation16,Citation17 The LINGO-1 ectodomain structure revealed that the protein self-associates to form a ring-shaped tetramer in which the Ig domain makes contacts with the N-terminal LRR sequences from an adjacent LINGO-1 to drive homotetramer formation (Figure S1A and S1B).Citation15
Immunoglobulin (Ig) G monoclonal antibodies (mAbs) are the most common drug platform of the biopharmaceutical industry, with over 85 antibody drugs approved and hundreds of others in clinical trials.Citation18,Citation19 IgG mAbs, which have two antigen-binding fragment (Fab) arms, can bind to one or two ligand molecules, leading to 1:1 and/or 1:2 antibody:ligand complexes. The anti-LINGO-1 Li81 mAb (opicinumab) (equilibrium dissociation constant KD = 20 pM for LINGO-1) is a human antibody discovered using Fab phage display technology,Citation12 engineered into a human IgG1 aglycosyl framework for reduced effector function.Citation12,Citation20 It is currently being investigated in clinical trials as a potential treatment to repair neuronal damage that occurs in the CNS of individuals with multiple sclerosis (MS) (AFFINITY: clinical trial.gov number NCT03222973).Citation3,Citation21,Citation22
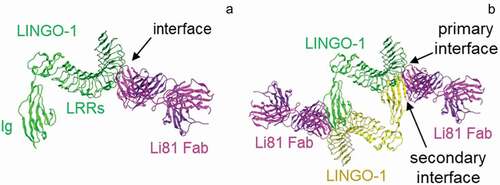
To investigate the mechanism of action of the Li81 antibody, we solved the crystal structure of the LINGO-1 ectodomain/Li81 Fab complex.Citation20 An unexpected feature of the structure was that the Li81 Fab contained two binding sites for LINGO-1, and this led to the formation of a heterotetrameric unit that contained 2 copies each of the Fab and LINGO-1, where the classical primary binding of the Fab through its complementarity-determining regions (CDRs) to LINGO-1 created a secondary binding site that recruited a second copy of LINGO-1 () vs. )). Indeed, a tetrameric LINGO-1/Li81 Fab complex was also observed by single particle tomography using electron microscopy and biochemical assessments.Citation20 The binding of Li81 blocks contacts that allow LINGO-1 to form its homotetramer, and somewhat obstructs the LINGO-1 Ig domain RKH sequence motif (residues 423–425), which is required for binding to Nogo receptor interacting protein-1 (NgR1).Citation20
Figure 1. Properties of the Li81 Fab–LINGO-1 ectodomain complex. Binding interfaces of the Li81 Fab-LINGO-1 ectodomain complex determined from the crystal structure.Citation20 Structural figures were rendered with MOE software.Citation23 (a) Contacts comprising the primary binding interface, between Li81 (pink) CDR residues and LINGO-1 (green) LRR domains 4–8. (b) Contacts comprising both the primary interface and the secondary binding interface, between Li81 (pink) light chain framework residues and the Ig domain of a separate molecule of LINGO-1 (yellow)
To explore the contribution of this secondary binding site on structure-function, we eliminated this site of Li81 by mutagenesis and compared the properties of the modified constructs with the parent antibody. Elimination of the cryptic site did not significantly affect the binding affinity of the mAb for LINGO-1, but prevented the assembly of LINGO-1 and the Li81 variant into higher order complexes, attenuated the ability of the antibody to drive the internalization and degradation of surface LINGO-1 in cell-based assays, and led to a loss of function in in vitro assays where Li81 treatment drives the differentiation of oligodendrocyte progenitor cells (OPCs) into mature myelin basic protein (MBP)-producing oligodendrocytes and myelination in OPC-dorsal root ganglia (DRG) neuron cocultures. This unique binding attribute of the Li81 antibody, where each Fab can simultaneously bind two copies of its ligand, is to our knowledge unprecedented. Concurrent engagement at both sites is key to the robust activity of the Li81 mAb.
Results
Treatment of LINGO-1 with Li81 mAb leads to concatemer formation
Previously we discovered that Li81 antibody has two distinct LINGO-1 binding sites.Citation20 Structural features of these sites are shown in . The primary binding site involves contacts between Li81 CDR residues and LINGO-1 LRR domains 4–8 ()), while the secondary site involves contacts between Li81 framework residues and the Ig domain of a second molecule of LINGO-1 ()). To assess the contributions of the two binding sites, we performed enzyme-linked immunosorbent assays (ELISA) to evaluate Li81 binding to chimeric versions of the LINGO-1 ectodomain that retained the secondary binding site and/or most of the primary binding site (Figure S2). In these constructs, portions of the LINGO-1 sequence were replaced with the corresponding sequences from LINGO-2, since Li81 does not bind LINGO-2. In the chimeric constructs named 1–2, 2–1, 1-2-1, and 2-1-2, LINGO-1 sequences were replaced with LINGO-2 LRR domains 7–12, 1–6, 5–8, or 1–4 plus 9–12, respectively. Constructs 1–2 and 2-1-2 also contain the LINGO-2 Ig domain and stalk region. Li81 bound only to the chimeric construct 2-1-2, which includes most of the LINGO-1 primary binding site epitope in LRRs 4–8. Li81 did not bind to constructs 2–1 and 1-2-1, which contain the Ig domain of LINGO-1, but for which most of LRRs 4–8 do not have their LINGO-1 sequences. To confirm that the lack of binding was not caused by improper folding of the LINGO-1 Ig domain, we tested binding of the same constructs to anti-LINGO-1 1A7 mAb that binds to the LINGO-1 Ig domain.Citation20 1A7 bound to constructs 2–1, 1-2-1, and LINGO-1, which all contain the LINGO-1 Ig domain, but not to constructs 1–2, 2-1-2, and LINGO-2, which contain the LINGO-2 Ig domain. Together these studies indicate that the Li81 antibody only binds the LINGO-1 Ig domain when the primary binding site is occupied.
To study the binding stoichiometry of LINGO-1/Li81 in solution, we analyzed complex formation of mixtures of LINGO-1 ectodomain and intact Li81 mAb at various ratios by size exclusion chromatography (SEC) using in-line multiangle light scattering (MALS) to calculate the apparent molecular weights (MWs) for each peak ()). LINGO-1 and Li81 alone eluted from the column at 14.2 and 13.7 min with 88 and 150 kDa MWs, respectively, consistent with their predicted molecular masses. When LINGO-1 and Li81 were mixed at a 2:1 molar ratio of LINGO-1:antibody (10 μg of each), all of the added LINGO-1 and Li81 formed complexes, thus no free LINGO-1 or Li81 was detected. At the 2:1 ratio, we observed a major ~900 kDa peak representing the predominant form, a minor earlier eluting peak of higher MW, and a minor later eluting peak of ~500 kDa. These three peaks were also observed when excess LINGO-1 was mixed with Li81 (10 µg LINGO-1 and 3 µg Li81) and when LINGO-1 was mixed with excess Li81 (10 µg LINGO-1 and 30 µg Li81). The ~900 kDa peak was the predominant peak with excess LINGO-1 added, whereas the ~500 kDa peak was the predominant peak with excess Li81. Free LINGO-1 or Li81 in these samples eluted at their expected MWs.
