ABSTRACT
Therapeutic monoclonal antibodies (mAbs) are highly complex proteins that must be exhaustively characterized according to the regulatory authorities' recommendations. MAbs display micro-heterogeneity mainly due to their post-translational modifications, but also to their susceptibility to chemical and physical degradations. Among these degradations, aggregation is quite frequent, initiated by protein denaturation and then dimer formation. Here, we investigated the nature and structure of the high molecular weight species (HMW) present at less than 1% in an unstressed formulated roledumab biopharmaceutical, as a model of high purity mAb. HMW species were first purified through preparative size-exclusion chromatography (SEC) and then analyzed by a combination of chromatographic methods (ion-exchange chromatography (IEX), SEC) coupled to native mass spectrometry (MS), as well as sodium dodecyl sulfate–polyacrylamide gel electrophoresis and capillary gel electrophoresis under non-reducing conditions. Both covalently and non-covalently bound dimers were identified at a proportion of 50/50. In-depth characterization of the HMW fraction by SEC and IEX hyphenated to native MS revealed the presence of three mAb dimer forms having the same mass, but differing by their charge and size. They were attributed to different compact and elongated dimers. Finally, high-resolution middle-up approaches using different enzymes (IdeS and IgdE) were performed to determine the mAb domains implicated in the dimerization. Our results revealed that the roledumab dimers were associated mainly by a single Fab-to-Fab arm-bound association.
Introduction
Roledumab is a human recombinant anti-D monoclonal antibody (mAb) used to prevent rhesus D (RhD)-related hemolytic disease of RhD (+) newborns, which occurs during the subsequent pregnancies of RhD (-) women. By binding to RhD (+), this intravenously administrated protein clears antigen, avoids B cell memory production, and allows thus feto-maternal alloimmunization. Roledumab is produced in Yb2/0 rat cell lines to provide effective binding to the cell effector related to the glycosylation pattern. Roledumab is a low-fucose containing glycoprotein of 1340 amino acids with 16 disulfide bridges (including 12 intrachain and 4 interchain bridges) and one glycosylation site (Asn306) on each heavy chain. Roledumab’s efficacy and safety profile has been studied in three clinical trials, in healthy volunteersCitation1 (clinicalTrials.gov NCT00952575), as well as in RhD-negative pregnant women carrying an RhD-positive fetus (clinicalTrials.gov NTCT02287896). The clinical results obtained show similar pharmacokinetics profile as plasma anti-D without immunogenic response or RhD immunization. This provided sufficient evidence for roledumab to enter into a Phase 3 trial, and confirms this new therapeutic agent as an alternative to plasma-derived anti-D.
MAbs generally exhibit a high level of heterogeneity, due to their post-translational modifications,Citation2 but also to their susceptibility to chemical and physical degradations. Chemical degradations include oxidation, deamidation, isomerization, racemization, glycation, fragmentation, pyroglutamate formation, disulfide bond modification and covalent oligomerization, whereas physical degradations entail denaturation, unfolding and aggregation.Citation3-Citation9 Aggregates result from different environment changes and stresses during the production, shipping and storage of mAbs.Citation10-Citation14 They entail small and larger oligomers, polymers or multimers called high molecular weight (HMW) species that can be formed via covalent bonds or non-covalent interactions.Citation15,Citation16 Even if soluble reversible or irreversible aggregates are generally removed during the downstream process, some may arise from degradation and partial unfolding of the mAbs during the product life cycle. They may be triggered by pH or ionic strength variation of dissolution media as well as thermal stresses, stirring or photo-oxidation.Citation17 Commonly, the level of aggregates, in formulated mAbs is a critical quality attribute (CQA),Citation18 as it may affect the pharmacokinetics and pharmacodynamics of the biopharmaceutical product or lead to adverse side effects and immunogenic responses.Citation19-Citation21 Roledumab is formulated at very low concentration (0.3 g. L−1), reducing thereby the concentration of potential dimers and aggregates that could appear in the pharmaceutical preparation. However, it is mandatory to better understand how they are formed to find out strategies to reduce their occurrence.
For decades, the detection of aggregates and dimers of therapeutic proteins, particularly mAbs, has been mostly achieved by biophysical techniques such as transmission electron microscopy,Citation22 circular dichroism, analytical ultra-centrifugation (AUC, for example, with two-dimensional spectrum analysis data treatment)Citation23,Citation24 or dynamic light scattering.Citation25 Except for AUC, which is currently used for HMW species quantification, these approaches provide information on global protein conformation and are mostly qualitative ones. However, they are not able to detect slight differences in protein conformation nor major ones when present at very low levels.Citation16 Besides these methods, liquid chromatography or electrophoresis-based methodsCitation26-Citation28 allow separation of oligomers or aggregates and their reliable quantification. Size-exclusion chromatography (SEC), and in particular SEC coupled to multi-angle light scattering, is widely used to this purpose in the biopharmaceutical context,Citation29 as well as asymmetric flow field-flow fractionation.Citation30 Advances in SEC column technology led to improved resolution and faster analyses. However, certain conditions used for SEC might produce oligomer dissociation or analyte adsorption to the stationary phase as shown by Arakawa et al.Citation31 Recently, efforts have been directed toward hyphenation of capillary electrophoresisCitation32-Citation36 and liquid chromatographyCitation37 with mass spectrometry using optimum native conditions (native MS) and specific mobile phasesCitation38 to maintain non-covalent interactions. The relevance of native MS was demonstrated for the evaluation of HMW and size variants of mAbs, with accurate mass and shape discrimination of the molecules after ion mobility mass spectrometry.Citation39,Citation40 Other separation methods such as ion-exchange chromatography (IEX) hyphenated with MS are also able to preserve conformational properties of the protein, even non-covalent complexes. Differences in charge state distribution observed for the separated variants can be used to determine their conformation and aggregation state, while UV detection allows their accurate quantification at very low levels.Citation41-Citation43 Hydrophobic interaction chromatography (HIC), which separates proteins based on hydrophobicity and folding state, can also be useful. HIC-MS allowed already the detection of dimers and trimers linked by non-covalent interactions, analyzed under non-denaturing separation and native MS conditions.Citation44
The use of hydrogen-deuterium exchange MS has also emerged as a powerful technique to detect small changes in the higher-order structure of mAbs.Citation45 Changes in hydrogen/deuterium atom exchange may give insight on specific mAbs domains by which aggregation/self-association occurs.Citation46,Citation47 However, this methodology has the disadvantage of being complex, requiring special chromatographic devices to avoid backbone hydrogen exchange during analysis.
