?Mathematical formulae have been encoded as MathML and are displayed in this HTML version using MathJax in order to improve their display. Uncheck the box to turn MathJax off. This feature requires Javascript. Click on a formula to zoom.
?Mathematical formulae have been encoded as MathML and are displayed in this HTML version using MathJax in order to improve their display. Uncheck the box to turn MathJax off. This feature requires Javascript. Click on a formula to zoom.ABSTRACT
As biologics have become a mainstay in the development of novel therapies, protein engineering tools to expand on their structural advantages, namely specificity, affinity, and valency are of interest. Antibodies have dominated this field as the preferred scaffold for biologics development while there has been limited exploration into the use of albumin with its unique physiological characteristics as a platform for biologics design. There has been a great deal of interest to create bispecific and more complex multivalent molecules to build on the advantages offered by protein-based therapeutics relative to small molecules. Here, we explore the use of human serum albumin (HSA) as a scaffold for the design of multispecific biologics. In particular, we describe a structure-guided approach to the design of split HSA molecules we refer to as AlbuCORE, that effectively and spontaneously forms a native albumin-like molecule, but in a heterodimeric state upon co-expression. We show that the split AlbuCORE designs allow the creation of novel fusion entities with unique alternate geometries. We also show that, apart from these AlbuCORE fusion entities, there is an opportunity to explore their albumin-like small hydrophobic molecule carrying capacity as a drug conjugate in these designs.
Introduction
Human serum albumin (HSA) is the most abundant protein in serum. Opportunities for the use of albumin-based therapeutic design stem from its natural physiology and function as a transporter molecule with the ability to pass through the endothelial barriers lining blood vessels and access extravascular space and the lymphatic system.Citation1–Citation5 Additionally, albumin also exhibits a long plasma half-life, which can be exploited for improving the pharmacokinetic (PK) properties of both small molecules and polypeptides that typically exhibit fast in vivo clearance. In normal epithelial cells, the neonatal Fc receptor (FcRn) is recognized for its role in recycling HSA following pinocytosis.Citation6 The binding and transport capacity of albumin in the tumor microenvironment has been used for the detection of sentinel lymph nodes during tumor biopsies using a variety of imaging techniques in malignant melanoma and early breast cancer detectionCitation7,Citation8 and to deliver chemotherapeutics, including nab-paclitaxel (a nanoparticle formulation of the chemotherapeutic drug paclitaxel with albumin), with fewer side effects.Citation9
The pathological hypoalbuminemia and cachexic condition observed in critically ill cancer patients has been linked to catabolic degradation of serum albumin in diseased cells.Citation10–Citation12 A drastic change in the metabolic state of the tumor microenvironment is a recognized hallmark of aggressive cancers,Citation13 and albumin is taken up by Ras-transformed pancreatic cancer cells by micropinocytosis for catabolic degradation to meet essential amino acid requirements.Citation14,Citation15 It was recently reported that wild-type Ras tumor cells downregulate the expression of FcRn, thus facilitating the uptake and breakdown of albumin by tumor cells for its metabolic needs.Citation16 A number of putative albumin-binding proteins and receptors, including FcRn, gp60, gp30, calreticulin, cubilin, megalin, and secreted protein acidic and rich in cysteine (SPARC), have been identified to have physiological roles in albumin transport and homeostasis.Citation3 Among these, overexpression of the extra-cellular SPARC protein in the tumor microenvironment and its potential relation to tumor growth as a positive or negative prognostic indicator have been proposed.Citation17,Citation18 A recent study suggests that caveolin-1 is a predictive biomarker of nab-paclitaxel activity, as well as albumin uptake across pancreatic and non-small cell lung cancer cells where nab-paclitaxel is clinically active.Citation19
Prior to the recent successes in manipulating immunomodulatory checkpoint targets, the success rate of antibodies against solid tumor cell surface targets had only been modest in the 20 years since the first approved antibody drug (trastuzumab) against a solid tumor target. There have been 10 new approvals for use in solid tumors (against 5 unique targets) of the total of 81 US FDA-approved antibody therapies.Citation20 Among other factors, the limited efficacy of monoclonal antibodies in treating solid tumors has been attributed to the barriers introduced by the tumor microenvironment and stroma that limit tumor.Citation21–Citation24 A number of alternate scaffolds have been developed with notably smaller molecular size than a full-length monoclonal antibody, but biologics based on such scaffolds are hindered by poor half-life.Citation25 Some of these designs have relied on hitchhiking with albumin to balance serum persistence without loss of the distribution advantage.Citation26 In a tumor imaging study, a bifunctional molecule targeting human epidermal growth factor receptor 2 (HER2) and albumin was shown to rapidly target tumors and result in lower overall tissue exposure and achieve better tumor penetration than trastuzumab.Citation27
The Fc domain of an antibody therapeutic also plays an important functional role in mediating various effector functions, including antibody-dependent cell-mediated cytotoxicity (ADCC), complement-dependent cytotoxicity (CDC), and antibody-dependent cellular phagocytosis (ADCP). However, in a number of contemporary biologics treatment modalities such as for immuno-oncology or in autoimmune settings, effector function might be best avoided.Citation28,Citation29 For example, both the recently successful PD1-targeting antibodies nivolumab and pembrolizumab are IgG4 molecules.Citation30 Using albumin as an alternate to Fc naturally avoids interactions with the immune effector system.
Rarely is targeting a single antigen or pathway sufficient to effectively treat complex diseases such as cancer, given the multitude of factors that contribute to disease progression and drug resistance. The substantial opportunity offered by novel multi-target engagement strategies involving biologics is conveyed by the plethora of approaches developed to make bispecifics.Citation31 Bispecific molecules introduce a spatio-temporal constraint during the engagement of the two targeted arms, leading to novel functional effects potentially not achievable with a combination of the component parental arms. For instance, there is a growing body of data showing that targeting multiple epitopes concurrently on the epidermal growth factor receptor family members can introduce unique effects, such as receptor downregulation and altered rate of endocytosis.Citation32–Citation36 Another example would be achieving agonistic activity by targeting tumor necrosis factor family receptors, which requires particular geometric clustering of the receptors, mediated by engagement of particular epitopes.Citation37 While a number of antibody Fc-based designs have been developed for bispecific therapeutics, we were interested in developing an alternate albumin-based scaffold for bispecific design.
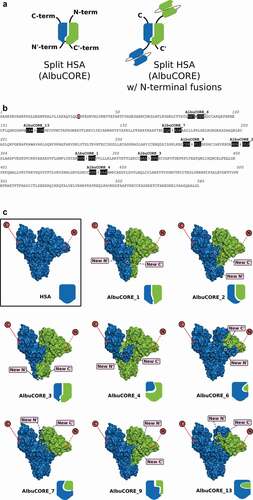
The use of albumin in a variety of therapeutic development efforts has been described in the literature. MM-111, a bispecific molecule that was evaluated clinically, was designed as a single polypeptide composed of HER2- and HER3-targeting single-chain variable fragments (scFvs) fused to HSA.Citation38 Albinterferon, composed of interferon alpha-2b fused to HSA, was evaluated in Phase 3 trials as part of hepatitis C treatment regimen.Citation39,Citation40 Albiglutide is a glucagon-like peptide 1 fusion with albumin approved for treatment of type 2 diabetes.Citation41 All these designs involve fusions to either the N or C-terminus of albumin, or both, as in the case of the bispecific MM-111. As the C-terminal domain of HSA is involved in binding to FcRn,Citation42 fusions to that terminus can have a negative impact in the binding affinity between HSA and FcRn,Citation43 leading to a putative reduction in serum half-life. There is limited opportunity to design bispecific molecules using albumin beyond creating fusions to the natural termini. Here, we present the engineering strategy and development of a novel HSA-derived multivalent molecular platform () we refer to as AlbuCORE, in which the HSA is split into two self-associating polypeptides, thus allowing for the introduction of up to four targeting or therapeutic fusion moieties at each N- or C-termini of the two polypeptides. The engineering is designed to utilize the intrinsic features of albumin as a core scaffold while enabling specific targeting opportunities with different alternate multivalent geometric design.
Figure 1. A. Cartoon representation of the AlbuCORE scaffold as a split Human Serum Albumin (left) and as a putative bispecific with two distinct N-terminal-fused scFv moieties (right). B. HSA sequence showing the eight AlbuCORE split points evaluated. The three residues on either side of the split position are shaded to help visualization. A simple split between two amino acids is indicated by an arrow. AlbuCORE_6 has a single amino acid deletion indicated by crossing that amino acid letter. The ubiquitous mutation C34S is indicated by a red-shaded box. C. Space-filling and cartoon depiction of the human serum albumin (HSA) and the eight engineered AlbuCORE scaffolds. The natural N- and C-termini are indicated by a red circle in the surface cartoon, while the newly created N’- and C’-termini are shown by pink boxes. Solid lines indicate the termini are facing the viewer and dotted lines indicate the termini are behind the molecule. The interchain disulfide, if present, is indicated by a black dash connecting the two chains in the block cartoons. AlbuCORE_2 and AlbuCORE_13 are the only ones without an interchain disulfide link.
Materials and methods
In silico design of AlbuCORE scaffolds
Most of the structural analysis were conducted using the published crystallographic model of native HSA (Protein Data Bank ID 3JRYCitation44) with Zymeworks’ proprietary molecular modeling and simulation platform ZymeCAD™, including molecular dynamics simulations, root mean square fluctuation (RMSF) analyses, residue solvent exposure, residue contact analysis, and interface and per-residue energy contributions. Temperature factors were derived directly from the crystal structure.
For the in-silico evaluation of putative split albumin (AlbuCORE) molecules we used the following guidelines:
The split site should reside on an exposed flexible loop. This loop should be relatively mobile and should not be involved in significant contacts with the rest of the protein, as determined by RMSF, solvent exposure, and residue contact analysis. This split site must not be part of one of the many secondary structure elements (α-helices) forming the albumin fold. That achieves two things: it minimizes interference with the native structure of the albumin molecule, and it exposes the engineered termini to allow for fusion of moieties.