Figure 2. Concentration-dependent oligomerization of Li81 mAb-LINGO-1 ectodomain complexes. Samples containing 10 µg LINGO-1 ectodomain and 0, 3 10, or 30 µg of Li81 mAb, or 10 µg Li81 mAb alone, were subjected to analytical SEC. The column effluent was analyzed for absorbance at 280 nm and in-line light scattering. (a) Chromatograms from the SEC column. Y-axis: milli-absorbance units (mAU). X-axis: time in minutes after injection. (b) Image of three representative 34 nm particles observed by EM from a sample containing 10 µg each of Li81 mAb and LINGO-1 ectodomain and model of the 34 nm particle. (c and d) SDS-PAGE/Western blot analysis of LINGO-1 in CHO cells expressing HA LINGO-1 following treatments for 2 days with serial dilutions of the Li81 mAb (c) and Li81 Fab (d). Apparent molecular weights in kiloDaltons (kDa), based on the positions of pre-stained markers, are shown at the left
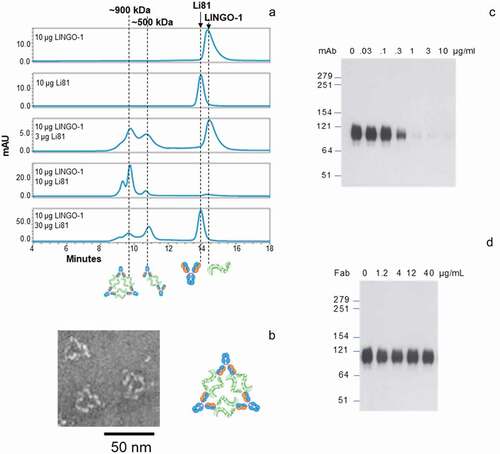
To assess the composition of the LINGO-1 ectodomain/Li81 complexes at a molecular level, we subjected samples to transmission electron microscopy (EM). Interestingly, the EM images contained ring-like structures with the most prevalent form having an average particle size of ~34 nm (), Figure S3). The geometry of the 34 nm particle, together with the molecular mass of ~900 kDa that we calculated in the SEC-MALS study, suggests the formation of a complex containing 6 copies of LINGO-1 and 3 copies of intact Li81 mAb. ) depicts a cartoon schematic representing the organization of the 6 LINGO-1 ectodomain/3 Li81 mAb complex. In the schematic, the 2 LINGO-1 ectodomain/2 Li81 Fab tetrameric complex seen in the crystal structure was used as a building block for assembly of the complex.Citation20 We refer to complexes containing multiple copies of this LINGO-1/Li81 unit as concatemers. The high representation of the 34 nm particles by EM suggests that the 6 LINGO-1 ectodomain/3 Li81 mAb concatemer is the preferred organization of the complex at this molar ratio. Other less common high MW particles seen in the EM image formed more varied assemblies (Figure S3). Dynamic light scattering (DLS) was used to monitor gross changes in the organization of the particles as a function of time. The complexes formed by LINGO-1 and Li81 maintained their steady state at room temperature or 37ºC and over a range of concentrations (data not shown). Distribution of the hydrodynamic radius (Rh) of the complexes formed by mixing 10 μg/ml LINGO-1 and 30 μg/ml Li81 did not change over time at 37ºC for 3 days. Only minor large aggregates were observed by the end of incubation and no aggregation was seen with LINGO-1 and Li81 individually under the same conditions (data not shown).
Designing mutants of Li81 to eliminate the secondary LINGO-1 binding site
To elucidate the contribution of the secondary binding site to Li81 function, we designed mutations in the light chain of Li81 to eliminate or greatly reduce binding at the secondary site while retaining the primary site binding. The designs targeted residues that make up the secondary binding site or are predicted to interfere with the interface of the LRR domains of the first LINGO-1 molecule with the Ig of the second LINGO-1 molecule. Our design was structure-guided, relying mostly on our pdb:4OQT crystal structure, whose triple interface of Li81/first LINGO-1 LRRs/second LINGO-1 Ig we assumed as the explanation for the concatemers observed in solution.Citation20 The structure indicated that only a few Li81 residues could possibly directly affect the secondary binding site. We based most of our mutations ()) on corresponding residues in Li113, another high affinity anti-LINGO-1 antibody that does not form higher order complexes.Citation24 Li113 was chosen because it shares both the binding geometry of Li81, based on epitope mapping/cross-blocking studies (Figure S2), and the conservation of key residues that contribute to LINGO-1 binding (); Figure S4).
Figure 3. The secondary LINGO-1 binding site of Li81 mAb. (a) Interfaces between Li81 and LINGO-1 LRR domains and the Ig domain of the second LINGO-1 molecule identified in the pdb:4OQT crystal structure, with the three mutations in CN1373 and the glycan on LINGO-1 N225 highlighted. (b) Close-up of Li81 VL R18 and LINGO-1 Ig domain contacts, with distances for the closest contacts shown in green (Å). (c) Close-up of Li81 VL R54 and LINGO-1 Ig domain contacts, and possible hydrogen bonds to backbone O atoms in each molecule. (d) Close-up of where Li81 VL S31 could, if mutated, contact the glycan at N225 of LINGO-1 LRR region and the second LINGO-1 molecule Ig domain. (e) Sequence of Li81 VL. Li81 residues with lines above indicate the primary binding site (i.e., interatomic distance within 4.5 Å of LINGO-1 LRR domains); lines below indicate the secondary binding site (i.e., within 4.5 Å of LINGO-1 Ig domain). Li113 VL, which inspired most of the mutations, is shown for comparison; S31R and 4 other differences in the secondary binding site are emboldened and highlighted in yellow. CDRs are highlighted in gray
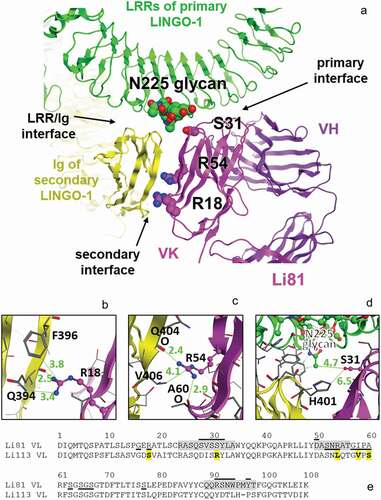
The secondary binding site, i.e., the interface of the Li81 light chain variable domain (VL) with the Ig domain of the second LINGO-1 molecule, buries 495 Å2 and involves 9 VL and 11 Ig residues. The closest contacts are where Li81 VL R18 and R54 contacts the LINGO-1 Ig domain ()). Additional contacts between the LINGO-1 LRR domain and the second LINGO-1 Ig domain in the vicinity of the secondary binding site bury 1012 Å2, involve 10 LRR and 10 Ig residues, have a more closely packed interface, and are predicted to be higher-affinity than the VL:Ig secondary binding site. One of the Li81 mutations in our designs, S31R, was chosen to interfere with binding near the LINGO-1 LRR domain/LINGO-1 Ig domain contacts.
Li81 VL R18 extends to contact the LINGO-1 Ig domain, possibly forming a hydrogen bond with the backbone O atom of Q394 within the LINGO-1 Ig domain ()), so mutation to most short amino acids was expected to reduce affinity. R18S was chosen because Li113 has a serine at this position (due to an unusual somatic hypermutation at this framework position) and because serine is an innocuous short hydrophile.
Li81 VL R54 makes a hydrogen bond with the backbone O atom of Li81 VL A60, and a possible second hydrogen bond with the backbone O atom of LINGO-1 Ig Q404 ()). Mutating this position required more care, as it could affect the structure of the CDR-L2 within which it resides, and therefore the affinity of the primary binding site. Two different approaches were tried. The first approach was to use R54T (in designs CN1372 and CN1373) because threonine is an innocuous short hydrophile. However, R54T would leave a gap in the buried structure near its turn, unlike either the semi-conserved germline sequences of humIGKV3-like Li81 or of humIGKV1-like Li113, and so was judged to have some risk of reducing the affinity of the primary binding site. Therefore we also tried a second approach (in designs CN1374, CN1375, CN1376, and CN1373) which involved mutating the R54 adjoining strand to match the Li113 humIGKV1-typical sequence, i.e., R54L A55Q I58V A60S. Modeling the structure on Li81 or on a humIGKV1 structure suggested that the overall secondary structure would not change because of these mutations, and neither would the fit at the primary binding site.
Notably, Li81 is nearest to the interface between LINGO-1 LRR domains and the second LINGO-1 Ig domain ()) where S31R in Li113 appears able to interact with the glycan on N225 of the LINGO-1 LRR domains, based on molecular modeling. Whether such an interaction would be favorable was uncertain, but the large arginine residue could interfere with the association between that glycan and the Ig domain of the second LINGO-1. Based on molecular modeling, we judged S31R unlikely to harm the affinity of the primary binding site, and possibly able to improve it. Since no mutations of these three residues, R18, R54, and S31, looked able to directly impede the secondary binding site, we did not generate constructs with single mutations, anticipating that combinations of two or three mutations would be needed ().