MAb aggregation is a complex process and many studies have been conducted to characterize residual HMW species generated after various stresses, including shaking, stirring, freeze-thawing cycles, exposure to extreme pH, temperature or to UV-light.Citation48-Citation50 Reported stress protocols used to generate HMW forms substantially vary from each other, leading to different species in terms of size, charge, hydrophobicity, nature (covalent or non-covalent) and site of association.Citation51 In addition, physical and chemical instabilities are not only dependent on the type of stresses but also on the primary amino acid sequence. Different mAbs, therefore, exhibit different aggregation proneness or pathways, which suggests that a general rule to predict mAb aggregation cannot be established. Remmele et al.Citation52 noticed that in a formulated epratuzumab (IgG1), the apparent homogenous nascent dimer was in fact heterogeneous and contained Fab-Fab, Fc-Fab and Fc-Fc bound dimers. Among these dimers, 70% were covalently linked forms. After thermal heat and aging, this proportion increased to 84%. They found that among those covalent dimers, not all were bound via disulfide bridges. Moreover, Iwura et al.Citation53 showed that palivizumab (IgG1) dimers were associated also with dityrosine covalent linkages. Other experiments performed on several IgG1 subjected to different stresses indicated that the aggregation pathway started with non-covalent association of Fab domains, and that two predominant elongated and compact dimeric structures were present.Citation51-Citation55 Interestingly, Iacob et al.Citation46 showed the probable involvement of the hinge-loop region in domain swapping, disulfide scrambling and surface interactions in the dimerization of mAbs.
We recently noticed the presence of a very small amount (<1%) of HMW species in roledumab. The goal of this study was therefore to isolate these species and characterize as much as possible their structure. A preparative SEC method was used to isolate this HMW fraction. Then, a combination of orthogonal techniques, such as SEC-UV, sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE) and capillary gel electrophoresis with sodium dodecyl sulfate (SDS-CGE), was used to separate and quantify the different HMW species. Native and middle-up approaches using SEC and IEX hyphenated to MS were then developed to identify these species and characterize the region involved in the dimerization as well as the nature of the chemical linkage.
Results
Isolation and structural characterization of the HMW species of roledumab
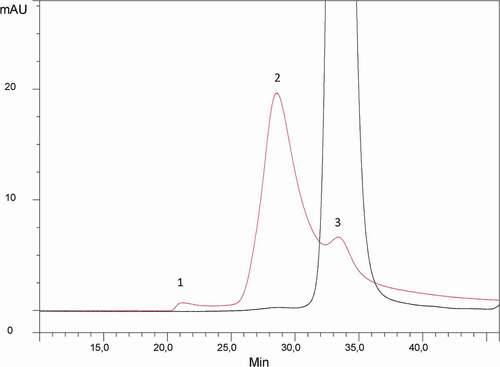
In order to investigate the nature of the HMW species, detected at trace level in roledumab, they were first isolated from the monomeric form of the antibody by a semi-preparative SEC performed under mild elution conditions (100 mM phosphate-buffered saline (PBS), pH 7.4). To ensure that non-artefactual species were formed or dissociated during this step, both isolated fractions were analyzed again by analytical high-performance SEC (HPSEC) using the same stationary and mobile phases. The HPSEC-UV profiles obtained are presented in . No HMW species were detected in the isolated monomer fraction. The isolated HMW fraction was found to contain 85% of HMW species (peak 2), 10% of monomer (peak 3) and less than 5% of very high MW oligomers (peak 1). The small percentage of monomer in this HMW fraction can be explained by the partial resolution obtained with this semi-preparative HPLC.
Figure 1. HPSEC-UV profile of the roledumab preparation (black trace) and the purified dimer fraction (red trace). Separation is performed on a Superdex 200 column at a flow rate of 0.4 mL/min using an isocratic-elution with PBS. Detection at 280 nm.
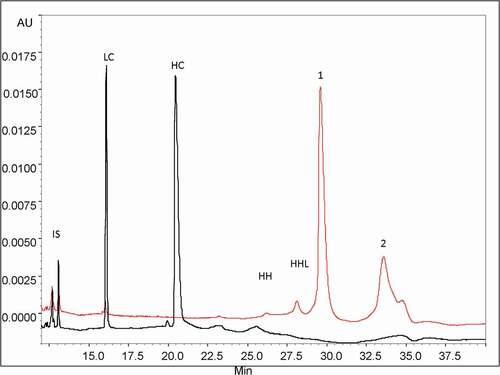
We analyzed thoroughly the HMW fraction using an intact mAb analysis approach. First, the proportion of covalently and non-covalently bound species was estimated using a denaturing CGE method under non-reducing (NR) and reducing (R) conditions. The SDS-CGE profiles obtained are presented in . Under non-reducing conditions, two minor peaks (26–28 min) were identified as mAbs, lacking either one or two light chains (HHL and HH, respectively). The major peak 1 (29.8 min) accounted for 53% of the total species and had the same migration time as the monomer, indicating that it corresponds to the dissociated form of the non-covalently bound HMW species. Assuming 10% of residual monomer in the purified fraction, these non-covalently bound HMW species may represent up to 43% of the total species. The last migrating peak 2 (34–35 min), presenting at least two shoulders, accounted for 40% of the total protein content and was attributed to covalently bound HMW species. The sum of those different forms (83%) is consistent with the 85% of HMW species previously determined by analytical HPSEC of the isolated HMW fraction. The analysis of the HMW fraction performed under reducing conditions showed two major peaks corresponding to the light (LC) and heavy (HC) chains, migrating at 16.1 and 20.5 min, respectively (). This clearly proves that the HMW species contain a mixture of non-covalent and covalent forms, the latter dissociated by the dithiothreitol (DTT) reduction are associated via disulfide bonds. Furthermore, we noticed that, under reducing conditions, the proportion of the HC and LC observed for the HMW species was similar to that of the monomer (data not shown), suggesting that no additional single HC or LC are implicated in the aggregation.
Figure 2. CE-SDS profiles of the purified HMW fraction with (black trace) and without (red trace) reduction by DTT. Peak assignments: LC: Light Chain (25 kDa), HC: Heavy Chain (50 kDa), mAbs without one LC: HHL (125 kDa) and without two LC: HH (75 kDa). Conditions: uncoated silica capillary, 50 µm I.D., 50/60 cm effective/total length, BGE: SDS-MW Gel Buffer at pH 8.0 with 0,2% SDS, −15 kV, 40°C, UV detection at 280 nm. IS: internal standard is a 10 kDa protein.
These results were confirmed by SDS-PAGE analysis of the HMW fraction. Under non-reducing conditions two major bands were observed, one at 150 kDa corresponding to the dissociation of the non-covalently bound HWM forms and one at 317 kDa corresponding to the covalently bound HWM species. After DTT reduction only two bands consistent with HC and LC were observed, as in SDS-CGE.
In order to identify the cysteines involved in the covalent dimerization, non-reducing peptide mapping experiments were performed using LC-MS on both the monomer and purified dimer fractions. The expected tryptic peptides, including disulfide-linked peptides, were detected. However, no significant difference between monomer and dimer fractions was identified, and notably, no scrambled disulfide bridges specific to mAb dimers were observed.