The interface between the two split parts should be rich in hotspot residues. Hotspots are well-connected residues as judged by our residue contacts analysis. We iterated through the HSA structure screening for loops satisfying the first requirement above (i) and generated models of split HSA for each of those putative split points, favoring interface rich in inter-chain contacts. We also looked for interface composed mainly by apolar residues to favor an obligatory heterodimer over homodimers or single chains. We calculated the interface buried area, carbon contribution, and ∆∆Gassoc using a MM-GBSA-likeCitation45 free energy decomposition approach.
The split scaffold should not interfere with FcRn binding. Schmidt et al.Citation46 and Oganesyan et al.Citation42 have described the crystallographic co-complex between HSA and FcRn, which largely comprises interactions in the domain III of HSA. None of the proposed split sites reside in domain III or anywhere near the putative HSA:FcRn interface.
Construct generation, protein production and purification
The genes encoding the full-length wild-type HSA, as well as the N- and C-terminal fragments of each AlbuCORE scaffold (), were constructed by gene synthesis (Genscript) using codons optimized for human/mammalian expression. The constructs were designed from known full-length Human Serum Albumin Preprotein (GENEBANK: NP_000468.1), after exclusion of the precursor sequence MKWVTFISLLFLFSSAYSRGVFRR. Fusion constructs between HSA/AlbuCORE and a binding moiety were generated by appending the sequence of the binding module to the N- and/or C-terminus of the relevant fragment, as appropriate. The final gene products were subcloned into the mammalian expression vector pTT5 (NRC-BRI, Canada)Citation47 with the signal peptide MAVMAPRTLVLLLSGALALTQTWAG appended to the N-terminal of the polypeptide sequence.
Wild-type HSA or any of the AlbuCORE scaffolds, with or without a fusion moiety, were expressed in CHO-3E7 cell line grown in suspension in FreeStyle F17 medium (Invitrogen) supplemented with 0.1% w/v pluronic and 4 mM glutamine. On the day of transfection cell density should be around 1.5–2 million cells/ml and viability must be greater than 97%. Transfection was carried out as described by Durocher and coworkersCitation48,Citation49 using a mixture of plasmid DNA made of 5% pTTo-GFP plasmid (green fluorescent protein to determine transfection efficiency), 15% pTT22-AKT plasmid, 21% HSA/AlbuCORE plasmids (10.63% of each), 68.37% of Salmon Sperm DNA. Following transfection, the shake flask containing cells was placed on an orbital shaker set to 120 rpm in a humidified incubator with 5% CO2 at 37°C. Twenty-four hours post-transfection, 1% w/v TN1, and 0.5 mM valproic acid were added to the cultures. The cultures were then transferred on an orbital shaker (120 rpm) placed in a humidified incubator with 5% CO2 set at 32°C. At 24–48 h, GFP positive cells should be between 30% and 60% as determined by flow cytometry. Cells were harvested 7–10 d post-transfection and spun at 4,000 rpm for 20 minutes. Supernatants were adjusted to pH 5.0 with 0.5 M sodium citrate, pH 5.0, filter-sterilized (clarified) using a 0.45 μm filter (Millipore), and frozen at −80°C.
HSA or AlbuCORE purifications were performed similar to the AlbuPure™ resin manufacturer’s suggestions (Prometic Bioseparations; product code 3151). Several conditions were tested to improve yields and purity of protein targets using batch binding strategies. A more detailed description of the method development can be found in the Supplementary Material. For the most part, proteins evaluated in this study were purified using the optimized protocol below. Prior to purification, the frozen supernatants were thawed in a lukewarm water bath. The pH of the supernatant was then adjusted to pH 5 by adding 0.5 M sodium citrate pH 3 buffer (approximately 3 ml per 100 ml supernatant or approximately 15 mM sodium citrate final). AlbuPureTM resin equilibrated in 50 mM sodium citrate pH 5 was then added to the supernatants and incubated with agitation (~140 rpm with orbital shaker) at 4°C overnight (typically 1 ml of resin per 20–40 mg target protein). The supernatant and resin samples were then transferred to glass columns, the flow-through collected by gravity flow, and the resin was washed with 2 × 5 column volumes (CV) 50 mM sodium citrate pH 5, and then 2 × 5 CV 50 mM ammonium acetate pH 8. Target proteins were eluted first in 50 mM ammonium acetate pH 7, 20 mM sodium octanoate (7 x 2 CV), followed immediately by 50 mM ammonium acetate pH 7, 100 mM sodium octanoate (7 x 2 CV). Fractions containing HSA or AlbuCORE (as per Bradford protein assay, A280 and/or SDS-PAGE analysis) were pooled and applied on a 16/60 HiLoad Superdex 200 prep grade gel filtration column coupled to an AKTA purification system (GE Healthcare; Express, FPLC, or Purifier system) using a flow rate of 1 ml/ml. Fractions were pooled based on SDS-PAGE or capillary electrophoresis analysis (LabChip GX/PerkinElmer). When necessary, samples were concentrated by centrifugation using an Amicon Ultra-filtration membrane (10 000 MWCO). In order to meet low endotoxin requirements, all systems, columns, and resin (where applicable) were depyrogenated using NaOH solutions with standard protocols prior to protein purification.
Biophysical characterizations
All DSC experiments were carried out using a MicroCal VP-Capillary instrument (Malvern) and measured with a scan rate of 1°C/min from 20°C to 100°C. SEC purified samples were prepared in phosphate-buffered saline (PBS), pH 7.4. For the samples with fatty acid, 20-fold molar excess of sodium octanoate was added to the samples and incubated at 60°C (simulated pasteurization) or 4°C overnight (~18 h). Data was analyzed using the Origin software with the appropriate buffer background subtracted. The reported melting temperature (Tm) was derived using a non-2-state fitting (see ref. 53).
UPLC-SEC analysis was performed using an Acquity BEH200 SEC column (Waters, 2.5 mL, 4.6 × 150 mm, stainless steel, 1.7 μm particles) at 30°C with a flow rate of 0.4 ml/min. Run times consisted of 7 min and a total volume per injection of 2.8 mL with running buffers PBS, pH 7.4, 0.02% Tween-20. Peak integration was done using Empower 3 software and the results reported are the average of two independent runs. A calibration curve was determined just prior to the sample injection using a set of Gel Filtration Standards (Bio-Rad, PN 151–1901) and the samples fit using the equation logMW = (retention time – 8.0313)/-0.9835.
CD measurements were carried out using a Jasco J-815 spectropolarimeter. Sample concentrations were measured by absorbance at 280 nm on a Nanodrop 2000 (SN P312). Samples were buffer exchanged into 10 mM phosphate, 150 mM NaF, pH 7.37 to reduce the interference by Cl− ions in the far UV. Buffer exchange was done using 0.5 ml Zeba spin columns (Thermo Scientific PN 89,882, lot #RD233862). Each column was washed with 3 × 300 µl buffer with centrifugation at 1500 x g for 1 minute. Sample volumes ranging from 37 to 222 µl totaling 100 µg each were then desalted with centrifugation at 1500 x g for 2 minutes. Desalted sample concentrations were again measured by absorbance at 280 nm on a Nanodrop 2000 (SN P312) and each sample was diluted to 0.15 mg/ml in the NaF-containing buffer. Far UV CD spectra were measured at 20°C in a 1 mm cuvette scanning from 260 to 185 nm with a data pitch of 0.5 nm, a scan rate of 50 nm/min, and a response time of 1 sec. Twenty-five acquisitions were collected. Raw data was smoothed and baseline subtracted using the spectrum of a buffer blank.
FcRn binding affinity
Binding between FcRn and HSA/AlbuCORE were determined by BLI on an Octet Red 96 system (PALL/ForteBio) using a modified method previously described.Citation50 Briefly, biotinylated human FcRn (ImmuniTrack) at 0.5 μg/mL was immobilized on Streptavidin (SA) Biosensors (PALL/ForteBio) in regeneration buffer (PBS, pH 7.4, 0.05% Tween-20). The sensors were blocked with HSA (Sigma A9511) at 5 mg/mL before and after loading with FcRn to minimize nonspecific interactions. HSA and AlbuCORE samples were prepared at a concentration of 2 μM into binding buffer (25 mM NaOAc, 25 mM NaH2PO4, pH 5.5, 150 mM NaCl, 0.05% Tween-20), and an eight-step 2-fold dilution series prepared in a 96-well plate. Equilibrium measurements were determined at 30°C with 360 s association and 150 s dissociation in binding buffer. Between each cycle of association/dissociation, sensors were regenerated for 120 s in regeneration buffer, and baselined for 120 s in binding buffer. All data were double-referenced with a sensor without sample and a serial dilution of HSA with an unloaded sensor. Processing and fitting of the raw data were performed with the Octet Data Analysis software (version: 9.0.0.12) using a 1:1 binding mode.
Antigen-binding affinity
All HER2-binding experiments were performed on a Biacore T200 (GE Healthcare) at 25°C. Running buffer was PBS with 0.05% P20 (Teknova) with ethylenediaminetetraacetic acid (EDTA) added to 3.4 mM. HER2 used in the binding assay was purchased from ThermoFisher and further purified by SEC to ensure a monomeric preparation. Assays were performed by indirectly capturing the AlbuCORE and Azymetric constructs using 2000 RU surfaces of anti-HSA goat polyclonal antibody (Abcam cat#ab18074) or goat anti-Fc polyclonal (Jackson ImmunoResearch cat#109-001-008) and flowing HER2 over top. The antibody capture surfaces were created using standard amine coupling to a CM-5 sensor chip using the Immobilization Wizard application within the Biacore T200 control software. For AlbuCORE and Azymetric analysis, 20–80 RUs were captured at 10 uL/min using 0.4 to 0.6 ug/mL solutions (Azymetric) or 2 to 4 ug/mL solutions (AlbuCORE) diluted in running buffer. HER2 was then injected at 100 uL/min over the captured surfaces at 2.5, 5, 10, 20, 40 nM concentrations using single-cycle kinetics with an association time of 200 s, and a dissociation time of 1800 s (Azymetric) or 3600 s (AlbuCORE). The capture surfaces were regenerated using a 120 injection of 10 mM Glycine pH 1.5 at 30 uL/min between each round of HER2 injections. Sensorgrams were double referenced and KD determined by fitting the sensorgrams to a 1:1 binding model using Biacore BiaEvaluation software (GE Healthcare).