Table 1. Thermal stability and binding affinity of Li81 light chain variant antibodies designed to reduce binding at the secondary site. Tm values were measured by differential scanning fluorimetry. EC50 values were calculated in an ELISA study assessing binding on LINGO-1-Fc coated plates
Purification and properties of Li81 variants targeting secondary binding site contacts
The seven new constructs designed to disrupt Li81-LINGO-1 contacts within the secondary binding site were expressed in Chinese hamster ovary (CHO) cells, and the resulting mAbs were purified on Protein A Sepharose. For all the purified mAbs, 50 and 25 kDa bands for the heavy and light chains, respectively, were observed under reducing conditions, and the characteristic 150 kDa band for the tetrameric complex of two heavy and two light chains was observed under nonreducing conditions ()). The apparent affinities of these new mAbs for LINGO-1 were analyzed by ELISA ()). The observed EC50 values () were similar to Li81, indicating that the mutations did not have a significant effect on the binding to LINGO-1. Because LINGO-1 is immobilized on the plate and is not free to assemble into the complexes seen in solution, we can infer that the ELISA method only measures binding at the primary site, and consequently that the mutations designed to disrupt the secondary binding site did not affect binding at the primary site. We then evaluated the effect of the mutations on the secondary binding site by monitoring the concatemer formation with LINGO-1 by analytical SEC ()). Three of the mutants, CN1373, CN1374, and CN1375, showed a loss of the concatemer peaks where the resulting complexes eluted as single peaks. While it was not possible to directly measure the affinity for binding at the secondary site because of the much lower affinity for it compared to the primary site,Citation20 based on the differences in their elution profiles and sharpness of the SEC peaks, we selected CN1373 as the lead for our study, and CN1375 as the back-up molecule.
Figure 4. Biochemical attributes of Li81 variants targeting secondary binding site contacts. (a) Samples (4 µg/lane) were subjected to SDS-PAGE and stained with Coomassie brilliant blue. Samples on the left were analyzed under reducing conditions, and samples on the right were analyzed under non-reducing conditions. Molecular weight markers and their apparent molecular masses are shown at the left of the panel. (b) The apparent affinities of the seven secondary binding site mutants for LINGO-1 were measured by a direct-binding ELISA. Data are plotted as absorbance at 450 nm versus concentration. (c) Samples, each containing 10 µg LINGO-1 ectodomain and 10 µg of of one of the mutant mAbs, were subjected to SEC on an analytical SEC column using PBS as the mobile phase. The column effluent was analyzed for absorbance at 280 nm. (d) Samples containing CN1373 mAb alone, or 3 µg LINGO-1 ectodomain and 1.6, 3, or 10 µg of CN1373 mAb, were subjected to SEC with in-line light scattering
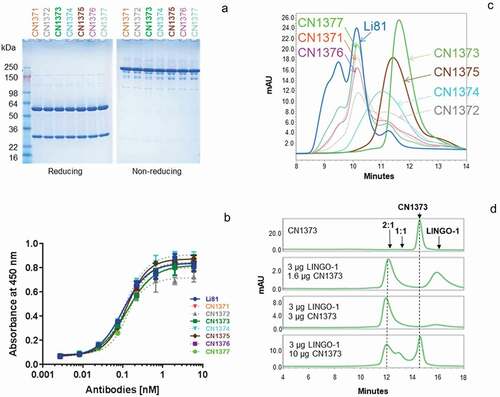
We next examined the CN1373 mAb at three different concentrations with a fixed concentration of LINGO-1 using SEC with in-line light scattering, as we had done for Li81 mAb ()). In the presence of molar excess and equivalent amounts of LINGO-1, we observed a single complex peak of the characteristic 2:1 LINGO-1:mAb stoichiometry, whereas in the presence of excess CN1373 we see peaks characteristic of 2:1 and 1:1 complexes. Under none of the conditions did we see evidence for concatemer formation for CN1373, indicating that the designed mutants successfully eliminated the secondary binding site without affecting the affinity of the antibody for LINGO-1. Similar results were observed for CN1374 and CN1375 (data not shown). The three residues mutated in CN1373 target all three potential contacts of Li81 with the second LINGO-1 Ig domain ()). Constructs CN1371 and CN1372 each contain two of the three mutations found in CN1373. Neither of the double mutants fully ablated secondary binding. The R18S and S31R mutations (CN1371) led to about a 25% reduction in the levels of the concatemer peaks ()). The R18S and R54T mutations (CN1372) were more significant and led to about a 50% reduction of the concatemer peaks. R18 and R54 are both at the interface between the Li81 light chain and LINGO-1 Ig domain ()), confirming the role these residues play in the creation of the secondary binding site.
The abilities of mAbs CN1373 and CN1375 to block LINGO-1 function were tested in an in vitro OPC assay by measuring their impact on the differentiation of OPCs into mature MBP-producing oligodendrocytes ()). As we expected, blockade of LINGO-1 function with Li81 promoted OPC differentiation in this assay, as evident by the dramatic increase in MBP expression, whereas treatment with an isotype control mAb had no impact on MBP expression. To our surprise, CN1373 and CN1375 did not lead to a significant increase in MBP expression. Compared to Li81, there were clear reductions in the level of MBP expression in cultures treated with CN1373 and CN1375 for 2 days (), left panel), and this effect was even more pronounced in a repeat study from cultures treated for 3 days (), right panel). These studies show that engagement of LINGO-1 through framework contact residues in the second binding site is required for the OPC differentiation activity of Li81. Like Li81, LINGO-1 ectodomain is a potent inducer of MBP expression in the OPC differentiation assay when it is added to the culture medium, where in soluble form the ectodomain acts as a decoy receptor to antagonize endogenous LINGO-1 activity.Citation2 Previously we showed that LINGO-1 ectodomain was inactive when complexed with Li81 before it was added to the assay cultures.Citation20 In contrast, when LINGO-1-CN1373 mAb complex was added to the OPC cultures, the presence of the antibody did not neutralize the exogenously added LINGO-1, further substantiating the role of the secondary binding site for function ()).
Figure 5. Activity of the Li81 secondary binding site mutants in the OPC differentiation and coculture assays. In A and B, samples were evaluated in the OPC differentiation bioassay using expression of MBP as a readout for differentiation. MBP expression was quantified by ICC and recorded as MFI per differentiated oligodendrocyte (Olig2+ cell). (a) Dose-response of Li81, CN1373, and CN1375 in the OPC differentiation assay. Data shown are from 2 independent studies. (b) Activity of Li81, LINGO-1, CN1373, and premixed CN1373/LINGO-1 complex in the OPC differentiation assay. C. DRG neuron/A2B5+ cell cocultures were treated with 3 µg/mL of Li81, CN1373, or 5C8 isotype control antibody. The cultures were fixed on day 10 and visually analyzed for axonal myelination by oligodendrocytes by ICC using anti-MBP (green, 488) and anti-tubulin (red, 594) antibodies for detection of myelinating oligodendrocytes and neurons, respectively
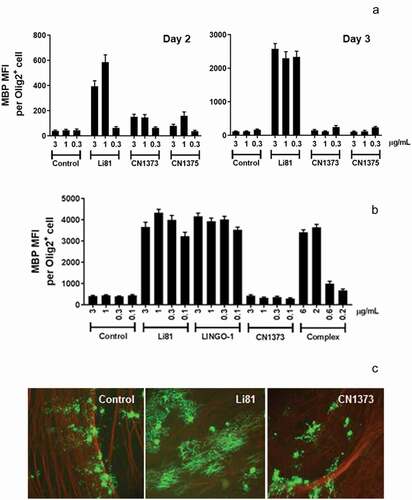
Myelination of axons in CNS by oligodendrocytes is tightly regulated. This process can be recapitulated in vitro in cocultures of DRG neurons and OPCs; however, spontaneous myelination is very inefficient. LINGO-1 is a known suppressor of this process and inhibition of LINGO-1 function leads to robust myelination. When we add Li81 to OPC/DRG neuron cocultures to block LINGO-1 activity, the treatment leads to robust myelination, as indicated by the formation of clusters of elongated MBP+ fibers by immunocytochemistry (ICC) ()). Each cluster represents myelination from a single differentiated oligodendrocyte. To understand the effect of the secondary binding site of Li81 on this process, we tested CN1373 in the OPC/DRG neuron coculture myelination assay. CN1373 treatment led to significantly reduced MBP+ fiber clusters, and the clusters were more disordered compared to Li81 treatment, similar to results we observed in cocultures treated with the control mAb ()). These findings reveal an important role of engaging the secondary binding site on Li81 to promote myelination.