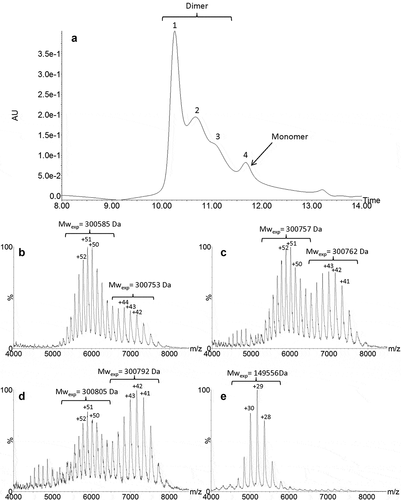
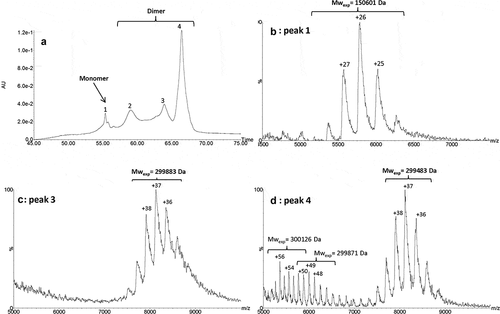
We then developed a native ultra-high-performance SEC (UPSEC)-UV-MS method using a resolutive-bridged ethylene hybrid (BEH) 450 column and an isocratic elution with volatile salts to analyze the HMW fraction. The percentage of ammonium acetate in the mobile phase (from 50 to 150 mM) and the sample cone voltage energy (60 V to 150 V) were optimized in order to: (1) minimize the dissociation of the non-covalent interactions, and (2) allow the efficient ion transfer of non-covalent oligomers to the analyzer. It was assumed that combining the use of a mobile phase composed of 0.1 M ammonium acetate and a low-flow rate (50 µL/min) would preserve the dimer structural integrity. Native MS allowed accurate identification of each separated species. The SEC analysis presented in showed four partially resolved peaks, the HMW species of roledumab being eluted as three peaks (1–3) at 10.2, 10.7 and 10.9 min. Relative abundance of peak 1 was 46% while peaks 2 and 3 accounted for 32% and 13%, respectively. The masses obtained for the three different HMW forms were 300,581 ± 28 Da, 300,757 ± 32 Da and 300,805 ± 27 Da (, , ) which correspond to mAb dimers. These experimental masses were close to the theoretical one (299,556 Da), with a relative error between these values below 0.4%. Moreover, the mass differences between the conformers were quite low (~ 224 Da) considering the broad charge state distribution and inherent mass heterogeneities of mAbs. Furthermore, the mass spectra of peaks 1–3 showed a bimodal profile. For peak 1 (), a first envelope centered at +51 charge state (mass range, 5200 to 6500 m/z) and a second at +43 (mass range, 6200 to 7800 m/z) were observed. For peak 2 (), the mass spectrum presented a decreased intensity of the first signal centered at +51 charge state, while the second one, centered at +43 was increased. For peak 3, the ratio between the two envelopes was reversed (). These two charge state distributions suggest the presence of two co-existing forms, which could be for instance elongated and compact dimers as previously reported by other groups.Citation51-Citation55 The elongated one appears as the main form eluting in peak 1, while the compact one is more abundant in peak 3. This can be explained by the lower apparent hydrodynamic volume of the compact form that elutes later according to the SEC mechanism, and enlightened by MS results, the compact form having the smaller number of charges with the higher m/z range. In addition, for peaks 2 and 3, a third envelope (mass range, 4300 to 5000 m/z) was also observed. This envelope likely corresponds to a monomer generated by in-source dimer dissociation. Peak 4 (11.5 min) corresponded to the monomer and represented 8% of the total species. The experimental mass was measured at 149,956 Da, which is close to the theoretical mass calculated for roledumab (), considering a G0/G0 N-glycan combination (theoretical MW: 149,778 Da), which is the smallest expected IgG N-glycan (pentasaccharide core + 2 GlcNac residues).
Middle-up analysis of HMW species of roledumab using UPSEC-UV-MS
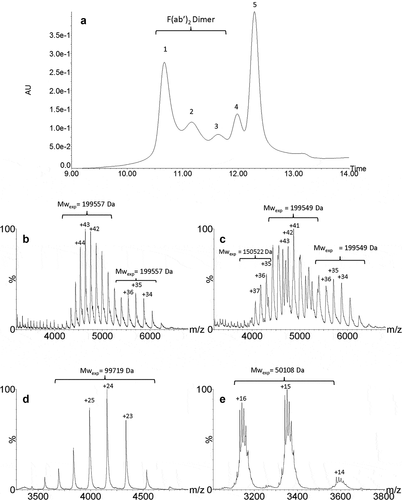
To determine the domains involved in the dimer formation, a middle-up approach using IdeS enzyme was performed to cleave the mAb under the hinge region releasing F(ab’)2 and (Fc/2)2 fragments. This strategy based on the analysis of mAb subunits provides better mass accuracy for protein characterization. The IdeS-digested HMW fraction was analyzed by UPSEC-UV-MS. The UV trace () displayed five peaks with relative abundances of 32%, 16%, 8%, 10% and 34% for peaks 1 to 5, respectively. The masses obtained for peaks 1 to 3 measured at 199,557 Da, 199,549 Da and 199,485 Da, respectively, correspond to the theoretical mass of F(ab’)2 dimers (theoretical MW: 199,412 Da). This result reveals the presence of three different F(ab’)2 dimers, as previously observed for the intact dimers, and shows that dimerization occurs mainly via the F(ab’)2 domain of the mAb. The masses for peaks 4 and 5 (,) determined at 99,719 Da and 50,108 Da correspond to F(ab’)2 and (Fc/2)2, respectively, this latter being characterized by the multi charge mass spectrum caused by Fc glycoform heterogeneity. Peak 2 also contained a small amount of a 150 kDa form with a charge state distribution centered at +36 (mass range, 4000 to 5200 m/z) that could correspond to residual undigested monomers (). However, as it eluted faster than the monomer that presented a different charge state distribution centered at +29 (mass range, 4600 to 6000 m/z) (), we considered that the 150 kDa form was rather an F(ab’)2-Fc dimer.
Figure 4. UPSEC-UV-MS chromatogram of the isolated HMW fraction submitted to IdeS enzymatic digestion (a). Native mass spectra of peaks 1 (b), 2 (c), 4 (d) and 5 (e) obtained by UPSEC-MS. See conditions in Figure 3. Theoretical masses of F(ab’)2, F(ab’)2 dimer and Fc are 199,412 Da, 99,706 Da and 50,108 Da, respectively.
As previously observed after MS analysis of the intact HMW fraction, the structural heterogeneity of the dimers was detected with two charge state distributions centered at +43 and +35 (), confirming the presence of two co-existing conformers. As dimerization was preserved even after IdeS treatment, we hypothesized that it occurred on the Fab domain. However, these experiments could not provide a clear indication about the nature of the association; head-to-head dimer orientations in a single arm-bound Fab-to-Fab or a head-to-head double arm-bound (Fab’)2-to-(Fab’)2.
To clearly identify the nature of the monomer association, the same UPSEC-UV-MS method was then applied to the analysis of the HMW fraction after IgdE digestion. This enzyme cleaves the antibody at the upper hinge region, releasing one Fc and two Fabs. IgdE treatment of single Fab-Fab arm-bound dimers releases Fc, Fab and Fab dimers, while cleavage of double Fab-Fab arm-bound dimers releases only Fab dimers and the Fc (Figure SI-1). UPSEC-UV-MS analysis of IgdE-digested HMW fractions revealed the presence of three different forms: 1) the Fab (48,541 Da), which represents the major form; 2) the Fc (52,802 Da); and 3) the Fab dimer (97,087 Da). These observations confirmed the results (e.g., dimerization on the Fab domain) obtained after IdeS treatment for which F(ab’)2 and (Fc/2)2 were rather detected. Considering the upper hinge IgdE cleavage, the presence of Fabs and Fab dimers strongly suggests that the dimers were of the single Fab-Fab arm-bound type. Furthermore, Fab dimer mass spectra showed only one distribution charge, indicating no conformational heterogeneity, and therefore the absence of coexisting compact and elongated structures.