Flow cytometry: Indirect cell binding
Cell Lines
SKOV-3 and BT-474 (ATCC) cancer cell lines were maintained in their recommended culture medium (McCoy’s 5A and DMEM, respectively) supplemented with L-glutamine and the 10% fetal bovine serum (FBS) (Hyclone) in a humidified +5% CO2 incubator at 37°C. Cells were routinely passaged two to three times a week using 0.05% trypsin (HyClone) and discarded after 15 passages.
Flow cytometry cell binding assay
Unless otherwise stated, all media were kept are 4°C and all incubations were performed on wet ice. On the day of the assay, exponentially growing cells were harvested using warm Cell dissociation solution (Sigma), centrifuged, counted, and resuspended in complete medium at a cell density of 2 × 106 cells/mL. Cells were distributed in microtubes (105 cells/tube). All variants were diluted as 2X master dilutions in complete medium followed by five 3-fold serial dilutions for a total of six concentration points. Equal volumes of 2X test variants or controls were added to cells and incubated for 2 h. Cells were washed twice (RPMI+ 2% FBS), centrifuged, and the supernatants removed. In some cases, rHSA (Albumin, human recombinant, expressed in Rice: Sigma Aldrich cat# A9731) was used as an additional control to measure nonspecific binding.
Detection of bound AlbuCORE or antibody was achieved by incubation with a fluorescently labeled FITC-Goat-anti-Human Albumin cross-adsorbed antibody (Bethyl cat# A80-229 F) or AF488-Donkey-anti-human IgG, FC-gamma specific, AffiniPure F(ab’)2 fragment (Jackson ImmunoResearch cat#709-546-098) for an hour. Cells were washed twice, centrifuged, and cell pellets were resuspended in complete medium with 1% propidium iodide, filtered through a Nitex membrane, and analyzed by flow cytometry (BD-LSRII). Two thousand alive/single-cell events were acquired.
Data analysis
Specific mean fluorescent intensity (MFI) was calculated for each sample point by subtracting the MFI value of negative control (background). Binding curves (specific MFI versus linear or log variant concentration) were fitted with GraphPad Prism software.
HER2-mediated internalization
AlbuCORE and antibody variants were conjugated to AlexaFluor-488 (AF488, Invitrogen Molecular Probes, Eugene, OR). Conjugation, purification and determination of dye/antibody ratio were carried out according to manufacturer’s specifications.
Direct internalization
BT474, SKOV3, and MDA-MB-231 cells were seeded separately in 12-well plates (1 X 105 cells/well) and incubated at 37ºC, 18 h. The media was removed and replaced with 250 µL media + 10% FBS containing 200 nM AlexaFluor-conjugated variant, followed by incubation at 37°C, 18 h. The cells were washed twice with Dulbecco’s PBS (D-PBS), dissociated in 250 µL Cell Dissociation Solution (Sigma-Aldrich) at 37ºC, and then dispensed into two equal volumes. The cells were collected by centrifugation and resuspended in 100 uL medium +10% FBS with or without 50 µg/mL anti-AlexaFluor-488 quenching antibody (Molecular Probes A11094) and incubated at 4°C, 30 min. The samples were then diluted by adding 300 µL medium +10% FBS and 4 µL Propidium Iodide solution (Life Technologies), filtered (SEFAR NITEX 03–50/37, Sefar Inc.), and analyzed via flow cytometry. Relative amounts of surface and internal fluorescence were calculated according to Schmidt et al.Citation51 Quenching efficiency (QE) was determined by incubating cells with 200 nM of AF488-conjugated samples at 4°C and measuring the MFI in the presence or absence of quenching Ab.
QE = 1 – (MFI+quenching)/(MFI-quenching).
At 37°C
Surface MFI = (total unquenched MFI – quenched MFI)/QE.
Internal MFI = total unquenched MFI – surface MFI.
Dose response internalization via ImageXpress. BT474 cells were seeded in 96-well plates (2.5 X 103 cells/well) and incubated at 37°C, 18 h. AF488-labeled variants were added to cells in 3-fold, 8-point dilution series (0.06–200 nM), followed by incubation at 37°C, 18 h. The cells were washed thrice with D-PBS at room temperature, and then treated with Hoechst 33,342 stain (Invitrogen Molecular Probes) and Lysotracker Red DND-99 (Thermo Fisher Scientific) for 1 h. Imaging was performed using ImageXpress Micro XLS (Molecular Devices, San Jose, CA) with the following channels and exposure times; TRITC (250 ms), FITC (70 ms), and DAPI (70 ms), and using a 20X objective. Measurement of colocalization was performed using the Multi-wavelength Translocation (Transfluor) module of the ImageXpress software, which quantifies AF488 fluorescence signal from small, intracellular punctate objects (which represents internalized molecules). A correlation coefficient is calculated from the measured area and intensity of AF488 (FITC channel) and Lysotracker red (TRITC channel) signals. Internalization was measured as total integrated fluorescence intensity, normalized to nuclear count (FI/nucleus).
In vivo pharmacokinetics
B6.Cg-Albem12Mvw Fcgrt(tm1Dcr) Tg(FCGRT)32Dcr/MvwJ (Alb-/- Tg32, JAX stock # 025201) mice (24 F + 24 M) at 6– 11 weeks of age were distributed into 8 groups with 3 female and 3 male mice per group. At 0 h, test articles were administered to Alb-/- Tg32 mice as IV injections at two dose levels in dose volumes of 10 ml/Kg. Blood samples were collected from each mouse at 5 m, 1 h, 6 h, 1d, 3d, 5d, 7d, 14d, and 21d. The blood samples were processed to plasma and were assessed using a human albumin ELISA using polyclonal goat anti-human albumin (Sigma #A1151) and goat anti-human albumin-alkaline phosphatase (Bethyl Lab, A80-229AP) as capture and detection antibodies, respectively. PK parameters were calculated using noncompartmental analysis (Phoenix WinNonlin 6.4. Certara).
Bioconjugation
AlbuCORE and dimeric Fc constructs were conjugated to N2ʹ-deacetyl-N2ʹ-(3-mercapto-1-oxopropyl)-maytansine (DM1) via surface accessible lysines via the NHS-ester group on the heterobifunctional cross linker succinimidyl trans-4-[maleimidylmethyl] cyclohexane-1-carboxylate (SMCC). The linker was purchased with the payload DM1 attached via the free thiol to the maleimide group on the cross-linker (SMCC-DM1) (Levena Biopharma, USA), thus allowing conjugation in 1-step and avoiding issues with heterogeneity in payload attachment efficiency that can be observed with a 2-step process. The degree of payload attachment or DAR was controlled by the molar ratio of drug-linker:protein. SMCC-DM1 was solubilized as a working stock in dimethylacetamide (DMA) and added to protein in reaction buffer (100 mM potassium phosphate, 20 mM NaCl, 2 mM EDTA, pH 7.2) with a final DMA co-solvent concentration of 5% v/v to maintain reagent solubility during the reaction. Reaction mixtures were incubated at 25°C ±2°C in the dark for 12 h. After incubation, reactions were terminated and samples purified and formulated by passing samples 2 times through Zeba spin columns 7 K MWCO (Thermo Fisher Scientific) pre-equilibrated in formulation buffer (20 mM sodium succinate, 0.02% w/v Polysorbate-20, pH 5.5). Sample volumes were determined gravimetrically and 1/5 volume of 36% w/v Trehalose (Sigma, USA) added to a final concentration of 6% w/v.
Conjugated samples were analyzed by HPLC-SEC using a Superdex 200 Increase 10/300 GL column (GE Lifesciences, USA) to assess colloidal stability, determine the monomeric purity, and calculate DAR. Measurements of the integrated areas for monomeric peaks at both 280 nm and 252 nm were determined using Empower 2 software and used in the following equation to determine the molar concentration of conjugated species and DAR.
where the ADC specific values of absorbance or extinction coefficient are at 252 nm for DM1.
ADC cytotoxicity assays
Antibody variants were tested for their effects on viability on HER2 cell lines, including SKOV3, BT474, and HER2-negative Jurkat cells. Cells were seeded overnight at 200, 500, and 1500 cells/well for the three cell lines, respectively, in 384-well plates (Corning® 384 Well White Flat Bottom Polystyrene TC-Treated Microplates, Cat. # 3570). Cells were grown for 5 d in the presence of serial dilutions of DM1-conjugated test antibodies ranging from 100 nM to 0.0017 nM. After 5 d (37°C, 5% CO2, humidified incubator), the number of viable cells was determined using CellTiterGlo (Promega, Madison, WI), based on quantitation of the adenosine triphosphate (ATP) present in each well, which signals the presence of metabolically active cells. Signal output was measured on a luminescence plate reader (Envision, Perkin Elmer, Shelton, CT) set at an integration time of 0.1 s. Integration time is adjusted to minimize signal saturation at high ATP concentration. Dose-response curves of % Survival Fraction vs. Log10 of concentration were fit using GraphPad Prism 6.0 with a four-parameter logistic model to estimate IC50 and maximum efficacy.
Results
We used a structure-guided approach to engineer split sites in the HSA sequence with the goal that the two derived fragments would preferentially associate to form a hetero-dimeric, but quasi-native HSA molecule. A detailed description of the parameters that guided the in-silico engineering is given in the Materials and Methods section and in Supplementary Table 1. Based on the in-silico analysis and evaluation, we proposed and tested a set of eight split quasi-native HSA that satisfied our design goals. We refer to these split HSA molecules as AlbuCORE scaffolds. The corresponding model structures and cut site details are presented in and . Each scaffold is composed of two cognate albumin-derived segments. The majority of the designed AlbuCORE models contain one of the natural disulfide links of albumin crosslinking the two cognate segments.