We also previously observed that, after binding of Li81 to the LINGO-1 ectodomain, formation of the 2:2 complex protected LINGO-1 from proteolytic cleavage with endoproteinase Lys-C at the junction of the LRR and Ig domain.Citation20 When the CN1373-LINGO-1 ectodomain complex was treated with endoproteinase Lys-C, digestion was indistinguishable from the control digest that contained LINGO-1 and a non-binding antibody, indicating that binding to the LINGO-1 primary site is not sufficient to protect the LINGO-1 from proteolysis (Figure S5).
Li81 mAb promotes the internalization and degradation of LINGO-1 ()). When CHO cells expressing LINGO-1 were treated with serial dilutions of Li81 mAb and Fab and analyzed by western blotting for LINGO-1, Li81 mAb treatment led to loss of LINGO-1 at concentrations greater than 0.1 µg/mL, with complete loss of the western signal at 1 µg/mL ()). LINGO-1 Fab treatment had no effect on LINGO-1 levels ()). To assess the impact of the secondary binding site on the internalization of LINGO-1, we performed a series of fluorescence-activated cell sorting (FACS) studies (). Treatment of CHO cells expressing LINGO-1 with Li81 for 23 h at 37°C led to a large decrease in mean fluorescence intensity (MFI) of fluorescently labeled anti-human detection antibody, indicating reduced surface levels of LINGO-1 ()). In contrast, when CHO-LINGO-1 cells were treated with CN1373 and CN1375, only a modest decrease in LINGO-1 surface levels resulted. Treatment with the isotype control mAb had no effect on surface LINGO-1. Similarly, treatment of murine hematopoietic Ba/F3 cells expressing LINGO-1 with Li81 for 3 h led to an ~40% reduction in the levels of surface LINGO-1 at 37°C versus 4°C, whereas treatment with CN1373 led to only a 15% decrease ()).
Figure 6. Internalization of LINGO-1 following treatment with Li81 and CN1373. (a) CHO cells expressing full-length LINGO-1 were treated with 5C8 isotype control antibody (Ctl), Li81, or CN1373 at 1 and 3 µg/mL for 23 h at 37°C and analyzed by FACS, staining for anti-human IgG. (b) Ba/F3 cells expressing full-length LINGO-1 were treated for 3 h at 3°C or 37°C with serial dilutions of 5C8, Li81, or CN1373 and analyzed by FACS. (c) Rat primary cortical neurons cultured for 12 days were treated with isotype control (5C8) and LINGO-1 specific antibodies for 90 min at 37°C, acid washed and fixed for with 1.5% paraformaldehyde for 5 min at room temperature. For assessing intracellular trafficking, the cells were treated with detergent, and then treated with Alexa-647 secondary antibody (red) for detection of Li81, CN1373, and 5C8, and Alexa-488 (green) for the lysosomal marker Rab7. Representative confocal middle sections of each condition are shown. Selected regions (stippled-line squares) were cropped and enlarged and are displayed using an inverted monochrome color scale to aid visualization. Scale bar, 5 µm
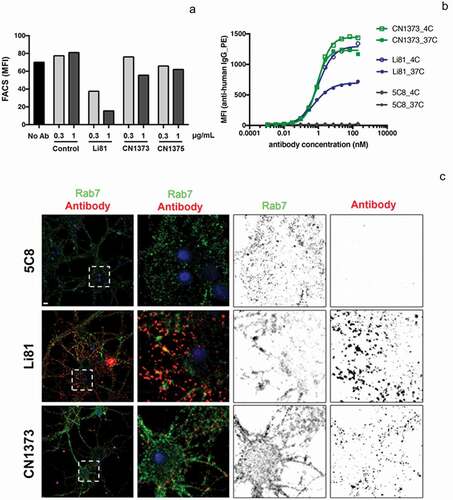
To explore the binding and internalization characteristics of Li81 and CN1373 in a more physiological setting, we performed studies on cultured rat cortical neurons. Both CN1373 and Li81 antibodies showed extensive surface staining of cultured neurons (Figure S6, surface) and less intense intracellular binding sites (Figure S6, internal), indicating that the binding pattern of both anti-LINGO-1 antibodies are very comparable. Minor staining was detected with the isotype control antibody. When cortical neurons were treated for 90 min with Li81 and CN1373, acid washed to release surface bound antibody, and probed for the intracellular distribution of the antibodies, extensive Li81 staining was observed for the Li81-treated culture, far less staining was detected following CN1373 treatment, and no staining was detected in the isotype control-treated culture ()). When the cultures were costained with the lysosomal marker Rab7, 30% of the antibody in the Li81-treated sample costained with Rab7, but only 6% of the antibody in the CN1373 culture costained with Rab7, and no staining was detected with the control antibody. Together these studies show that engagement of LINGO-1 through both binding sites on Li81 enhances the internalization of LINGO-1 compared to that seen with CN1373. A similar series of ICC studies were performed on rat primary OPC cultures. Li81 treatment led to internalization of LINGO-1 and targeting to lysosomes whereas internalization was greatly attenuated with CN1373 treatment (data not shown).
Multimeric CN1373 complexation increases antibody internalization but not activity
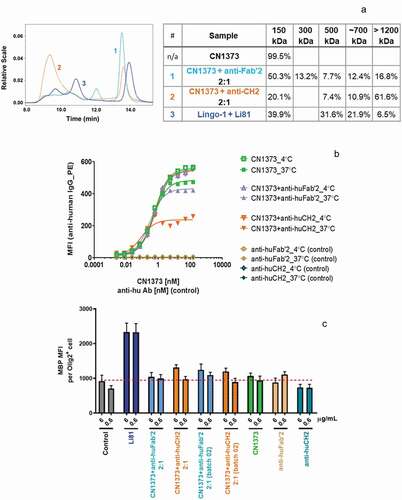
To test if internalization of LINGO-1 contributes to the potent activity of Li81 in the OPC differentiation assay, we reasoned that we could force CN1373 to cluster LINGO-1 and promote internalization by first treating it with an antibody directed against constant regions in the CN1373 framework, and then studying the biological activities of the complex. In these studies, CN1373 was mixed with anti-human CH2 or Fab′2 antibody at a ratio of 2:1. Treatment of CN1373 with the anti-CH2 domain antibody led to extensive formation of high order CN1373/anti-Fc complexes, while treatment with anti-Fab′2 antibody was less effective ()). The complexes maintained their steady state at 37ºC for 24 h when monitored by DLS (data not shown). The size distribution of the CN1373 complexes formed by the anti-human antibodies was determined by SEC-MALS ()). Anti-human CH2 shifted ~80% of CN1373 to dimer or larger clusters with a main population of ~1500 kDa containing 70% of the total mass, whereas the anti-human Fab′2 was only able to shift ~35% of CN1373 to a more heterogenous mixture of complexes. The sizes of the complexes and their proportion to total mass did not change when comparing the same sample before and after the 24 h incubation at 37°C. When these complexes were added to LINGO-1-expressing Ba/F3 cells, we observed extensive internalization of LINGO-1 with the CN1373 clustered by the anti-CH2 antibody approaching levels observed with Li81 and no significant change with the CN1373 treated with the anti-Fab′2 antibody ()). However, when the same samples were tested in the OPC assay, there was no gain of function for the CN1373/anti-Fc complex ()). Thus, we were able to decouple internalization from activity, showing that internalization is not a key driver for Li81 activity in the OPC differentiation assay.