Orthogonal analysis of HMW species of roledumab using IEX-UV-MS
Additionally, an IEX-MS analysis was used to identify the charge variants of our purified mAb dimers. The elution gradient developed by Leblanc et al.Citation41 for monomer analysis was adapted by increasing both ionic strength and pH of the mobile phase. Indeed, dimers exhibit a higher affinity to the stationary phase than monomers. The IEX-UV profile of the intact HMW fraction showed four distinct peaks (). Peak 1 presented a relative abundance of 7%, with two shoulders corresponding to three monomeric forms of the antibody with 0 to 2 C-term Lys residues on the HC. The mass spectrum presented in showed a major charge state centered at +26 corresponding to the residual monomer at 150,601 Da. The three other peaks (peaks 2–4) displayed a relative abundance of 20%, 13% and 65%, respectively. Those three HMW species differ from their local charge distribution, the major one being eluted toward retention times of the most basic species. The related MW of peaks 2 to 4 was measured at 299,898 Da, 299,883 Da and 299,483 Da, respectively, corresponding to mAb dimers. shows a mass spectrum with three envelopes, the one with charge state distribution centered at +37 (mass range, 7400 to 9000 m/z) is similar to that observed for peaks 2 and 3 (c). These envelopes correspond therefore to different conformers. For instance, charge state distributions centered at +49 (mass range, 5400 to 6600 m/z) and +54 (mass range, 5200 to 6200 m/z) for peak 4 could correspond to covalent structures detected by SDS-CGE (compact and/or elongated conformers) differing from the major non-covalent ones (at +37).
Figure 5. IEX-UV profile of the isolated HMW fraction (a). Native mass spectra of peaks 1 (b), 3 (c) and 4 (d). Conditions: MAbPac SCX-10 column, mobile phases were 50 mM ammonium formate, at pH 3.9 (Buffer A) and 500 mM ammonium acetate at pH 7.4 (Buffer B). Gradient elution at a flow rate of 0.4 mL/min: 15% of B for 5 min, 15% to 31% in 40 min, 31% to 85% of B in 30 min. UV detection was set at 280 nm.
A middle-up approach, using IdeS or IgdE treatment was then performed to understand the differences between charge variants and to confirm the hypothesis that a single Fab-Fab arm-bound was involved in dimerization. Three peaks were observed on the UV profile of the IdeS-digested HMW fraction (). Their relative abundances were 22% for (Fc/2)2 fragments, 16% for the F(ab’)2 and 54% for F(ab’)2 dimer with a mass of 199,442 Da. MS spectrum of peak 3 exhibited three different envelopes (). The charge states at +31 (mass range, 5900 to 7200 m/z) and +40 (mass range, 4400 to 6000 m/z) corresponded to F(ab’)2 dimers, which could be compact and elongated conformers. The envelopes with charge state distribution centered at +30 correspond rather to F(ab’)2 monomer probably generated by partial in-source dissociation. The two shoulders observed for peak 3 () assigned to F(ab’)2 dimers were consistent with the three separated peaks (2–4) observed on the intact roledumab dimer in .
Figure 6. IEX-UV elution profile of the IdeS-digested HMW fraction (a) with insets representing native mass spectrum of peak 2. Native mass spectrum of peak 3 (b). IEX-UV elution profile of the IgdE-digested HMW fraction (c) with insets representing native mass spectrum of peak 2. Native mass spectrum of peak 3 (d). See conditions in Figure 5. Theoretical masses of F(ab’)2 dimer and Fab dimer are 199,411 Da and 99,076 Da, respectively.
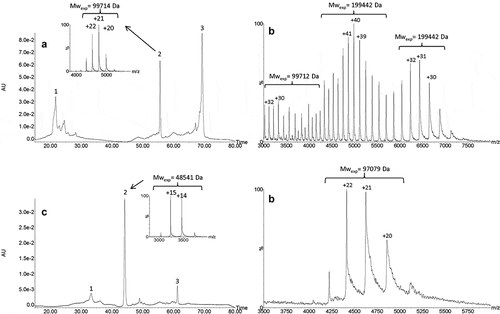
IEX separation was performed after IgdE treatment (). Three main peaks were observed with a relative abundance of 11% for the Fc, 75% for the Fab and 10% for Fab dimers with a mass at 97,079 Da. Considering the upper hinge IgdE clipping, the presence of Fabs and Fab dimer confirmed the single Fab-Fab arm-bond. Moreover, we noticed that, contrary to the results observed after IdeS treatment, the mass spectrum of the Fab dimers presented only one envelope (). The conformational and structural heterogeneities were gone after IgdE treatment, suggesting that the hinge region is implicated in the minor dimeric forms previously evidenced after IdeS cleavage and suspected to be covalent forms.
Discussion
Therapeutic mAbs are highly complex proteins that are susceptible to aggregation. The level of aggregates in formulated mAbs is a CQA that must be closely monitored. SEC and IEX methods hyphenated to native MS in association with different middle-up approaches were developed to further characterize the aggregates present in the unstressed roledumab product.
We successfully purified, with a purity close to 85%, stable HMW species present at a very low level (<1%) in roledumab. This allowed in-depth structural investigation of their nature and conformation. Most of the isolated HMW species were covalently and non-covalently bound dimers with different ways of association. We demonstrated that our purified dimers included 40% of covalent species linked by disulfide bonds. The covalent/non-covalent ratio of dimers for the unstressed roledumab was 50/50, which is consistent with the results obtained by Iwura et al.Citation52 for palivizumab. However, in the case of palivizumab, 15% of covalent species were not reduced by DTT and corresponded to di-tyrosine bounds whereas our purified dimers were all linked by disulfide bridges. Remmele et al.Citation52 reported a slightly higher level of covalent dimer measured at 70% for unstressed epratuzumab, while for the 37°C-aged mAb, it increased up to 84%. In contrast, Paul et al.Citation51 reported exclusively non-covalent hydrophobic interactions. These discrepancies in the covalent/non-covalent ratios or in the nature of covalent bonding highlight the complexity of dimer formation and are probably related to the nature of the mAb (e.g., primary structure, protein concentration), formulation composition, and, when tested, type of the stress. Indeed, heat stress-induced dimerization has been reported to occur mainly via covalent linkages,Citation51,Citation55 while pH and light stresses would generate rather non-covalent dimers.Citation52
SEC-native MS analysis revealed the presence of three size variants with two envelopes under each peak that could correspond to elongated and compact forms. This is the first time that three different dimers are resolved and identified by this method. Considering their elution volume by SEC and relative peak areas, the elongated forms were found to be more abundant in solution than compact ones. The presence of both conformers was confirmed by sedimentation velocity (SV)-AUC experiments (Table SI-2). We indeed observed in the purified dimer fraction two main peaks, one with a high sedimentation coefficient (s20 w) of 11.0 S and a frictional ratio (f/f0) of 1.2 and another peak with an s20 w of 9.1 S and an f/f0 of 1.7. The peak with the lowest s20 w and the highest f/f0 was attributed to the elongated forms, which were found to be the most abundant. Roledumab containing mostly the monomer presented one main peak with an s20 w of 6.6 S and an f/f0 of 4.5. These results confirmed by an orthogonal structural method the presence in an unstressed product of elongated and compact dimer forms.