Table 1. Description of the split Human Serum Albumin (AlbuCORE) scaffolds.
Production and biophysical characterization of AlbuCORE scaffolds
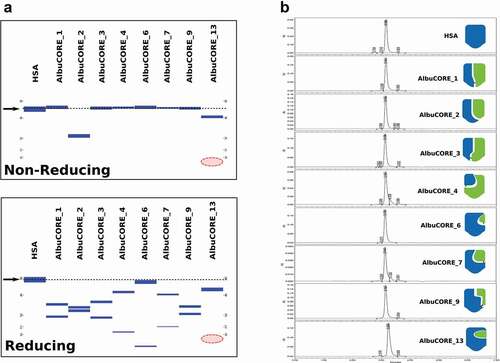
All of the AlbuCORE scaffolds, as well as the control wild-type HSA, were produced by transient expression in Chinese hamster ovary (CHO) cells. Each of the AlbuCORE scaffolds had the C34S mutation to remove the free cysteine naturally present in HSA and avoid potential cysteine oxidation issues during evaluation. Plasmids containing the nucleic acid sequence for each of the two segments comprising an AlbuCORE scaffold were co-transfected, with the two co-expressed proteins forming a heterodimeric pair, the quasi-native albumin structure. AlbuPURE™, an albumin-selective affinity chromatography absorbent resin, was used as the initial affinity step, followed by preparative size-exclusion chromatography (SEC). Capillary electrophoresis of the purified products showed that the majority of the scaffolds elute at the same molecular weight as intact HSA (~68 kDa) under non-reducing conditions (). The exceptions are AlbuCORE_2 and AlbuCORE_13, consistent with their lack of an interchain disulfide bond. Under reducing conditions, the two individual chains can be observed with the exception of AlbuCORE_13, which is missing its low molecular weight (MW) chain. This, along with the non-reducing electrophoresis data, indicates that in all AlbuCORE scaffolds, with the exception of AlbuCORE_13, the complementary segments are able to effectively pair and form a quasi-native albumin (AlbuCORE) molecule. All AlbuCORE scaffolds are homogenous and appear to be monodisperse, as indicated by analytical SEC (). AlbuCORE scaffolds 1–9 have the same elution profile and retention as HSA, including AlbuCORE_2, which lacks an interchain disulfide. AlbuCORE_13, however, has a lower apparent MW than HSA (see ), consistent with the capillary electrophoresis result.
Table 2. Biophysical characterization of HSA and AlbuCORE scaffolds.
Figure 2. Analysis of SEC purified HSA and the eight AlbuCORE scaffolds. A. Microfluidic Capillary Electrophoresis under non-reducing (top) and reducing (bottom) conditions. The migration of the intact HSA molecule is given as a reference point and is marked by an arrow and dotted line in both panels. The red-shaded area marks the region where the AlbuCORE_13 lower MW band (13.4KDa) was expected to elute but is not visible. B. UPLC-SEC chromatograms of all samples. The elution times were aligned for comparison, and the cartoon representation for each AlbuCORE construct is shown for easy identification. The actual elution time and calculated molecular weight for each of the UPLC-SEC runs is shown in
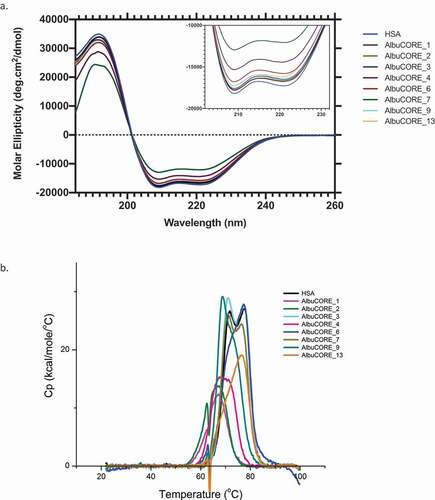
To validate the structural integrity of the AlbuCORE heterodimers, we compared the Far UV circular dichroism (CD) spectra of HSA with each of the AlbuCORE scaffolds, in particular the wavelength minima for the typical alpha-helical bands (, inset). There is a high degree of overlap among the HSA and the AlbuCORE spectra, with the exception of AlbuCORE_7, which shows a lower ellipticity signal at the expected alpha-helix range. Finally, we also compared the thermostability of each AlbuCORE construct with that of HSA using differential scanning calorimetry (DSC). Under these assay conditions, the HSA DSC thermograms can be deconvoluted into three transitions. The majority of the Tm1 transitions for the AlbuCORE constructs are within 5°C of HSA Tm1 (lowest transition temperature), as shown in . Exceptions are the AlbuCORE_1 and 2 constructs, with Tm1 reduced by about 10°C. We initially observed a high degree of variability in the transition melting temperatures among independent replicates for the recombinant HSA and AlbuCORE constructs. Reproducible thermal stability results were obtained by adding excess fatty acid (20 molar equivalents octanoic acid) and heat treating (simulating pasteurization) the samples prior to DSC measurements.Citation52,Citation53 DSC thermograms of the untreated samples can be seen in Supplementary Figure 2.
Figure 3. A. Far UV CD spectra (a) and DSC thermograms (b) of HSA and the eight AlbuCORE scaffolds. A. The inset shows a zoomed in view of the characteristic CD alpha-helix double bands between 200–230 nm. B. DSC thermograms for samples treated with 20 molar equivalents of sodium octanoate at 60°C O/N. The first transition at lower temperature (Tm1) was used to compare each of the AlbuCORE scaffolds with HSA, as reported in
Binding of HSA or AlbuCORE to human FcRn at pH 5.5 was determined by biolayer interferometry (BLI) and the results are shown in and Supplementary Figure 4. Seven of the eight AlbuCORE scaffolds bind to FcRn with affinities comparable to HSA, with AlbuCORE_13 being the only exception. The reported equilibrium KDs is very closely matched by the KDs derived from kinetic fitting (data not shown).
Influence of geometry and valence on affinity and avidity
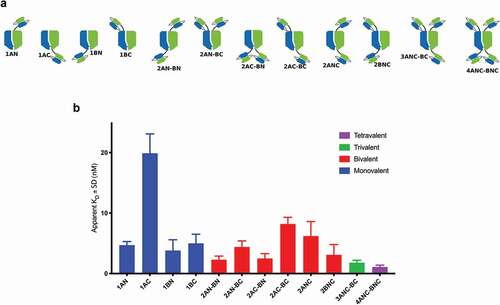
In order to assess the influence of valency, fusion point, and overall geometrical relationship between paratopes, we generated a series of fusion constructs with the anti-HER2 scFv B1D2Citation54,Citation55 and the AlbuCORE_1 scaffold, as shown in . Binding affinities were determined by fluorescence-activated cell sorting (FACS) using the HER2-positive SKOV3 cell line. shows apparent KD for each of the constructs tested. The monovalent fusion 1AC shows the lowest KD in the series, 4-5-fold lower than the other three monovalent fusions. The tetravalent construct 4ANC-BNC had the highest affinity in the series. The bivalent constructs show a distribution of KDs with a ~3.5-fold spread among them, depending on the specific arrangement of the scFv paratope.
Figure 4. FACS binding of anti-HER2 scFv-fused AlbuCORE_1 constructs to HER2 positive cell line SKOV3. The constructs depicted in the top panel were devised to demonstrate the influence of valency, geometry, and fusion site on binding affinity. Affinities are depicted as bar graphs for each construct, colored according to valency. The constructs are named according to the valency (numbers 1 through 4 representing the number of scFv moieties) and according to the chain and termini used (chains A and B, N or C termini). For example, the construct 2AC-BN contains two scFv fusions (bivalent), one to chain A, C-terminal and another one to chain B, N-terminal.
Construction and characterization of anti-HER2 Biparatopic AlbuCORE fusions
We built targeted fusion molecules using two quasi-native AlbuCORE scaffolds for further functional testing. AlbuCORE_3 and AlbuCORE_6 scaffolds were chosen based on their desirable thermostability and distinct spatial organization of their termini, and were fused to two unique HER2-binding modules as shown in . A single pertuzumab antigen-binding fragment (Fab) (2C4)Citation56 and a single trastuzumab scFv (4D5)Citation57 were N-terminally fused to either AlbuCORE_3 or AlbuCORE_6 to generate bispecific (biparatopic) species termed TP-AlbuCORE_3 and TP-AlbuCORE_6. An equivalent construct using a heterodimeric IgG1 Fc was used as a control.Citation58 Because pertuzumab and trastuzumab have non-overlapping epitopes in the same target antigen (HER2), both arms of these bispecific constructs are, in principle, capable of co-engaging the receptor.
Figure 5. Internalization properties of AlbuCORE and antibody Fc fusions A. Cartoon depiction of Trastuzumab-Pertuzumab (TP) bispecific fusions with AlbuCORE and Azymetric Fc. B. Direct Internalization of AF488-labeled bispecific fusions and HSA at 200 nM in two HER positive cell lines (BT-474 and SKOV3) and a triple negative (HER2 negative) cell line (MDA-MB-231) as determined by flow cytometry. The number in parenthesis in the x-axis indicate the HER2 expression levels. C. Orthogonal method showing dose response internalization of AF488-labeled samples of bispecific, one-armed (OA-) monospecific, and unfused constructs in HER2 positive cell line BT-474, as determined using ImageXpress detection and high content imaging. The one-armed (OA) fusions were designed to preserve the format used in the bispecific fusions: scFv for Trastuzumab (OA-T) and Fab for Pertuzumab (OA-P).