Figure 7. Internalization of LINGO-1 following treatment with complex forms of CN1373. (a) SEC chromatograms and tabulated summary of the data from samples containing CN1373 that were clustered with anti-human CH2 or Fab′2 antibody. (b) Ba/F3 cells expressing full length LINGO-1 were treated for 3 h at 4°C or 37°C with serial dilutions of CN1373, anti-CH2-1373 complexes, and anti-Fab′2-1373 complexes and analyzed by FACS. (c) Activity of CN1373, anti-CH2-1373 complexes, and anti-Fab′2-1373 complexes in the OPC differentiation assay. MBP expression was quantified by ICC and recorded as MFI per differentiated oligodendrocyte (Olig2+ cell)
Discussion
MS is an inflammatory demyelination disease of the CNS that affects over 2 million individuals worldwide. While there are many approved drugs that target pathologic immune responses and provide significant relief from relapses, none directly target CNS repair.Citation25 Li81, an anti-LINGO-1 mAb, was the first drug in clinical trials for remyelination in MS patients. LINGO-1 is highly conserved evolutionarily, with the human and rat/mouse genes sharing 99.5% sequence identity. Preclinical studies in rodent models provided biological rationale for targeting MS with anti-LINGO-1 antagonists.Citation8,Citation10 The remyelination effects of Li81 were assessed in a Phase 2 human acute optic neuritis trial by measuring visual evoked potential. The study showed a trend toward improvement in the affected eye and significant improvement in the fellow eye.Citation22 However, there is a lack of qualified methods to directly assess CNS remyelination in humans.
The Li81 antibody opicinumab functions through an unprecedented mechanism of action where contacts between CDR residues on Li81 and LRR residues on LINGO-1 create a secondary binding site composed of Li81 light chain framework residues that recruits a second molecule of LINGO-1. From our studies comparing Li81 to the Li81 variant CN1373, which only retained the primary binding site, was attenuated in its ability to internalize LINGO-1, and was inactive in functional studies, we conclude that engagement at both sites is crucial to the activity of Li81. In theory, it is possible that the internalization of LINGO-1 induced by clustering upon Li81 binding was required for the antibody’s activity. However, this possibility was not supported by our findings since the regained LINGO-1 clustering activity achieved by treatment of CN1373 with anti-Fc antibody to create multimeric CN1373 complexes indeed promoted internalization, but did not increase OPC differentiation activity. Town et al. also explored internalization of LINGO-1 by Li81, but in a different context – an in vivo demonstration of a potential therapy using Li81 conjugated to the toxin doxorubicin to target Ewing sarcoma tumor cells, which have high LINGO-1 expression.Citation26 Their study did not rely on blocking the functional activity of LINGO-1, but simply used the antibody to selectively target tumor cells that specifically express LINGO-1 on surfaces in the periphery, but not the healthy tissues.Citation26 Taken together with our other findings, this is evidence that the increase of OPC differentiation activity by Li81 is not caused merely by its promotion of LINGO-1 clustering, but instead requires more specific aspects of the complexation geometry created when it engages LINGO-1 at both of its binding sites.
Previously we discovered that the RKH motif at positions 423–425 of the LINGO-1 Ig domain was critical and that mutations in the tripeptide led to loss of function. We proposed that the recruitment of the second LINGO-1 through its Ig domain after binding of the primary site of Li81 led to loss of LINGO-1 function by rendering the RKH motif inaccessible, presumably by preventing an interaction with NgR1 or other cellular targets. This mechanism was supported by three observations: 1) the positioning of the RKH motif in the heterotetrameric unit of our crystal structure, which appears to make it inaccessible to NgR1;Citation20 2) the reduced binding of the altered RKH→EKV version of the soluble LINGO-1 ectodomain to NgR1 compared to the wild-type ectodomain and loss of function in the OPC assay;Citation16,Citation20 and 3) the Ig domain is critical for LINGO-1 function, while truncation of the LRR domain did not affect function.Citation17 Since CN1373 only binds at the primary site on the LRR domain and presumably leaves the Ig domain of LINGO-1 accessible to bind its cellular targets, we predicted its binding would not affect function. Indeed, CN1373 failed to block LINGO-1 activity in the OPC assay. The mixing experiments from our current study provided further support for this mechanism. Exogenously added LINGO-1 ectodomain leads to a positive readout in the OPC assay.Citation20 When exogenously added LINGO-1 was mixed with Li81 under conditions where there was no free LINGO-1 or Li81, the complex was inactive in the OPC assay, but when CN1373 was similarly mixed with LINGO-1, CN1373 failed to block the function of the exogenously added LINGO-1, leading to a positive readout in the OPC assay. We speculate the key to the block of LINGO-1 activity in the Li81 mixing experiment is masking of the Ig domain from the second LINGO-1 following engagement in the secondary binding site, and not from the LINGO-1 that was bound with Li81 through the primary site. The slight reduction in activity of the CN1373/LINGO-1 complex versus LINGO-1 alone may result from partial interference with the activity of the Ig domain following CN1373 binding.
An additional unique feature of Li81 is that it induces a conformation change within the Ig domain, leading to a 15° rotation from its orientation in the LINGO-1 structure in the absence of Li81 (Figure S1C, S1D, and S1E). Li81 binding protects LINGO-1 against proteolysis by endoproteinase Lys-C, whereas CN1373, which no longer binds to this critical region of LINGO-1, is susceptible. It is unclear if the conformation change is in part responsible for the inactivation of LINGO-1 function or simply contributes to the stabilization of the 2:2 LINGO-1/Li81 Fab complex. In studies assessing complex formation over a wide range of LINGO-1 and Li81 Fab concentrations, we were unable to detect a 1:1 complex in association and dissociation studies.Citation20
Bothwell and coworkers published a series of papers suggesting LINGO-1 was intracellular and not on the cell surface.Citation27,Citation28 To explore this possibility, we performed ICC studies on rat cortical neurons with Li81. When cortical neuron cultures were treated with Li81 without permeabilization, we saw robust staining of the cells. After permeabilization we also saw intracellular LINGO-1 staining, although a significant amount or a majority of the LINGO-1 was on the cell surface. As part of the analysis we tested the same commercial antibody described by Meabon et al., Citation27,Citation28 PA5-77544, and saw a much higher percentage of intracellular-stained LINGO-1. While the source of the disparity in the staining is unclear, one possibility is that the commercial antibody only recognizes an intracellular form of LINGO-1. The peptide that was used as an immunogen for the commercial antibody, CHVRSYSPDWPHQPNK, is (except for the initial C) the sequence at the transition of the Ig domain to the stalk of LINGO-1; this epitope location makes it plausible that an antibody raised against it could bind an intracellular form, but not its conformation when on the cell surface. Also, the asparagine residue is actually an N-linked glycosylation site and therefore it is possible that glycosylation at this site could affect antibody recognition.
To explore cell trafficking events following engagement of surface LINGO-1, we treated the cells with Li81 and then acid washed them to remove residual surface-bound LINGO-1. Under these conditions, about 30% of the Li81 was detected in lysozymes, consistent with the data published monitoring internalization of LINGO-1 on Ewing sarcoma cells.Citation26 Similar studies were performed with the CN1373 mAb. Treatment with CN1373 mAb led to robust staining of the cortical neurons, but only 6% of the antibody was detected in lysozymes, consistent with the less efficient internalization seen on CHO and Ba/F3 cells expressing LINGO-1. We extended our analysis to study Li81 and CN1373 binding to OPCs. Both antibodies bound OPCs, and as seen on cortical neurons, Li81 treatment led to more efficient trafficking into lysozymes (data not shown).