Identification of compact and elongated structures agrees well with Paul’sCitation51 and Plath’sCitation55 observations on the mAb dimerization with or without induced stresses. Plath et al.Citation55 described elongated antiparallel structures and more compact propeller-like dimers. Paul et al.Citation51 reported elongated bone-like dimers for unstressed product and more compact forms for pH and light stressed dimers.
Literature dealing with the characterization of stressed and unstressed mAbs showed a wide range of dimeric mAb structures, with head-to-head single arm-bound Fab-to-Fab,Citation51,Citation52,Citation54,Citation55 double arm-bound F(ab’)2-to-F(ab’)2,Citation52,Citation54,Citation55 Fab-FcCitation52,Citation53 or Fc-FcCitation52 dimers, depending on the physicochemical properties of each individual mAb. Our results obtained by middle-up approaches using SEC-MS, allowed identification of, after IdeS treatment, F(ab’)2 dimers as major forms that could originate from single or double Fab-Fab arm bonds. In addition, the presence of very small amounts of F(ab’)2-Fc dimer forms has been suspected. After IgdE treatment, SEC-MS and IEX-MS analysis confirmed the presence of Fab dimers, indicating that dimerization mostly occurs through the Fab-to-Fab, head-to-head single-arm bond.
Finally, contrary to the results obtained after IdeS treatment, no conformational heterogeneity was detected after IgdE digestion, indicating that another interaction through the hinge region could be also involved in the dimerization. Plath et al.Citation55 reported a propeller-like F(ab’)2-F(ab’)2 dimeric forms also connected by the hinge region that was lost after papain digestion. Regarding this hinge interaction, disulfide bonds have different reduction susceptibilities.Citation56 The upper one between the two inter HC is the first to be reduced, but disulfide bonds in the CH2 domain are also prone to reduction. Reduction involving the hinge and/or CH2 regions could lead to a swapping phenomenon as described by Iacob et al.Citation46 Our non-reducing peptide maps obtained for the monomer and dimers were similar and the cysteines issued from covalent dimers were not detected. These results are consistent with the swapping hypothesis previously reported to explain mAbs dimerization.
To conclude, we investigated the nature and structure of the HMW species of roledumab. After purification of the nascent roledumab dimer, intact analysis and middle-up approach pointed out one major interaction occurring in the Fab domain. The difference of bioactivity in the case of double Fab and single Fab arm-bound dimers regarding antigen interaction should be investigated.
Materials and methods
Materials
A roledumab (molecular weight of 149,978 Da) solution at 2 g/L was produced at LFB in Yb2/0 rat cell lines. The endo-proteases IdeS (FabRICATOR) and IgdE (FabALACTICA) were provided by Genovis (Lund, Sweden). All chemical reagents (MS grade ammonium formate, sodium phosphate dibasic, sodium chloride, PBS) were obtained from Sigma-Aldrich (St. Louis, MO, USA), except formic acid, which was bought from Merck Biosciences (Darmstadt, Germany). All buffers were prepared with ultra-high-performance liquid chromatography (UPLC)/MS grade water purchased from Biosolve (Dieuze, France).
Purification of roledumab dimer
A roledumab solution was separated by preparative SEC to purify HMW species that constituted less than 1% of the total injected drug substance. Preparative SEC separations were performed at ambient temperature on an ÄKTA PURIFIER UPC System (GE Healthcare, Chicago, USA) using a Superdex 200 prep grade column (26 mm x 600 mm, 34 μm particle diameter, GE Healthcare) at room temperature and a flow rate of 2 mL/min. An isocratic-elution was used with the mobile phase consisting of PBS (0.01 M phosphate, 0.138 M NaCl, 0.0027 M KCl, at pH 7.4). Different collected fractions were pooled and concentrated by centrifugation using a 30 kDa cut off Amicon filter (Millipore, Burlington, USA) to a final concentration of ~0.7 g/L of isolated dimer. Finally, purified dimers were stored in aliquots at −80°C.
Sample preparation and enzymatic digestions
Roledumab and purified dimer samples were concentrated using a centrifugal concentrator (10,000 Da cut off Vivaspin (Sartorius, Goettingen, Germany)). Protein concentration was thus determined with nanoDrop. mAb samples were digested using two enzymes. IdeS, a cysteine protease from Genovis cleaves mAbs at a specific site below the hinge region (CPAPELLG/GPSVF) generating a homogenous pool of F(ab)’2 and (Fc/2)2. IgdE cleaves mAbs at one specific site above the hinge (KSCDKT/HTCPPC) and generates intact Fab and Fc domains.Citation57,Citation58 One hundred micrograms of mAbs was incubated with 100 UI of enzymes following the instructions of the enzyme kits, at 37°C and for 4 hours for IdeS and overnight for IgdE.
HPSEC analysis
HPSEC analysis was performed on a Dionex U3000 System with a Superdex 200 column (10 mm i.d. x 300 mm, GE Healthcare) at a flow rate of 0.4 mL/min using an isocratic-elution with PBS as mobile phase. 50 µg of purified HMW species and starting material were loaded onto the column and quantification was performed based on UV-detection at 280 nm.
SDS-PAGE analysis
The isolated dimers were analyzed by SDS-PAGE under non-reducing conditions. Five micrograms of samples was diluted in 10 µL of buffer composed of Tris 50 mM with 1% SDS. The obtained sample solution was mixed and heated for 10 min at 70°C before dropped on a GE homogenous gel 7.5%. Separation was performed at 200 V for 50 min on a Multiphor system (GE Healthcare). A Coomassie Brilliant Blue (CCB) staining method was used (0.1% CCB in water, 30 min) then gels were washed with a mix of methanol/acetic acid. Gels were read on an Image Scanner III imaging system and images were processed with the Quantity One software (Biorad, Hercules, USA).
SDS-CGE analysis
Capillary electrophoresis experiments were conducted on a System ProteomLab PA800+ (SCIEX, Brea, CA, USA), coupled to a UV detection. Software 32 Karat version 7.0 (SCIEX) was used to control the instrument and to collect the data. Silica capillary (50 cm effective length, 60 cm total length, 50 µm internal diameter) purchased from SCIEX was used. Before the first use, the capillary was rinsed at 70 psi for 5 min with 1 M NaOH, 0.1 M HCl and deionized water. Samples were injected electrokinetically at 20 kV for 10 s. Analysis was performed using SDS-Gel buffer (0.2% SDS, pH 8) (SCIEX) at −15 kV and 40°C, with a typical current of 40 μA. UV detection was set at 280 nm.