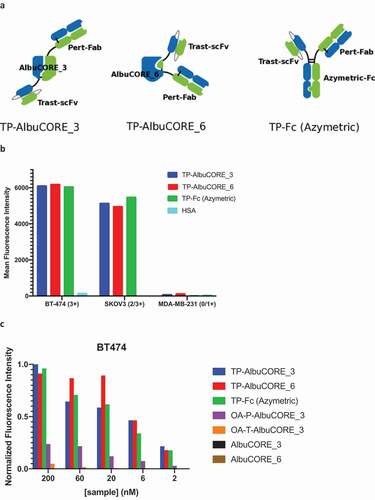
Samples were efficiently expressed and purified according to the previously established protocol and retained the favorable monodispersity and thermostability of the unfused controls (Supplementary Figure 1). HER2 binding was determined by surface plasmon resonance (SPR) and FACS experiments, as shown in . The Fc-based and the AlbuCORE-based biparatopic constructs show small affinity differences by SPR. All biparatopic constructs have comparable affinities as determined by FACS on HER2-positive cell lines. HSA, a negative control for HER2 binding, did not show any detectable binding.
Table 3. In vitro HER2-binding affinities derived by SPR and FACS.
We also measured the HER2-mediated internalization of a panel of constructs, as shown in and . All three biparatopic constructs, including the control Fc-based design, show comparable internalization in the two HER2-positive cell lines (), without significant internalization in a HER2-negative cell line. HSA does not exhibit appreciable internalization in any of the three cell lines at the concentration tested. Dose response internalization experiments in a HER2-positive cell line () confirmed that the biparatopic fusion constructs display comparable concentration-dependent internalization. As expected, the one-armed AlbuCORE_3 constructs, fused to either trastuzumab (OA-T-AlbuCORE_3) or pertuzumab (OA-P-AlbuCORE_3), showed greatly reduced internalization. The untargeted (unfused) AlbuCORE scaffolds did not show any significant internalization under these experimental conditions.
Pharmacokinetic properties of AlbuCORE constructs
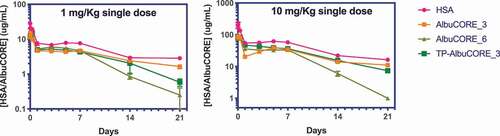
The biophysical characterization and cell-based functional assessment of AlbuCORE scaffolds indicated we could generate agents that were stable and biologically active in vitro. In order to determine if the engineered AlbuCORE retains the favorable attributes of HSA in vivo, its PK properties were evaluated in Tg32/AlbKO mice.Citation59 These mice carry both a human FcRn knock-in (mouse FcRn null) and a mouse albumin knock out, and have been demonstrated to accurately model the PK of human albumin. Following administration of a single intravenous (IV) dose of 1 or 10 mg/Kg, AlbuCORE_3 and anti-HER2 TP-AlbuCORE_3 biparatopic had largely similar PK profiles (t1/2, clearance, and AUC) to HSA ( & ). AlbuCORE_6 at both dose levels tested appeared to have a reduced half-life compared to HSA. The PK for all test articles was approximately dose proportional between the 1 and 10 mg/Kg doses.
Table 4. PK parameters derived from the PK profiles shown in . The values in parenthesis represent the standard error of the mean.
AlbuCORE drug conjugation
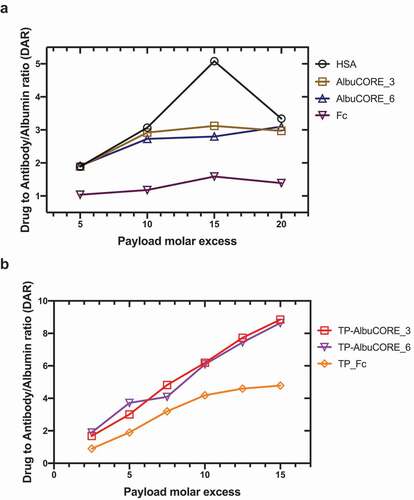
Antibody-drug conjugates (ADCs) are showing increasing clinical impact. In an effort to understand the relative ability of AlbuCORE to conjugate to hydrophobic small molecules, we conjugated the maytansinoid cytotoxic agent DM1 via the heterobifunctional SMCC linker to lysine residues of HSA, AlbuCORE, and IgG1 Fc.Citation60 Under similar conjugation conditions, the drug to albumin or antibody ratio (DAR) is about 2-fold higher for HSA or the AlbuCORE constructs relative to an IgG1 Fc lacking a Fab or scFv at the N-terminus of the hinge (). Further, conjugating to the HER2-binding biparatopic TP-AlbuCORE constructs previously described, we observe that the molecules conjugate to a DAR of approximately 9 with linear conjugation efficiency under the conditions we used (). On the other hand, a similar biparatopic construct with an antibody Fc saturates at a DAR of approximately 4.5 under the same conditions. We were able to confirm that the binding affinity for the target HER2 as determined by SPR was maintained regardless of DAR in these constructs (data not shown). Also, the drug conjugated molecules are well behaved as observed in analytical high-performance liquid chromatography (HPLC)-SEC, without significant changes in aggregation propensity with higher DAR, evaluated as the percentage of monomeric species by HPLC-SEC (Supplementary Table 8).
Figure 7. Stochastic conjugation of DM1 to lysine residues. Drug to protein (antibody, Albumin or AlbuCORE) ratio achieved using different molar ratio of the linker drug (SMCC-DM1) relative to the protein. DARs determined using spectroscopic techniques. A Conjugation of the scaffolds Albumin, AlbuCORE or homodimeric IgG1 Fc fragment, all constructs lacking fused paratopes (scFv or Fab units). See Figure 2A for cartoon representation of the scaffolds. B Conjugation of the anti-HER2 biparatopic AlbuCORE or Fc-based constructs (Trastuzumab scFv and Pertuzumab Fab, TP-), as shown in Figure 5A.
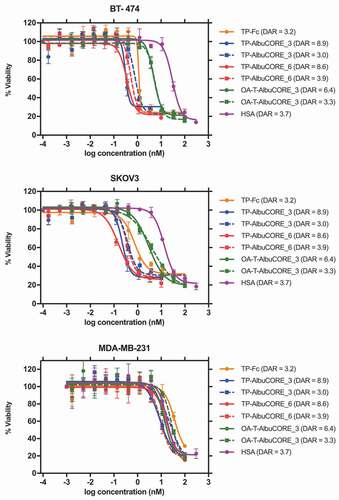
Cytotoxicity screen of these drug conjugated molecules in multiple cell lines with diverse expression levels of HER2 showed that the TP-AlbuCORE biparatopics have similar efficacy to the equivalent Fc-based construct when the drug conjugation levels are comparable ( and Supplementary Table 9). TP-AlbuCORE variants with higher DARs appear to display a small increase in cytotoxic potency. Importantly, the AlbuCORE variants do not exhibit off-target effects in cell lines that do not over-express HER2 (MDA-MB-231, HUVEC, WI-38), with no significant difference in the IC50 values for all variants irrespective of the drug conjugation ratio.
Figure 8. Cytotoxicity profile of DM1-conjugated IgG1 Antibody or AlbuCORE constructs (see Figure 5A) targeting HER2 in three different cell lines SKOV3 (HER2 high), BT-474 (HER2 high) and MDA-MB-231 (HER2 negative). DAR values are indicated in parenthesis in the figure legend. For comparison reasons, a sample with DAR ~ 3 was tested for each one of the constructs. In addition, for each AlbuCORE construct the sample with the highest DAR obtained was also tested. The drug conjugated HSA (untargeted) is presented as a control. OA-T-AlbuCORE_3 is the one-armed version of AlbuCORE_3 fused to a Trastuzumab scFv.
Discussion
The primary goal of this engineering effort was to create a series of quasi-native split HSA molecules to allow design of novel albumin fusion molecules (). The creation of this self-complementing pair by splitting albumin results in the formation of a novel but natural heterodimeric protein pair with very high complementary selectivity between the segments upon co-expression. A number of these new self-complementing split albumin scaffolds, which we refer to as AlbuCORE, retain intact HSA’s native properties, as judged by thermostability, secondary structure content, FcRn binding, and purification profile. The AlbuCORE constructs introduced approximately 3000 Å2 of buried surface area at the interface of the two split segments (supplementary Table 1), well in the range of 1910– 3880 Å2 observed in a number of other natural cognate protein-protein heterodimeric and homodimeric co-complex structures.Citation61 To the best of our knowledge, this is the first time that albumin has been split to generate new termini and new geometries for molecular design. Heaney-Kieras and King have shown that limited pepsin digestion of HSA caused cleavage at residue 307 of the albumin molecule to yield two fragments which remain associated quite strongly when studied using chromatography,Citation62 but that has never been explored to produce a split scaffold. HSA’s α-helix rich structure with a number of interconnected linkers provides a great opportunity for pursuing such designs. By splitting on different loops around the natural albumin structure, the geometrical relationship between the natural N- and C-termini and the new N’- and C’-termini changes, allowing for exploration of different fusion geometries.
AlbuCORE modules retain wild-type albumin biophysical properties
A primary requirement of such engineered scaffolds was that they retain the natural biophysical properties of the HSA both in vitro and in vivo. All eight constructs were produced and purified using the same standard techniques used for wild-type recombinant HSA. We have demonstrated that the majority of the AlbuCORE constructs associated into an HSA-like heterodimer and retained HSA’s monodispersity (UPLC-SEC, ), secondary structure (CD, ), thermostability (DSC, ), and FcRn binding (BLI, ). One of the eight AlbuCORE constructs tested, AlbuCORE_13, did not stably assemble and was the only exception. Overall, given that the AlbuCORE scaffolds purified similarly to HSA using the AlbuPureTM resin, the manufacturability of the AlbuCORE scaffolds and its feasibility as a pharmaceutical therapeutic platform is strengthened.