In summary, we demonstrated that the Li81 mAb has two LINGO-1 binding sites and that binding at both sites is required for Li81 activity. Primary binding via the CDRs, though high affinity, was not sufficient for function. Our results provide valuable insights into the mechanism of action of the anti-LINGO-1 opicinumab antibody, and how it affects the physiological function of LINGO-1.
Materials and methods
Antibody production
Li81 mAb (opicinumab) is a human antibody engineered into a human IgG1 aglycosyl framework. Seven variants of Li81 () designed to impede binding to the Ig domain of a second molecule of LINGO-1 were expressed in CHO cells with titers ranging from 65–80 mg/L. The mAbs were purified from 300 mL of clarified and filtered culture supernatants on 1.2 mL recombinant Protein A Sepharose Fast Flow (GE Healthcare) columns. The protein content of the eluted samples was estimated from absorbance spectra using an extinction coefficient at 280 nm of 1.43 per cm for a 1 mg/mL solution. Details for the purification of Li81 and generation of its Fab were previously described.Citation12
LINGO-1 production
LINGO-1-Fc (extracellular portion of human LINGO-1 (residues 1–488) fused to the hinge and Fc region of human IgGl was expressed in CHO cells and purified from clarified and filtered cell culture medium on recombinant Protein A Sepharose Fast Flow (GE Healthcare). The free ectodomain was generated by limited proteolysis with chymotrypsin (Roche) and purified by sequential chromatography steps, first removing undigested protein and free Fc on Protein A Sepharose, followed by SEC of the Protein A flow through fraction on a Superdex 200 column (GE Healthcare). Peak fractions were pooled, aliquoted, and stored at −70°C. Details for the purification of LINGO-1-Fc and generation of the LINGO-1 ectodomain fragment were previously described.Citation20
Analytical size exclusion chromatography and light scattering
SEC was carried out on an Agilent 1260 Infinity II high-performance liquid chromatography system equipped with a BioSep-SEC-S3000 column (7.8 x 300 mm, Phenomenex) at a flow rate of 0.6 mL/min with 0.1 M sodium phosphate, 0.2 M NaCl, pH 6.8 buffer as the mobile phase. The column effluent was monitored by ultraviolet detection at 280 nm. In-line static light scattering was performed using Wyatt tREX and Dawn HeliosII detectors. Average molecular weights were calculated using ASTRA v6.1 software.
Negative stain EM
LINGO-1 and Li81 were mixed at a 2:1 molar ratio and SEC purified as described above. A 5 μL aliquot of the peak fraction containing the LINGO-1 ectodomain/Li81 mAb complex (8 μg/mL) was adsorbed for 1 min on a glow-discharged carbon-coated copper grid. After blotting with filter paper, the grid was washed 3 times in water before being stained with 0.75% (w/v) uranyl formate solution. Specimens were examined using a Philips CM10 electron microscope (FEI) equipped with a tungsten filament and operated at an acceleration voltage of 100 kV. Micrographs were collected at a calibrated magnification of 41,513× (nominal magnification of 52,000×) with an XR16L-ActiveVu camera (AMT). For each sample, 30 micrographs were collected of areas that showed a consistent level of complex particles in the background.
SDS-polyacrylamide gel electrophoresis
Samples were subjected to sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) on 4-20% Tris-glycine gradient gels (Invitrogen) and stained with Coomassie brilliant blue. Non-reduced samples were diluted with Laemmli non-reducing sample buffer, and heated at 75°C for 5 min prior to analysis. Reduced samples were treated with sample buffer containing 2% 2-mercaptoethanol and heated at 95°C for 2 min.
Analysis of function by direct binding ELISA
MaxiSorp 96-well ELISA plates (Thermo Scientific) were coated with LINGO-1-Fc (10 µg/mL), blocked, and incubated with a 3-fold dilution series of each test compound starting at 10 µg/mL (8 dilutions). Bound mAb was detected using horseradish peroxidase (HRP)-conjugated goat-anti-human Fab′2 (Jackson ImmunoResearch #109-035-097) with 3,3′,5,5′-tetramethylbenzidine HRP substrate. Plates were read at 450 nm on a Molecular Devices plate reader. EC50 values of binding were calculated from the titration curve using Prism software.
Differential scanning fluorimetry
Five µL of 100x SYPRO Orange Protein Gel Stain (ThermoFisher Scientific #S6650) diluted in phosphate-buffered saline (PBS) buffer was added to 50 µL of antibody at 0.2 mg/mL in PBS, pH 7.0. 50 µL was delivered to each well of an ABI Prism 96-Well Optical Reaction Plate (Applied Biosystems #4306737) and analyzed with MXPro qPCR software on a Stratagene Mx3005P Real-time System (Agilent Technologies). The thermal denaturation method involved ramping temperature from 25–95°C in 0.5°C increments for 142 cycles.
Cell surface LINGO-1 expression levels as a function of Li81 fab and mAb treatment
Stable CHO cells expressing hemagglutinin (HA)-tagged full-length LINGO-1 were seeded at 1.2 × 105 cells/well in 12-well plates and treated with the indicated serial dilutions of Li81 Fab and mAb for 2 days in Alpha plus Eagle Minimum Essential Medium containing 10% fetal bovine serum without Geneticin. Cells were washed twice with buffer (2 mM HEPES, pH 7.5, 150 mM NaCl, 1 mM CaCl2, 1 mM MgCl2, and 25 μg/mL human serum albumin) and lysed in 350 μL of cold RIPA lysis buffer (50 mM Tris-HCl, pH 7.2, 1% Triton X-100, 0.5% sodium deoxycholate, 0.1% SDS, 150 mM NaCl, 10 mM MgCl2, 5% glycerol). The samples were clarified by centrifugation in an Eppendorf centrifuge. Cell supernatants (7.5 μL/sample) were analyzed by SDS-PAGE followed by Western blotting using HRP-rat-anti-HA antibody (Roche #10654300) for detection of the LINGO-1. The LINGO-1 CHO cells were also analyzed for surface LINGO-1 by FACS following a 23 h treatment at 37ºC with Li81, CN1373, CN1375, and 5C8 mAbs.
Oligodendrocyte assays
Enriched populations of A2B5+ OPCs isolated from the forebrain of female Sprague Dawley post-natal day 2 rats were grown in culture in a defined Dulbecco’s modified Eagle medium (DMEM) containing 4 mM glutamine, 1 mM sodium pyruvate, 0.1% w/v bovine serum albumin, 50 μg/ml apo-transferrin, 5 μg/ml insulin, 30 nM sodium selenite, 10 nM D-biotin and 10 nM hydrocortisone plus fibroblast growth factor/platelet-derived growth factor (Peprotech) (10 ng/ml). To assess differentiation of the rat A2B5+ progenitor cells into mature MBP-positive oligodendrocytes, A2B5+ cells were plated into 12-well poly-D-lysine coated culture plates in the defined DMEM medium supplemented with 15 nM T3 (Sigma) and 10 ng/ml ciliary neurotrophic factor (Peprotech), and treated with anti-LINGO-1 antibodies, soluble LINGO-1 reagents, or controls for 48 or 72 h. OPC differentiation was measured by the expression of MBP protein, visualized by ICC. Cultures were fixed by 4% paraformaldehyde and labeled with anti-MBP antibodies (SMI-94 and SMI-99, BioLegend) to visualize differentiated oligodendrocytes and an anti-Olig2 antibody (EMD Millipore #Ab9610) to quantify total cell numbers. The cultures were scanned by IncuCyte Zoom (Sartorius). Sixteen images were taken in each well and analyzed by a custom algorithm in IncuCyte. Data were presented as the fluorescent intensities of MBP staining normalized by the number of Olig2 cells.
Samples were also tested for function in the DRG neuron/A2B5+ cell coculture assay as previously described.Citation2 The cells were fixed on day 10 and analyzed by ICC using anti-MBP (green) antibodies (BioLegend #808402 and #836504 in a 1:1 mixture) and anti-tubulin (red) antibody (Covance #PRB-435P) both at a 1:500 dilution.