Peptide mapping (LC-MS/MS analysis)
Non-reducing peptide mapping experiments were performed on a UPLC Waters system coupled to a quadrupole time-of-flight mass spectrometer (Waters, Milford, MA, USA). Separations were performed on a C18 BEH 300 column (1.7 µm, 150 × 2.1 mm) (Waters Corp. CA, USA) at 60°C and a flow rate of 0.3 mL/min with 0.1% trifluoroacetic acid (TFA) in water as buffer A and 0.1% TFA in acetonitrile as buffer B, using a gradient of 12–37% buffer B in 83 min. Samples (100 µg) were dissolved in 8 M urea, 0.2 M ammonium acetate (pH 7.2) and submitted to trypsin digestion (Promega, Madison, USA) following enzyme provider instructions under non-reducing conditions with alkylated reagent (170 mM iodoacetamide). Twenty micrograms of tryptic peptides was injected per run. LC-MS measurements were acquired and analyzed under MassLynx software (Waters). The mass spectrometer was operated in the positive resolution mode and data were recorded from 200 to 3000 m/z. Calibration was carried out according to manufacturer’s procedure, on the acquisition range using NaI cluster ions (and Lockspray reference for internal calibration). MS/MS data were acquired with the instrument operating in the fast data-dependent acquisition fragmentation mode.
UPSEC-MS analysis
UPSEC-MS experiments were performed on an Acquity system (Waters) coupled to a UV detector and an electrospray mass spectrometer (Synapt G2 S, Waters) using a BEH 450 column with dimensions of 4.6 × 300 mm, porosity of 450 Ǻ and 2.5 μm particle size (Waters). An isocratic elution with 100 mM of ammonium acetate at 0.3 mL/min was applied. UV detection was set at 280 nm. The mass spectrometer was operated in the positive resolution mode and data were recorded from 2000 to 6000 m/z. Calibration was achieved on the acquisition range using cluster ions generated by a NaI solution diluted at 2 g/L in a mixture of water/isopropanol 50/50 (v/v) according to manufacturer’s procedure. Source parameters were optimized to get the best transmission and desolvation of ions. Source and desolvation temperature were set at 80°C and 200°C, respectively. Proteins were infused at 50 µL/min flow rate. The cone voltage was at 130 V and the source offset at 65 V. The nebulizer gas flow was set at 6 bars and the source pressure of 5 mbar was not modified and corresponded to the standard operating pressure of the stepwave-based interface of the mass spectrometer. Quadrupole MS profile settings were set as automatic. Data were analyzed using the MassLynx software (Waters) and processed with component analysis.
IEX-MS analysis
Separation of HMW species with or without enzymatic digestions was performed on the same equipment as that used for SEC-MS analysis. Fifty micrograms of sample was injected on a MAbPac SCX-10 column (4 mm x 150 mm, ThermoScientific, Waltham, MA, USA) equilibrated at 30°C and operated at a flow rate of 0.4 mL/min. MS-compatible mobile phases used for IEX were 50 mM ammonium formate buffered with formic acid at pH 3.9 (Buffer A) and 500 mM ammonium acetate at pH 7.4 (Buffer B). After an isocratic elution at 15% of B for 5 min, a linear gradient was applied from 15% to 31% in 40 min thus to 85% of B in 30 min. The column was then washed for 5 min at 90% of B and further equilibrated during 15 min at 15% of B. Mass spectrometer parameters were similar to those used for UPSEC-MS analysis.
AUC analysis
SV-AUC was performed on a Beckman Coulter XL-I analytical ultracentrifuge (Beckman Coulter, Palo Alto, USA) using an AN-50 TI rotor. The rotor speed and temperature were set at 42,000 rpm and 20°C, respectively. A sample volume of approximately 100 μl of the sample (the roledumab or the purified dimer fraction) was loaded into 3 mm path-length two-channel cells equipped with sapphire windows (Nanolytics, Potsdam, DE). A PBS buffer without protein was loaded in the reference channel. Sedimentation profiles were acquired using absorbance at 280 nm and interference optics every 7 minutes.
Abbreviations
| AUC | = | Analytical ultra-centrifugation |
| BEH | = | Bridged ethylene hybrid |
| IEX | = | Ion exchange chromatography |
| CQAs | = | Critical quality attributes |
| DTT | = | DL-dithiothreitol |
| HIC | = | Hydrophobic interaction chromatography |
| HMW | = | High molecular weight |
| HPSEC | = | High performance size exclusion chromatography |
| IEX | = | Ion exchange chromatography |
| kDa | = | Kilodaltons |
| HC | = | Heavy chain |
| LC | = | Light chain |
| LC-MS | = | Liquid chromatography-mass spectrometry |
| mAb | = | Monoclonal antibody |
| MS | = | Mass spectrometry |
| PBS | = | Phosphate-buffered saline |
| RhD | = | Rhesus D |
| SDS-CGE | = | Capillary gel electrophoresis sodium dodecyl sulfate |
| SDS PAGE | = | Sodium dodecylsulfate polyacrylamide gel electrophoresis |
| SECsize | = | Exclusion chromatography TFA trifluoroacetic acid |
| UPLC | = | Ultra-high-performance liquid chromatography |
| UV | = | Ultraviolet |
Disclosure of potential conflicts of interest
G. ROUBY, Y. LEBLANC and N. BIHOREAU are employees of LFB, the Company that develops roledumab. Beyond this, the authors are not aware of any affiliations, memberships, funding or financial holdings that might perceive as affecting the objectivity of this article.
Supplemental Material
Download Zip (806.3 KB)Acknowledgments
The authors would like also to express their gratitude to M. Andre, C. Groseil, N. Tilly, C. Ramon, V. Faid and G. Chevreux (LFB, Les Ulis, France), F. Halgand and G. Van der Rest (Université Paris-Saclay, France), for helpful discussions about structural characterization of antibody, L. Guilbert and M. LeBoedec for purification process support. We thank also M. Berger for valuable discussions.
Supplementary material
Supplemental data for this article can be accessed on the publisher’s website.
References
- Yver A, Homery MC, Fuseau E, E G, Dhainaut F, Quagliaroli D, Beliard R, Prost JF. Pharmacokinetics and safety of roledumab, a novel human recombinant monoclonal anti-RhD antibody with an optimized Fc for improved engagement of FCγRIII, in healthy volunteers. Vox Sanguinis. 2012;103:213–11. doi:https://doi.org/10.1111/j.1423-0410.2012.01603.x.
- Hmiel LK, Brorson KA, Boyne IIMT. Post-translational structural modifications of immunoglobulin G and their effect on biological activity. Anal Bioanal Chem. 2015;407:79–94. doi:https://doi.org/10.1007/s00216-014-8108-x.
- Chumsae CM. Discovery of unknown modifications recombinant monoclonal antibodies. Chemistry Dissertations. 2015:Paper;99.
- Manning MC, Chou DK, Murphy BM, Payne RW, Katayama DS. Stability of protein pharmaceuticals: an update. Pharm Res. 2010;27:544–75. doi:https://doi.org/10.1007/s11095-009-0045-6.
- Fischer S, Hoernschemeyer J, Mahler HC..Glycation during storage and administration of monoclonal antibody formulations. Eur J Pharm Biopharm. 2008;70:42–50. doi:https://doi.org/10.1016/j.ejpb.2008.04.021.
- LW D Jr, Kim C, Qiu D, Cheng KC. Determination of the origin of the N-terminal pyro-glutamate variation in monoclonal antibodies using model peptides. Biotechnol Bioeng. 2007;97:544–53. doi:https://doi.org/10.1002/bit.21260.
- Vlasak J, Ionescu R. Fragmentation of monoclonal antibodies. MAbs. 2011;3:253–63. doi:https://doi.org/10.4161/mabs.3.3.15608.