All of the split loops in the proposed AlbuCORE scaffolds connect two helical elements. If a particular split destabilizes the adjacent α-helices, it could cause them to partially unfold, likely reflecting in lower ellipticity in a CD spectra in comparison with the native structure, as observed for AlbuCORE_7. This putative loss of secondary structure does not seem to negatively affect this scaffold as judged by the other tested biophysical parameters. All other AlbuCORE scaffolds tested showed CD spectra indistinguishable from HSA, further indicating their quasi-native albumin structure. For AlbuCORE_13, loss of the N-terminal domain does not significantly change its calculated α-helical content when compared to the full-length HSA, which explains the lack of change in molar ellipticity. Surprisingly, the remaining fragment gives rise to a relatively stable construct and elutes in a single peak by UPLC-SEC, besides the change in retention time. AlbuCORE_13 lacks an interchain disulfide crosslinking the two chains, and the N-terminal fragment appears to be either lost during purification or not expressed. While the presence of an interchain disulfide most likely contributes to the overall stability of the construct, it does not seem to be strictly necessary for a quasi-native association. AlbuCORE_2, the other construct that lacks an interchain disulfide, associates into a stable heterodimer. The structure of HSA comprises 17 native disulfide bonds, which lends itself to the design of such quasi-native species with interchain covalent links ().
The presence of an interchain disulfide alone does not seem to correlate well with thermostability and is most likely a function of the stability of the newly formed interface (, ). For example, AlbuCORE_13 (with no interchain disulfide and loss of N-terminal chain) has a Tm1 transition within 4°C of HSA, while AlbuCORE_1 (with an interchain disulfide) has a Tm1 loss of ~10°C compared to HSA. The majority of the AlbuCORE constructs show transition temperatures within 5°C of HSA. It is possible that the lower melting temperatures for AlbuCORE_1 and AlbuCORE_2 are due to overall instability of those constructs rather than altered fatty acid-binding properties of the C-terminal domains.Citation53 This is evidenced by the decreased thermostability of all transitions, not only the Tm1 associated with fatty acid binding.
Binding of HSA to FcRn is a pH-dependent process that significantly contributes to the extended half-life of albumin in vivo.Citation6 Binding of HSA to human FcRn at pH <6 has been previously reported with KD in the order of ~0.5 μM.Citation55 Equilibrium KD determined in the present study for both serum-derived and recombinant HSA showed comparable binding affinity of 0.89 μM and 0.68 μM, respectively (). The lower affinity for AlbuCORE_13 likely stems from the loss of the N-terminal domain. All other AlbuCORE scaffolds bind FcRn with affinities comparable to HSA.
Affinity and avidity are geometry- and valence-dependent
In order to expand our understanding of parameters that influence fusion molecules, we designed a series of fusion constructs between the anti-HER2 scFv B1D2Citation54 and the AlbuCORE_1 scaffold as a representative ( top panel). Parameters tested with these constructs include the number of binding epitopes (valency), the fusion point (each one of the four termini), the orientation of the epitope (N- or C-terminal fusion), and the spatial distribution of the paratopes. shows an overall trend of increased affinities with increased valency, indicating some degree of additivity among the paratopes. As expected, the tetravalent construct 4ANC-BNC has the highest affinity in the series, ~3.5-fold higher than the best monovalent construct 1AN, followed by the trivalent 3ANC-BC and the bivalent 2AN-BN. Beyond valency, the orientation of the scFv can have a significant impact on the observed apparent KD, as demonstrated by the monovalent constructs. Fusion to the C’-terminal (construct 1AC) causes up to 5-fold reduction in apparent KD over the other three monovalent fusions, probably due to occlusion of target binding in that particular orientation. This is in line with our initial hypothesis that not all C-terminal fusions will be viable, being highly dependent on the paratopes and targeted epitopes.
The spatial distribution of the epitopes in the bivalent fusions are particularly revealing when it comes to the unpredictability of the avidity effect. Contrary to expectations, the affinity of the construct 2AC-BC is not hindered by using the lowest affinity monovalent binder 1AC fusion, and is in fact one of the best binders among the bivalent constructs. This demonstrated that the avidity effect, brought about by a particular spatial relationship between the paratopes and their epitopes, can overpower the simple additive effect of individual monomeric affinities.
Anti-HER2 biparatopic AlbuCORE fusions are functional in vitro
The AlbuCORE heterodimers with four termini enable the design of multimeric fusion molecules, including bispecific antibody-like constructs with distinct relative orientation of the cognate arms (). As a proof of concept, we generated bispecific anti-HER2 AlbuCORE constructs fused to paratopes derived from pertuzumab (P) and trastuzumab (T) (). The biparatopic AlbuCORE (TP-AlbuCORE) retains the general properties of a wild-type HSA molecule, including purification properties and monodispersity (Supplementary Figure 1), as well as thermostability (Supplementary Figure 3). HER2-binding affinities for the biparatopic AlbuCORE match the Fc-based control ().
With regard to functional characteristics, we have shown that the TP-AlbuCORE_3 and TP-AlbuCORE_6 show comparable, dose-dependent internalization to the biparatopic TP-Fc construct tested (). We have previously observed that biparatopic TP-Fc molecules have increased HER2-mediated internalization when compared to mono-specific versions of these molecules. As a consequence, these TP-Fc molecules promote higher tumor growth inhibition as a naked construct, and higher cytotoxicity as drug conjugates, when compared with their monoparatopic equivalents.Citation36 We hypothesize that it is the biparatopic molecule’s ability to bind in trans and form daisy-chain, crosslinked clusters of receptors that promote higher HER2 internalization, leading to receptor down-regulation and tumor growth inhibition as reported previously.Citation63–Citation65 A priori, internalization of HER2 by TP-AlbuCORE constructs could differ from that of TP-Fc, given the putative differences in orientation and conformation between anti-HER2 paratopes among these constructs. As expected, one-armed constructs, unfused AlbuCORE scaffolds or HSA, showed little to no internalization. It demonstrates that the biparatopic AlbuCORE fusions retained the functional properties of the biparatopic Fc construct (internalization, trans binding) as well as their affinity and biophysical properties.
PK Characteristics of AlbuCORE
Evaluating HSA PK in preclinical mouse models is not straightforward due to three significant species differences: (1) Mice and humans have a very different turnover rate of native albumin, with respective half-lives of 35 h and 19–21 d,Citation59 and the short half-life of albumin in mice make assessments of stability and PK unreliable; (2) Human albumin has a lower affinity to murine FcRn compared to murine albumin, and, as a consequence, HSA t1/2 in mice is shorter than mouse albumin (21 and 35 h, respectively), which further complicates PK assessments;Citation6,Citation43 and (3) Mouse serum albumin has a high affinity to human FcRn.Citation43 While HSA t1/2 is improved in transgenic human FcRn mice, the t1/2 remains lower than expected due to competition with native mouse albumin. As a result of these complications the PK evaluation of AlbuCORE could not be fairly assessed in conventional mouse strains and, therefore, we utilized the Tg32/AlbKO.Citation59 The PK analysis in these mice demonstrates that the AlbuCORE_3 scaffold can be modified as a bispecific biologic without sacrificing the long-circulating half-life of native albumin ( and ). However, the AlbuCORE_6 scaffold showed a reduced half-life compared to AlbuCORE_3 and HSA. It remains unclear why AlbuCORE_6 clearance appears to be faster than HSA, since both HSA and AlbuCORE_6 bind to human FcRn with comparable equilibrium KDs.
AlbuCORE as a Small Molecule Drug Carrier
Given the natural physiological role of albumin as a carrier of small hydrophobic molecules, there is an opportunity to consider the application of the albumin scaffold for drug delivery in a therapeutic setting. Crystallographic studies have revealed as many as 11 fatty acid binding sites in albumin.Citation66 At 585 residues in length, native albumin is about 25% longer than the 464 residues in the IgG1 Fc homodimer sequence (including two copies of the hinge, CH2, and CH3 domains), but it is interesting that we were able to conjugate about 2-fold higher on albumin or the AlbuCORE constructs relative to IgG1 Fc () without noticeably altering the biophysical properties of the drug-conjugated protein. As one may expect, cytotoxicity analysis with these drug-conjugated molecules suggests a drug-load dependent potency of the molecules (). The benefit of this approach has been demonstrated by Zhang et al. who have shown that a drug-conjugated HER2-binding scFv fused to albumin induces strong and rapid tumor tissue penetrability and distribution compared to trastuzumab in animal models.Citation67 The albumin-based scaffold we have presented here adds an additional layer of flexibility through its ability to fine tune target therapeutic response based on valency, geometry of binding, and stoichiometry of drug loading.
Final remarks
We demonstrated that the human serum albumin sequence can be split so as to generate a self-complementary pair of heterodimeric protein that upon co-expression forms a quasi-native albumin construct. Five of the tested split-HSA scaffolds retain HSA’s biophysical properties, with native-like purification, monodispersity, folding, thermostability, and FcRn binding affinity. Testing two of these scaffolds in a mouse model showed that the AlbuCORE can retain recombinant wild-type HSA-like PK properties. Evaluating multiple variants with distinct valency and geometries, we showed that the effect of these parameters on avidity should be determined empirically. The availability of such an albumin-based scaffold opens up the opportunity for design of novel multi-specific multi-functional biologics in order to explore complex but rational drug design paradigms for a number of disease conditions.
Disclosure of interests
MS, SL, GW, NW, DP, and SD are employees of Zymeworks, Inc. ID and GN were Zymeworks’s employees at the time this work was conducted. Research conducted at the National Research Council of Canada was funded by Zymeworks, Inc.
Supplemental Material
Download Zip (7.3 MB)Acknowledgments
We thank Yves Durocher, Alaka Mullick, Anna Moraitis, Karine Brault, Brian Cass, Louis Bisson, Sylvie Perret, Roseanne Tom, Nancy Remillard, Cassio Baptista, Richard Gingras, Marie-Joseé Massariol, Martine Pagé, Florence Do for support in production; Allan Matte, Anna Cunningham, Marie-France Goneau for purification support; The-Minh Tu for QC analytics support; Suzanne Grothe for SPR; Myriam Banville, Julie Lippens, and Luc Meury for support with flow cytometry binding and functional assays. We thank Normand Joliceour for help with bioconjugation activities. We thank Anthony Polvorino for his comments on the manuscript.
Supplementary material
Supplemental data for this article can be accessed on the publisher’s website.
References
- Peters T Jr., Stewart AJ. Albumin research in the 21st century. Biochim Biophys Acta. 2013;1830(12):5351–17. doi:https://doi.org/10.1016/j.bbagen.2013.05.012.