Binding and internalization studies on Ba/f3 cells expressing LINGO-1
Test proteins were diluted at 2x concentration (final concentration ranging from 1 pM to 150 nM) in cold FACS buffer (1% fetal calf serum, 20 mM Na2HPO4 pH 7.0, 150 mM NaCl, 0.05% NaN3) in a Nunc 96-well conical-bottom polypropylene plate (ThermoFisher Scientific) and eleven 3-fold serial dilutions were generated. Human LINGO-1-expressing Ba/F3 cells (200,000/well) suspended in cold FACS buffer were distributed into each well and incubated at 4°C or 37°C for 3 h. Cells were washed twice with cold FACS buffer after centrifugation at 1500 rpm for 2 min at 4°C. Cell pellets were re-suspended in goat anti-human kappa-phycoerythrin (Southern Biotech #2062-09) diluted 1:200 in cold FACS buffer and incubated at 4°C for 1 h. Cells were pelleted by centrifugation at 1500 rpm for 2 min and washed once with cold FACS buffer. Cells were fixed with fixation buffer (1% paraformaldehyde, 20 mM Na2HPO4 pH 7.0, 150 mM NaCl) for 10 min at room temperature and then pelleted by centrifugation. Cells were re-suspended in FACS buffer for analysis on a FACSCalibur Cell Analyzer (BD Biosciences).
Trafficking of LINGO-1 antibody complexes to late endosomes/lysosomes
Rat primary cortical neurons were dissociated from cortex of embryonic day 18 Sprague Dawley rats (BrainBits) and cultured on 12 mm diameter glass coverslips (no.1.5, Electron Microscopy Sciences, Hatfield, PA) coated with poly-D lysine in 24 well-plates and cultured for 12–14 days. The samples were washed with warmed PBS and further incubated with Alexa 568-labeled 5C8 isotype control antibody (prepared in-house), Li81 and CN1373 antibodies at 10 µg/ml in culture medium at 37°C for 90 min. Samples were acid washed with ice-cold PBS/glycine (pH 3) buffer for 1 min to remove surface-bound antibodies. Samples were then fixed with 4% paraformaldehyde at room temperature for 30 min and blocked with blocking buffer (PBS, 5% bovine serum albumin, and 1%goat serum) with detergent (0.2% Saponin) at room temperature for 15 min. The late endosomes/lysosomal compartment was detected by incubating samples with anti-Rab7 polyclonal antibodies (Cell Signaling Technology #9367) followed by Alexa 647-anti rabbit antibodies (ThermoFisher Scientific #A-21245) both for 1 hour incubations at room temperature in blocking buffer. All samples were then washed with PBS and mounted using ProLong Gold (ThermoFisher Scientific) with 4′,6-diamidino-2-phenylindole (Thermo Scientific, USA) and imaged as described below.
Imaging acquisition, processing, and analysis
Representative three-dimensional (3D) confocal images were acquired using a CSU-W1 spinning-disk confocal head (Yokogawa, Japan) coupled to a fully motorized inverted Zeiss AxioObserver Z1 imaging system (Carl Zeiss, Jena, Germany) equipped with an oil immersion 63X objective lens (Pan Apochromat, 1.4 numerical aperture) and an X-Cite XLED1 fluorescence illuminator (Excelitas Technologies, USA) for wide-field illumination. Solid-state laser stack (405, 488, 561 and 647 nm; Crystal Lase, Reno, NV) with fiber switcher technology (Intelligent Imaging Innovations, USA) were coupled to the spinning head through a fiber optic. The imaging system was operated under the control of SlideBook 6 (Intelligent Imaging Innovations, USA), which was used to acquire 3D confocal stacks of images spaced 0.27 µm apart with the aid of a piezoelectric Z motorized stage (Applied Scientific Instrumentation, USA) and a Hamamatsu ORCA-Flash4.0 v2 sCMOS Camera.
An image analysis application was developed with MATLAB 9 (Mathworks, USA) and named IMAB;Citation29 this was used to calculate the Manders colocalization coefficients.Citation30 Cellular regions containing LINGO-1 antibody complexes and early, late or recycling endosomes were masked by segmentation of the corresponding marker, with or without uniform background correction, by a defined pixel intensity that was typically greater than 2-fold of the local background. Masks or regions of interest were further refined by eliminating small objects (less than 10 voxels in volume and occupying fewer than 3 consecutive z-sections). Colocalization masks were obtained by a logical AND operation between the LINGO-1 receptor and the endosomal marker masks. The colocalization masks were further refined by eliminating small objects as described above. Manders coefficients were calculated by dividing the integrated fluorescence intensity under the colocalization mask (overlapping signal) by the integrated fluorescence intensity under the primary mask (total signal).
Abbreviations
| CHO | = | Chinese hamster ovary |
| CNS | = | central nervous system |
| CDR | = | complementarity-determining region |
| DLS | = | dynamic light scattering |
| DRG | = | dorsal root ganglion |
| ELISA | = | enzyme-linked immunosorbent assay |
| FACS | = | fluorescence-activated cell sorting |
| HRP | = | horseradish peroxidase |
| ICC | = | immunocytochemistry |
| Ig | = | immunoglobulin |
| LINGO | = | leucine-rich repeat and Ig containing Nogo receptor interacting protein |
| LRR | = | leucine-rich repeat |
| MALS | = | multiangle light scattering |
| MBP | = | myelin basic protein |
| MFI | = | mean fluorescence intensity |
| MS | = | multiple sclerosis |
| NgR1 | = | Nogo receptor interacting protein-1 |
| OPC | = | oligodendrocyte progenitor cell |
| PBS | = | phosphate-buffered saline |
| SDS-PAGE | = | sodium dodecyl sulfate polyacrylamide gel electrophoresis |
| SEC | = | size exclusion chromatography |
Disclosure of potential conflicts of interest
Biogen funded these studies. All authors are current or former employees and shareholders of Biogen. No potential conflicts of interest were reported to the authors.
Supplemental Material
Download Zip (4.1 MB)Acknowledgments
We thank our Biogen colleagues, particularly Christilyn Graff for phage display selections that led to the several relatives of Li81 discussed herein, and Chioma Nwankwo for work on LINGO-1-expressing cell lines and engineering of the constructs for the Li81 variants.
Supplementary metrial
Supplemental data for this article can be accessed on the publisher’s website.
References
- Mi S, Lee X, Shao Z, Thill G, Ji B, Relton J, Levesque M, Allaire N, Perrin S, Sands B, et al. LINGO-1 is a component of the Nogo-66 receptor/p75 signaling complex. Nat Neurosci. 2004;7(3):221–14. doi:https://doi.org/10.1038/nn1188.
- Mi S, Miller RH, Lee X, Scott ML, Shulag-Morskaya S, Shao Z, Chang J, Thill G, Levesque M, Zhang M, et al. LINGO-1 negatively regulates myelination by oligodendrocytes. Nat Neurosci. 2005;8:745–51. doi:https://doi.org/10.1038/nn1460.
- Mi S, Pepinsky RB, Cadavid D. Blocking LINGO-1 as a therapy to promote CNS repair: from concept to the clinic. Mol Cell Neurosci. 2013;60:36–42. doi:https://doi.org/10.1016/j.mcn.2014.02.006.
- Andrews JL, Fernandez-Enright F. A decade from discovery to therapy: lingo-1, the dark horse in neurological and psychiatric disorders. Neurosci Biobehav Rev. 2015;56:97–114. doi:https://doi.org/10.1016/j.neubiorev.2015.06.009.
- Inoue H, Lin L, Lee X, Shao Z, Mendes S, Snodgrass-Belt P, Sweigard H, Engber T, Pepinsky B, Yang L, et al. Inhibition of the leucine-rich repeat protein LINGO-1 enhances survival, structure, and function of dopaminergic neurons in Parkinson’s disease models. Proc Natl Acad Sci U S A. 2007;104(36):14430–35. doi:https://doi.org/10.1073/pnas.0700901104.