- Liu H, Gaza-Bulseco G, Faldu D, Chumsae C, Sun J. Heterogeneity of monoclonal antibodies. J Pharm. Sci. 2008;97:2426–47. doi:https://doi.org/10.1002/jps.21180.
- Liu H, May K. Disulfide bond structures of IgG molecules: structural variations, chemical modifications and possible impacts to stability and biological function. MAbs. 2012;4:17–23. doi:https://doi.org/10.4161/mabs.4.1.18347.
- Arosio P, Barolo G, Müller-Späth T, Wu H, Morbidelli M. Aggregation stability of a monoclonal antibody during downstream processing. Pharm Res. 2011;28:1884–94. doi:https://doi.org/10.1007/s11095-011-0416-7.
- Cromwell MEM, Hilario E, Jacobson F. Protein aggregation and bioprocessing. AAPS J. 2006;8:572–79. doi:https://doi.org/10.1208/aapsj080366.
- Vermeer AWP, Norde W. The thermal stability of immunoglobulin: unfolding and aggregation of a multi-domain protein. Biophys J. 2000;78:394–404. doi:https://doi.org/10.1016/S0006-3495(00)76602-1.
- Vázquez-Rey M, Lang DA. Aggregates in monoclonal antibody manufacturing processes. Biotechnol Bioeng. 2011;108:1494–508. doi:https://doi.org/10.1002/bit.23155.
- Brader ML, Estey T, Bai S, Alston RW, Lucas KK, Lantz S, Landsman P, Maloney KM. Examination of thermal unfolding and aggregation profiles of a series of developable therapeutic monoclonal antibodies. Mol Pharmaceutics. 2015;12:1005–17. doi:https://doi.org/10.1021/mp400666b.
- Wang W. Protein aggregation and its inhibition in biopharmaceutics. Int J Pharm. 2005;289:1–30. doi:https://doi.org/10.1021/mp400666b.
- Mahler HC, Friess W, Grauschopf U, Kiese S. Protein aggregation: pathways, induction Factors and analysis. J of Pharm Sc. 2009;59:407–17. doi:https://doi.org/10.1002/jps.21566.
- Chi EY, Krishnan S, Randolph TW, Carpenter JF. Physical stability of proteins in aqueous solution: mechanism and driving forces in non-native protein aggregation. Pharm Res. 2003;20:1325–36. doi:https://doi.org/10.1023/A:1025771421906.
- Alt N, Zhang TY, Motchnik P, Taticek R, Quarmby V, Tilman S, Beck H, Emrich T, Harris RJ. Determination of critical quality attributes for monoclonal antibodies using quality by design principles. Biologicals. 2016;44:291–305. doi:https://doi.org/10.1016/j.biologicals.2016.06.005.
- Dingman R, Balu-Iyer Sathy V. Immunogenicity of protein pharmaceuticals. J of Pharm Sc. 2019;108:1637–54. doi:https://doi.org/10.1016/j.xphs.2018.12.014.
- Ratanji KD, Derrick JP, Dearman RJ, Kimber I. Immunogenicity of therapeutic proteins: influence of aggregation. J Immunotoxicol. 2014;11:99–109. doi:https://doi.org/10.3109/1547691X.2013.821564.
- Rosenberg AS. Effects of protein aggregates: an immunologic perspective. AAPS J. 2006;8:501–07. doi:https://doi.org/10.1208/aapsj080359.
- Tran BN, Chan SL, Ng C, Shi J, Correia I, Radziejewski C, Matsudaira P. Higher order structures of Adalimumab, Infliximab and their complexes with TNFa revealed by electron microscopy. Protein Science. 2017;26:2392–98. doi:https://doi.org/10.1002/pro.3306.
- Berkowitz SA. Role of analytical ultracentrifugation in assessing the aggregation of protein biopharmaceuticals. AAPS J. 2006;8:590–605. doi:https://doi.org/10.1208/aapsj080368.
- Gabrielson JP, Arthur KK. Measuring low levels of protein aggregation by sedimentation velocity. Methods. 2011;54:83–91. doi:https://doi.org/10.1016/j.ymeth.2010.12.030.
- Arakawa T, Philo JS, Ejima D. Aggregation analysis of therapeutic proteins, part 1. General Aspects and Techniques for Assessment. Bioprocess Int. 2006;4:42–43.
- Beck A, Wagner-Rousset E, Ayoub D, Van Dorsselaer A, Sanglier-Cianférani S. Characterization of therapeutic antibodies and related products. Anal Chem. 2013;85:715–36. doi:https://doi.org/10.1021/ac3032355.
- Fekete S, Gassner AL, Rudaz S, Schappler J, Guillarme D. Analytical strategies for the characterization of therapeutic monoclonal antibodies. Trends Anal Chem. 2013;42:74–83. doi:https://doi.org/10.1016/j.trac.2012.09.012.
- D’Atri V, Fekete S, Clarke A, Veuthey JL, Guillarme D. Recent advances in chromatography for pharmaceutical analysis. Anal Chem. 2019;91:210–39. 10.1021/acs.analchem.8b05026.
- Goyon A, Fekete S, Beck A, Veuthey JL, Guillarme D. Unraveling the mysteries of modern size exclusion chromatography - the way to achieve confident characterization of therapeutic proteins. J Chrom B. 2018;1092:368–78. doi:https://doi.org/10.1016/j.jchromb.2018.06.029.
- Arakawa T, Philo JS, Ejima D. Aggregation analysis of therapeutic proteins, part 3: principles and optimization of field-flow fractionation. Bioprocess Int. 2007;5:52–70.
- Arakawa T, Ejima D, Li T, Philo JS. The critical role of mobile phase composition in size exclusion chromatography of protein pharmaceuticals. J Pharm Sc. 2010;99(4):1674–92. doi:https://doi.org/10.1002/jps.21974.
- Haselberg R, De Vijlder T, Heukers R, Smit MJ, Romijn EP, Somsen GW, Domínguez-Vega E. Heterogeneity assessment of antibody-derived therapeutics at the intact and middle-up level by low-flow sheathless capillary electrophoresis-mass spectrometry. Anal Chim Acta. 2018;1044:181–90. doi:https://doi.org/10.1016/j.aca.2018.08.024.
- Belov AM, Viner R, Santos MR, Horn DM, Bern M, Karger BL, Ivanov AR. Analysis of proteins, protein complexes, and organellar proteomes using sheathless capillary zone electrophoresis - native mass spectrometry. J Am Soc Mass Spectr. 2017;28:2614–34. doi:https://doi.org/10.1007/s13361-017-1781-1.
- Belov AM, Zang L, Sebastiano R, Santos MR, Bush DR, Karger BL, Ivanov AR. Complementary middle-down and intact monoclonal antibody proteoform characterization by capillary zone electrophoresis-mass spectrometry. Electrophoresis. 2018;39:2069–82. doi:https://doi.org/10.1002/elps.201800067.
- Le-Minh V, Tran NT, Makky A, Rosilio V, Taverna M, Smadja C. Capillary zone electrophoresis-native mass spectrometry for the quality control of intact therapeutic monoclonal antibodies. J Chrom A. 2019;1601:375–84. doi:https://doi.org/10.1016/j.chroma.2019.05.050.