- Fanali G, Di Masi A, Trezza V, Marino M, Fasano M, Ascenzi P. Human serum albumin: from bench to bedside. Mol Aspects Med. 2012;33(3):209–90. doi:https://doi.org/10.1016/j.mam.2011.12.002.
- Merlot AM, Kalinowski DS, Richardson DR. Unraveling the mysteries of serum albumin-more than just a serum protein. Front Physiol. 2014;5:299. doi:https://doi.org/10.3389/fphys.2014.00299.
- Liu H, Moynihan KD, Zheng Y, Szeto GL, Li AV, Huang B, Van Egeren DS, Park C, Irvine DJ. Structure-based programming of lymph-node targeting in molecular vaccines. Nature. 2014;507(7493):519–22. doi:https://doi.org/10.1038/nature12978.
- Chen N, Brachmann C, Liu X, Pierce DW, Dey J, Kerwin WS, Li Y, Zhou S, Hou S, Carleton M, et al. Albumin-bound nanoparticle (nab) paclitaxel exhibits enhanced paclitaxel tissue distribution and tumor penetration. Cancer Chemother Pharmacol. 2015;76(4):699–712. doi:https://doi.org/10.1007/s00280-015-2833-5.
- Chaudhury C, Mehnaz S, Robinson JM, Hayton WL, Pearl DK, Roopenian DC, Anderson CL. The major histocompatibility complex–related Fc receptor for IgG (FcRn) binds Albumin and prolongs its lifespan. J Exp Med. 2003;197(3):315–22. doi:https://doi.org/10.1084/jem.20021829.
- Wang Y, Lang L, Huang P, Wang Z, Jacobson O, Kiesewetter DO, Ali IU, Teng G, Niu G, Chen X, et al. In vivo albumin labeling and lymphatic imaging. Proc Natl Acad Sci U S A. 2015;112(1):208–13. doi:https://doi.org/10.1073/pnas.1414821112.
- Peek MC, Charalampoudis P, Anninga B, Baker R, Douek M. Blue dye for identification of sentinel nodes in breast cancer and malignant melanoma: a systematic review and meta-analysis. Future Oncology. 2017;13(5):455–67. doi:https://doi.org/10.2217/fon-2016-0255.
- Ma WW, Hidalgo M. The winning formulation: the development of paclitaxel in pancreatic cancer. Clin Cancer Res. 2013;19(20):5572–79. doi:https://doi.org/10.1158/1078-0432.CCR-13-1356.
- Maeda H, Matsumura Y. EPR effect based drug design and clinical outlook for enhanced cancer chemotherapy. Adv Drug Deliv Rev. 2011;63(3):129–30. doi:https://doi.org/10.1016/j.addr.2010.05.001.
- Porporato PE. Understanding cachexia as a cancer metabolism syndrome. Oncogenesis. 2016;5:e200. doi:https://doi.org/10.1038/oncsis.2016.3.
- Stehle G, Sinn H, Wunder A, Schrenk HH, Stewart JCM, Hartung G, Maier-Borst W, Heene DL. Plasma protein (albumin) catabolism by the tumor itself–implications for tumor metabolism and the genesis of cachexia. Crit Rev Oncol Hematol. 1997;26(2):77–100. doi:https://doi.org/10.1016/S1040-8428(97)00015-2.
- Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144(5):646–74. doi:https://doi.org/10.1016/j.cell.2011.02.013.
- Commisso C, Davidson SM, Soydaner-Azeloglu RG, Parker SJ, Kamphorst JJ, Hackett S, Grabocka E, Nofal M, Drebin JA, Thompson CB, et al. Macropinocytosis of protein is an amino acid supply route in Ras-transformed cells. Nature. 2013;497(7451):633–37. doi:https://doi.org/10.1038/nature12138.
- Davidson SM, Jonas O, Keibler MA, Hou HW, Luengo A, Mayers JR, Wyckoff J, Del Rosario AM, Whitman M, Chin CR, et al. Direct evidence for cancer-cell-autonomous extracellular protein catabolism in pancreatic tumors. Nat Med. 2017;23(2):235–41. doi:https://doi.org/10.1038/nm.4256.
- Swiercz R, Mo M, Khare P, Schneider Z, Ober RJ, Ward ES. Loss of expression of the recycling receptor, FcRn, promotes tumor cell growth by increasing albumin consumption. Oncotarget. 2017;8(2):3528–41. doi:https://doi.org/10.18632/oncotarget.13869.
- Chlenski A, Cohn SL. Modulation of matrix remodeling by SPARC in neoplastic progression. Semin Cell Dev Biol. 2010;21(1):55–65. doi:https://doi.org/10.1016/j.semcdb.2009.11.018.
- Yu IS, Cheung WY. A contemporary review of the treatment landscape and the role of predictive and prognostic biomarkers in pancreatic adenocarcinoma. Can J Gastroenterol Hepatol. 2018;2018:1863535. doi:https://doi.org/10.1155/2018/1863535.
- Chatterjee M, Ben-Josef E, Robb R, Vedaie M, Seum S, Thirumoorthy K, Palanichamy K, Harbrecht M, Chakravarti A, Williams TM, et al. Caveolae-mediated endocytosis is critical for albumin cellular uptake and response to albumin-bound chemotherapy. Cancer Res. 2017;77(21):5925–37. doi:https://doi.org/10.1158/0008-5472.CAN-17-0604.
- Lu RM, Hwang YC, Liu IJ, Lee CC, Tsai HZ, Li HJ, Wu HC. Development of therapeutic antibodies for the treatment of diseases. J Biomed Sci. 2020;27(1):1. doi:https://doi.org/10.1186/s12929-019-0592-z.
- Jain RK. Barriers to drug delivery in solid tumors. Sci Am. 1994;271(1):58–65. doi:https://doi.org/10.1038/scientificamerican0794-58.
- Beckman RA, Weiner LM, Davis HM. Antibody constructs in cancer therapy: protein engineering strategies to improve exposure in solid tumors. Cancer. 2007;109(2):170–79. doi:https://doi.org/10.1002/cncr.22402.
- Thurber GM, Schmidt MM, Wittrup KD. Factors determining antibody distribution in tumors. Trends Pharmacol Sci. 2008;29(2):57–61. doi:https://doi.org/10.1016/j.tips.2007.11.004.
- Choi IK, Strauss R, Richter M, Yun C-O, Lieber A. Strategies to increase drug penetration in solid tumors. Front Oncol. 2013;3:193. doi:https://doi.org/10.3389/fonc.2013.00193.
- Kintzing JR, Filsinger Interrante MV, Cochran JR. Emerging strategies for developing next-generation protein therapeutics for cancer treatment. Trends Pharmacol Sci. 2016;37(12):993–1008. doi:https://doi.org/10.1016/j.tips.2016.10.005.
- Sand KM, Bern M, Nilsen J, Noordzij HT, Sandlie I, Andersen JT. Unraveling the Interaction between FcRn and Albumin: opportunities for design of Albumin-based therapeutics. Front Immunol. 2014;5:682.
- Dennis MS, Jin H, Dugger D, Yang R, McFarland L, Ogasawara A, Williams S, Cole MJ, Ross S, Schwall R, et al. Imaging tumors with an albumin-binding Fab, a novel tumor-targeting agent. Cancer Res. 2007;67(1):254–61. doi:https://doi.org/10.1158/0008-5472.CAN-06-2531.
- Escobar-Cabrera E, Lario P, Baardsnes J, Schrag J, Durocher Y, Dixit S. Asymmetric Fc engineering for bispecific antibodies with reduced effector function. Antibodies. 2017;6(2):7. doi:https://doi.org/10.3390/antib6020007.
- Tam SH, McCarthy SG, Armstrong AA, Somani S, Wu SJ, Liu X, Gervais A, Ernst R, Saro D, Decker R, et al. Functional, biophysical, and structural characterization of human IgG1 and IgG4 Fc variants with ablated immune functionality. Antibodies. 2017;6(3):12.
- Almagro JC, Daniels-Wells TR, Perez-Tapia SM, Penichet ML. Progress and challenges in the design and clinical development of antibodies for cancer therapy. Front Immunol. 2018;8:1751.
- Brinkmann U, Kontermann RE. The making of bispecific antibodies. MAbs. 2017;9(2):182–212. doi:https://doi.org/10.1080/19420862.2016.1268307.
- Spangler JB, Manzari MT, Rosalia EK, Chen TF, Wittrup KD. Triepitopic antibody fusions inhibit cetuximab-resistant BRAF and KRAS mutant tumors via EGFR signal repression. J Mol Biol. 2012;422(4):532–44. doi:https://doi.org/10.1016/j.jmb.2012.06.014.
- Li JY, Perry SR, Muniz-Medina V, Wang X, Wetzel LK, Rebelatto MC, Hinrichs MJM, Bezabeh BZ, Fleming RL, Dimasi N, et al. A biparatopic HER2-targeting antibody-drug conjugate induces tumor regression in primary models refractory to or ineligible for HER2-targeted therapy. Cancer Cell. 2016;29(1):117–29. doi:https://doi.org/10.1016/j.ccell.2015.12.008.
- Jost C, Schilling J, Tamaskovic R, Schwill M, Honegger A, Plückthun A. Structural basis for eliciting a cytotoxic effect in HER2-overexpressing cancer cells via binding to the extracellular domain of HER2. Structure. 2013;21(11):1979–91. doi:https://doi.org/10.1016/j.str.2013.08.020.
- Tamaskovic R, Schwill M, Nagy-Davidescu G, Jost C, Schaefer DC, Verdurmen WPR, Schaefer JV, Honegger A, Plückthun A. Intermolecular biparatopic trapping of ErbB2 prevents compensatory activation of PI3K/AKT via RAS–p110 crosstalk. Nat Commun. 2016;7(1):11672. doi:https://doi.org/10.1038/ncomms11672.