- Fu QL, Hu B, Wu W, Pepinsky RB, Mi S, So KF. Blocking LINGO-1 function promotes retinal ganglion cell survival following ocular hypertension and optic nerve transection. Invest Ophthalmol Vis Sci. 2008;49:975–85. doi:https://doi.org/10.1167/iovs.07-1199.
- Mi S, Hu B, Hahm K, Luo Y, Kam Hui ES, Yuan Q, Wong WM, Wang L, Su H, Chu TH, et al. LINGO-1 antagonist promotes spinal cord remyelination and axonal integrity in MOG-induced experimental autoimmune encephalomyelitis. Nat Med. 2007;13:1228–33. doi:https://doi.org/10.1038/nm1664.
- Mi S, Miller RH, Tang W, Lee X, Hu B, Wu W, Zhang Y, Shields CB, Zhang Y, Miklasz S, et al. Promotion of central nervous system remyelination by induced differentiation of oligodendrocyte precursor cells. Ann Neurol. 2009;65:304–15. doi:https://doi.org/10.1002/ana.v65:3.
- Zhang ZH, Li JJ, Wang QJ, Zhao WQ, Hong J, Lou SJ, Xu XH. WNK1 is involved in Nogo66 inhibition of OPC differentiation. Mol Cell Neurosci. 2015;65:135–42. doi:https://doi.org/10.1016/j.mcn.2015.03.003.
- Gresle MM, Liu Y, Kilpatrick TJ, Kemper D, Wu QZ, Hu B, Fu QL, So KF, Sheng G, Huang G, et al. Blocking LINGO-1 in vivo reduces degeneration and enhances regeneration of the optic nerve. Mult Scler J Exp Transl Clin. 2016;2:2055217316641704. doi:https://doi.org/10.1177/2055217316641704.
- Ji B, Li M, Wu WT, Yick LW, Lee X, Shao Z, Wang J, So KF, McCoy JM, Pepinsky RB, et al. LINGO-1 antagonist promotes functional recovery and axonal sprouting after spinal cord injury. Mol Cell Neurosci. 2006;33:311–20. doi:https://doi.org/10.1016/j.mcn.2006.08.003.
- Pepinsky RB, Shao Z, Ji B, Wang Q, Meng G, Walus L, Lee X, Hu Y, Graff C, Garber E, et al. Exposure levels of anti-LINGO-1 Li81 antibody in the central nervous system and dose-efficacy relationships in rat spinal cord remyelination models after systemic administration. J Pharmacol Exp Ther. 2011;339:519–29. doi:https://doi.org/10.1124/jpet.111.183483.
- Cen J, Wu H, Wang J, Ren X, Zhang H, Wang J, Wan Y, Deng Y. Local injection of lentivirus encoding LINGO-1 shRNA promotes functional recovery in rats with complete spinal cord transaction. Spine. 2013 Sep 1;38(19):1632–39. doi:https://doi.org/10.1097/BRS.0b013e31829dd58f.
- Youssef AEH, Dief AE, El Azhary NM, Abdelmonsif DA, El-Fetiany OS. LINGO-1 siRNA nanoparticles promote central remyelination in ethidium bromide-induced demyelination in rats. J Physiol Biochem. 2019;75(1):89–99. doi:https://doi.org/10.1007/s13105-018-00660-6.
- Mosyak L, Wood A, Dwyer B, Buddha M, Johnson M, Aulabaugh A, Zhong X, Presman E, Benard S, Kelleher K, et al. The structure of the Lingo-1 ectodomain, a module implicated in central nervous system repair inhibition. J Biol Chem. 2006;281:36378–90. doi:https://doi.org/10.1074/jbc.M607314200.
- Mi S, McCoy J, Pepinsky RB, Lee DHS, Inventors; Biogen Idec, Inc., Assignee. 2010. Nogo receptor binding protein. United States patent US 7,785,829.
- Bourikas D, Mir A, Walmsley AR. LINGO-1-mediated inhibition of oligodendrocyte differentiation does not require the leucine-rich repeats and is reversed by p75(NTR) antagonists. Mol Cell Neurosci. 2010;45:363–69. doi:https://doi.org/10.1016/j.mcn.2010.07.009.
- Kaplon H, Muralidharan M, Schneider Z, Reichert JM. Antibodies to watch in 2020. MABS. 2020;12(1):e1703531. doi:https://doi.org/10.1080/19420862.2019.1703531.
- Ecker DM, Jones SD, Levine HL. The therapeutic monoclonal antibody market. MAbs. 2015;7(1):9–14. doi:https://doi.org/10.4161/19420862.2015.989042.
- Pepinsky RB, Arndt JW, Quan C, Gao Y, Quintero-Monzon O, Lee X, Mi S. Structure of the LINGO-1-anti-LINGO-1 Li81 antibody complex provides insights into the biology of LINGO-1 and the mechanism of action of the antibody therapy. J Pharmacol Exp Ther. 2014;350:110–23. doi:https://doi.org/10.1124/jpet.113.211771.
- Cadavid D, Balcer L, Galetta S, Aktas O, Ziemssen T, Vanopdenbosch L, Frederiksen J, Skeen M, Jaffe GJ, Butzkueven H, et al. Safety and efficacy of opicinumab in acute optic neuritis (RENEW): a randomised, placebo-controlled, phase 2 trial. Lancet Neurol. 2017;16(3):189–99. doi:https://doi.org/10.1016/S1474-4422(16)30377-5.
- Klistorner A, Chai Y, Leocani L, Albrecht P, Aktas O, Butzkueven H, Ziemssen T, Ziemssen F, Frederiksen J, Xu L, et al. Assessment of opicinumab in acute optic neuritis using multifocal visual evoked potential. CNS Drugs. 2018;32(12):1159–71. doi:https://doi.org/10.1007/s40263-018-0575-8.
- Molecular Operating Environment (MOE) software. 2016. 0801; Chemical Computing Group ULC, 1010 Sherbrooke St. West, Suite #910, Montreal, QC, Canada, H3A 2R7, 2018.
- Mi S, Pepinsky RB, Graff C, Inventors; Biogen Idec, Inc., Assignee. 2017. Compositions comprising antibodies to LINGO or fragments thereof. United States patent US 9,745,375.
- Baldassari LE, Feng J, Clayton BLL, Oh SH, Sakaie K, Tesar PJ, Wang Y, Cohen JA. Developing therapeutic strategies to promote myelin repair in multiple sclerosis. Expert Rev Neurother. 2019;19(10):997–1013. doi:https://doi.org/10.1080/14737175.2019.1632192.
- Town J, Pais H, Harrison S, Stead LF, Bataille C, Bunjobpol W, Zhang J, Rabbitts TH. Exploring the surfaceome of ewing sarcoma identifies a new and unique therapeutic target. Proc Natl Acad Sci U S A. 2016;113(13):3603–08. doi:https://doi.org/10.1073/pnas.1521251113.
- Meabon JS, De Laat R, Ieguchi K, Wiley JC, Hudson MP, Bothwell M. LINGO-1 protein interacts with the p75 neurotrophin receptor in intracellular membrane compartments. J Biol Chem. 2015;290(15):9511–20. doi:https://doi.org/10.1074/jbc.M114.608018.
- Meabon JS, de Laat R, Ieguchi K, Serbzhinsky D, Hudson MP, Huber BR, Wiley JC, Bothwell M. Intracellular LINGO-1 negatively regulates Trk neurotrophin receptor signaling. Mol Cell Neurosci. 2016;70:1–10. doi:https://doi.org/10.1016/j.mcn.2015.11.002.
- Massol RH, Boll W, Griffin AM, Kirchhausen T. A burst of auxilin recruitment determines the onset of clathrin-coated vesicle uncoating. Proc Natl Acad Sci U S A. 2006;103(27):10265–70. doi:https://doi.org/10.1073/pnas.0603369103.
- Manders EMM, Stap J, Brakenhoff GJ, van Driel R, Aten JA. Dynamics of three-dimensional replication patterns during the S-phase, analysed by double labelling of DNA and confocal microscopy. J Cell Sci. 1992;103:857–62.