- Marie AL, Dominguez-Vega E, Saller F, Plantier JL, Urbain R, Borgel D, Taverna M. Characterization of conformers and dimers of antithrombin by capillary electrophoresis-quadrupole-time-of-flight mass spectrometry. Analytica Chimica Acta. 2016;947:58–65. doi:https://doi.org/10.1016/j.aca.2016.10.016.
- Haberger M, Leiss M, Heidenreich AK, Pester O, Hafenmair G, Hook M, Bonnington L, Wegele H, Haindl M, Reusch D, et al. Rapid characterization of biotherapeutic proteins by size-exclusion chromatography coupled to native mass spectrometry. MAbs. 2015;8:331–39. doi:https://doi.org/10.1080/19420862.2015.1122150.
- Ventouri IK, Malheiro DBA, Voeten RLC, Kok S, Honing M, Somsen GW, Haselberg R. Probing protein denaturation during size-exclusion chromatography using native mass spectrometry’. Anal Chem. 2020;92:4292–300. doi:https://doi.org/10.1021/acs.analchem.9b04961.
- Ehkirch A, Hernandez-Alba O, Colas O, Beck A, Guillarme D, Cianférani S. Hyphenation of Size exclusion chromatography to native ion mobility mass spectrometry for the analytical characterization of therapeutic antibodies and related product. J Chrom B. 2018;1086:176–83. doi:https://doi.org/10.1021/acs.analchem.8b03333.
- Ehkirch A, Goyon A, Hernandez-Alba O, Rouviere F, D’Atri V, Dreyfus C, Cianferani S. A novel online four-dimensional SECxSEC-IMxMS methodology for characterization of monoclonal antibody size variants. Anal Chemis. 2018;23:13929–37. doi:https://doi.org/10.1021/acs.analchem.8b03333.
- Leblanc Y, Ramon C, Bihoreau N. Charge variants characterization of a monoclonal antibody by ion exchange chromatography coupled on-line to native mass spectrometry: case study after a long-term storage at+5 degrees C. J of Chrom B. 2017;1048:130–39. doi:https://doi.org/10.1016/j.jchromb.2017.02.017.
- Yan Y, Liu AP, Wang S, Daly TJ, Li N. Ultrasensitive characterization of charge heterogeneity of therapeutic monoclonal antibodies using strong cation exchange chromatography coupled to native mass spectrometry. Anal Chem. 2018;90(21):13013–20. doi:https://doi.org/10.1021/acs.analchem.8b03773.
- Lau H, Pace D, Yan B, McGrath T, Smallwood S, Patel K, Latypov RF. Investigation of degradation processes in IgG1 monoclonal antibodies by limited proteolysis coupled with weak cation-exchange HPLC. J chrom B. 2010;878(11–12):868–76. doi:https://doi.org/10.1016/j.jchromb.2010.02.003.
- Chen B, Lin Z, Alpert AJ, Fu C, Zhang Q, Pritts WA, Ge Y. Online hydrophobic interaction chromatography–mass spectrometry for the analysis of intact monoclonal antibodies. Anal Chem. 2018;90:7135–38. doi:https://doi.org/10.1021/acs.analchem.8b01865.
- Houde D, Arndt J, Domeier W, Berkowitz S, Engen JR. Characterization of IgG1 conformation and conformational dynamics by hydrogen/deuterium exchange mass spectrometry. Anal Chem. 2009;81:2644–51. doi:https://doi.org/10.1021/ac802575y.
- Iacob RE, Bou-Assaf GM, Makowski L, Engen JR, Berkowitz SA, Houde D. Investigating monoclonal antibody aggregation using a combination of H/DX-MS and other biophysical measurements. J Pharm Sci. 2013;102:4315–29. doi:https://doi.org/10.1002/jps.23754.
- Zhang A, Singh SK, Shirts MR, Kumar S, Fernandez EJ. Fernandez distinct aggregation mechanisms of monoclonal antibody under thermal and freeze-thaw stresses revealed by hydrogen exchange. Pharm Res. 2012;29:236–50. doi:https://doi.org/10.1007/s11095-011-0538-y.
- Kiese S, Papppenberger A, Friess W, Mahler HC. Shaken, not stirred: mechanical stress testing of an IgG1 antibody. J Pharm Sci. 2008;97(10):4347–66. doi:https://doi.org/10.1002/jps.21328.
- Hawe A, Kasper JC, Friess W, Jiskoot W. Structural properties of monoclonal antibody aggregates induced by freeze thawing and thermal stress. Eur J Pharm Sc. 2009;38:79–87. doi:https://doi.org/10.1016/j.ejps.2009.06.001.
- Joubert MK, Luo Q, Nashed-Samuel Y, Wypych J, Narhi LO. Classification and characterization of therapeutic antibody aggregates. J Biol Chem. 2011;286:25118–33. doi:https://doi.org/10.1074/jbc.M110.160457.
- Paul R, Graff-Meyer A, Stahlberg H, Lauer ME, Rufer AC, Beck H, Briguet A, Schnaible V, Buckel T, Boeckle S. Structure and function of purified monoclonal antibody dimers induced by different stress conditions. Pharm Res. 2012;29:2047–59. doi:https://doi.org/10.1007/s11095-012-0732-6.
- Remmele RL, Callahan WJ, Krishnan S, Zhou LD, Bondarenko PV, Nichols AC, Kleemann GR, Pipes GD, Park S, Fodor S, et al. Active dimer of epratuzumab provides insight into the complex nature of an antibody aggregate. J Pharm Sci. 2007;98:126–45. doi:https://doi.org/10.1002/jps.20515.
- Iwura T, Fukuda J, Yamazaki K, Kanamaru S, Arisaka F. Intermolecular interactions and conformation of antibody dimers present in igG1 biopharmaceuticals. J Biochem. 2013;155:63–71. doi:https://doi.org/10.1093/jb/mvt095.
- Deperalta G, Alvarez M, Bechtel C, Dong K, McDonald R, Ling V. Structural analysis of a therapeutic monoclonal antibody dimer by hydroxyl radical footprinting. MAbs. 2013;5:86–101. doi:https://doi.org/10.4161/mabs.22964.
- Plath F, Ringler P, Graff-Meyer A, Stahlberg H, Lauer ME, Rufer Arne C, Graewert MA, Svergun D, Gellermann G, Finkler C, et al. Characterization of mAb dimers reveals predominant dimer forms common in therapeutic. MAbs. 2016;8:928–40. doi:https://doi.org/10.1080/19420862.2016.1168960.
- Liu H, Chumsae C, Gaza-Bulseco G, Hurkmans K, Radziejewski CH. Ranking the susceptibility of disulfide bonds in human IgG1 antibodies by reduction, differential alkylation, and LC-MS analysis’. Anal Chem. 2010;82(12):5219–26. doi:https://doi.org/10.1021/ac100575n.
- Chevreux G, Tilly N, Bihoreau N. Fast analysis of recombinant monoclonal antibodies using IdeS proteolytic digestion and electrospray mass spectrometry. Anal Biochem. 2011;415:212–14. doi:https://doi.org/10.1016/j.ab.2011.04.030.
- Spoerry C, Seele J, Valentin-Weigand P, Baums CG, Von Pawel-Rammingen U. Identification and characterization of IgdE, a novel IgG-degrading protease of streptococcus suis with unique specificity for porcine IgG. J Biol Chem. 2016;291(15):7915–25. doi:https://doi.org/10.1074/jbc.M115.711440.