- Weisser N, Wickman G, Davies R, Rowse G. Abstract 31: preclinical development of a novel biparatopic HER2 antibody with activity in low to high HER2 expressing cancers. Cancer Res. 2017;77(13 Supplement):31–31. doi:https://doi.org/10.1158/1538-7445.AM2017-31
- de Miguel D, Lemke J, Anel A, Walczak H, Martinez-Lostao L. Onto better trails for cancer treatment. Cell Death Differ. 2016;23(5):733–47. doi:https://doi.org/10.1038/cdd.2015.174.
- McDonagh CF, Huhalov A, Harms BD, Adams S, Paragas V, Oyama S, Zhang B, Luus L, Overland R, Nguyen S, et al. Antitumor activity of a novel bispecific antibody that targets the ErbB2/ErbB3 oncogenic unit and inhibits heregulin-induced activation of ErbB3. Mol Cancer Ther. 2012;11(3):582–93. doi:https://doi.org/10.1158/1535-7163.MCT-11-0820.
- Nelson DR, Benhamou Y, Chuang WL, Lawitz EJ, Rodriguez–Torres M, Flisiak R, Rasenack JWF, Kryczka W, Lee C, Bain VG, et al. Albinterferon Alfa-2b was not inferior to pegylated interferon-alpha in a randomized trial of patients with chronic hepatitis C virus genotype 2 or 3. Gastroenterology. 2010;139(4):1267–76. doi:https://doi.org/10.1053/j.gastro.2010.06.062.
- Zeuzem S, Sulkowski MS, Lawitz EJ, Rustgi VK, Rodriguez–Torres M, Bacon BR, Grigorescu M, Tice AD, Lurie Y, Cianciara J, et al. Albinterferon Alfa-2b was not inferior to pegylated interferon-alpha in a randomized trial of patients with chronic hepatitis C virus genotype 1. Gastroenterol. 2010;139(4):1257–66. doi:https://doi.org/10.1053/j.gastro.2010.06.066.
- Matthews JE, Stewart MW, De Boever EH, Dobbins RL, Hodge RJ, Walker SE, Holland MC, Bush MA. Pharmacodynamics, pharmacokinetics, safety, and tolerability of Albiglutide, a long-acting glucagon-like peptide-1 mimetic, in patients with Type 2 diabetes. J Clin Endocrinol Metab. 2008;93(12):4810–17. doi:https://doi.org/10.1210/jc.2008-1518.
- Oganesyan V, Damschroder MM, Cook KE, Li Q, Gao C, Wu H, Dall’Acqua WF. Structural insights into neonatal Fc receptor-based recycling mechanisms. J Biol Chem. 2014;289(11):7812–24. doi:https://doi.org/10.1074/jbc.M113.537563.
- Andersen JT, Dalhus B, Viuff D, Ravn BT, Gunnarsen KS, Plumridge A, Bunting K, Antunes F, Williamson R, Athwal S, et al. Extending serum half-life of albumin by engineering neonatal Fc receptor (FcRn) binding. J Biol Chem. 2014;289(19):13492–502. doi:https://doi.org/10.1074/jbc.M114.549832.
- Hein KL, Kragh-Hansen U, Morth JP, Jeppesen MD, Otzen D, Møller JV, Nissen P. Crystallographic analysis reveals a unique lidocaine binding site on human serum albumin. J Struct Biol. 2010;171(3):353–60. doi:https://doi.org/10.1016/j.jsb.2010.03.014.
- Kollman PA, Massova I, Reyes C, Kuhn B, Huo S, Chong L, Lee M, Lee T, Duan Y, Wang W, et al. Calculating structures and free energies of complex molecules: combining molecular mechanics and continuum models. Acc Chem Res. 2000;33(12):889–97. doi:https://doi.org/10.1021/ar000033j.
- Schmidt MM, Townson SA, Andreucci AJ, King B, Schirmer E, Murillo A, Dombrowski C, Tisdale A, Lowden P, Masci A, et al. Crystal structure of an HSA/FcRn complex reveals recycling by competitive mimicry of HSA ligands at a pH-dependent hydrophobic interface. Structure. 2013;21(11):1966–78. doi:https://doi.org/10.1016/j.str.2013.08.022.
- Shi C, Shin YO, Hanson J, Cass B, Loewen MC, Durocher Y. Purification and characterization of a recombinant G-protein-coupled receptor, Saccharomyces cerevisiae Ste2p, transiently expressed in HEK293 EBNA1 cells. Biochemistry. 2005;44(48):15705–14. doi:https://doi.org/10.1021/bi051292p.
- Delafosse L, Xu P, Durocher Y. Comparative study of polyethylenimines for transient gene expression in mammalian HEK293 and CHO cells. J Biotechnol. 2016;227:103–11. doi:https://doi.org/10.1016/j.jbiotec.2016.04.028.
- Raymond C, Robotham A, Spearman M, Butler M, Kelly J, Durocher Y. Production of alpha2,6-sialylated IgG1 in CHO cells. MAbs. 2015;7(3):571–83. doi:https://doi.org/10.1080/19420862.2015.1029215.
- Viuff D, Antunes F, Evans L, Cameron J, Dyrnesli H, Thue Ravn B, Stougaard M, Thiam K, Andersen B, Kjærulff S, et al. Generation of a double transgenic humanized neonatal Fc receptor (FcRn)/albumin mouse to study the pharmacokinetics of albumin-linked drugs. J Control Release. 2016;223:22–30. doi:https://doi.org/10.1016/j.jconrel.2015.12.019.
- Schmidt MM, Thurber GM, Wittrup KD. Kinetics of anti-carcinoembryonic antigen antibody internalization: effects of affinity, bivalency, and stability. Cancer Immunol Immunother. 2008;57(12):1879–90. doi:https://doi.org/10.1007/s00262-008-0518-1.
- Lang BE, Cole KD. Unfolding properties of recombinant human serum albumin products are due to bioprocessing steps. Biotechnol Prog. 2015;31(1):62–69. doi:https://doi.org/10.1002/btpr.1996.
- Michnik A. Thermal stability of bovine serum albumin DSC study. J Therm Anal Calorim. 2003;71:509–19. doi:https://doi.org/10.1023/A:1022851809481.
- Schier R, McCall A, Adams GP, Marshall KW, Merritt H, Yim M, Crawford RS, Weiner LM, Marks C, Marks JD, et al. Isolation of picomolar affinity anti-c-erbB-2 single-chain Fv by molecular evolution of the complementarity determining regions in the center of the antibody binding site. J Mol Biol. 1996;263(4):551–67. doi:https://doi.org/10.1006/jmbi.1996.0598.
- Schier R, Bye J, Apell G, McCall A, Adams GP, Malmqvist M, Weiner LM, Marks JD. Isolation of high-affinity monomeric human anti-c-erbB-2 single chain Fv using affinity-driven selection. J Mol Biol. 1996;255(1):28–43. doi:https://doi.org/10.1006/jmbi.1996.0004.
- Franklin MC, Carey KD, Vajdos FF, Leahy DJ, de Vos AM, Sliwkowski MX. Insights into ErbB signaling from the structure of the ErbB2-pertuzumab complex. Cancer Cell. 2004;5(4):317–28. doi:https://doi.org/10.1016/S1535-6108(04)00083-2.
- Cho HS, Mason K, Ramyar KX, Stanley AM, Gabelli SB, Denney DW, Leahy DJ. Structure of the extracellular region of HER2 alone and in complex with the Herceptin Fab. Nature. 2003;421(6924):756–60. doi:https://doi.org/10.1038/nature01392.
- Von Kreudenstein TS, Escobar-Carbrera E, Lario PI, D’Angelo I, Brault K, Kelly JF, Durocher Y, Baardsnes J, Woods RJ, Xie MH, et al. Improving biophysical properties of a bispecific antibody scaffold to aid developability: quality by molecular design. MAbs. 2013;5(5):646–54. doi:https://doi.org/10.4161/mabs.25632.
- Roopenian DC, Low BE, Christianson GJ, Proetzel G, Sproule TJ, Wiles MV. Albumin-deficient mouse models for studying metabolism of human albumin and pharmacokinetics of albumin-based drugs. MAbs. 2015;7(2):344–51. doi:https://doi.org/10.1080/19420862.2015.1008345.
- Lambert JM, Chari RV, Emtansine A-T. (T-DM1): an antibody-drug conjugate (ADC) for HER2-positive breast cancer. J Med Chem. 2014;57(16):6949–64. doi:https://doi.org/10.1021/jm500766w.
- Bahadur RP, Zacharias M. The interface of protein-protein complexes: analysis of contacts and prediction of interactions. Cell Mol Life Sci. 2008;65(7–8):1059–72. doi:https://doi.org/10.1007/s00018-007-7451-x.
- Heaney-Kieras J, King TP. Limited pepsin digestion of human plasma albumin. J Biol Chem. 1977;252:4326–29.
- Ben-Kasus T, Schechter B, Lavi S, Yarden Y, Sela M. Persistent elimination of ErbB-2/HER2-overexpressing tumors using combinations of monoclonal antibodies: relevance of receptor endocytosis. Proc Natl Acad Sci U S A. 2009;106(9):3294–99. doi:https://doi.org/10.1073/pnas.0812059106.
- Friedman LM, Rinon A, Schechter B, Lyass L, Lavi S, Bacus SS, Sela M, Yarden Y. Synergistic down-regulation of receptor tyrosine kinases by combinations of mAbs: implications for cancer immunotherapy. Proc Natl Acad Sci U S A. 2005;102(6):1915–20. doi:https://doi.org/10.1073/pnas.0409610102.
- Kelton C, Wesolowski JS, Soloviev M, Schweickhardt R, Fischer D, Kurosawa E, McKenna SD, Gross AW. Anti-EGFR biparatopic-SEED antibody has enhanced combination-activity in a single molecule. Arch Biochem Biophys. 2012;526(2):219–25. doi:https://doi.org/10.1016/j.abb.2012.03.005.
- Bhattacharya AA, Grune T, Curry S. Crystallographic analysis reveals common modes of binding of medium and long-chain fatty acids to human serum albumin. J Mol Biol. 2000;303(5):721–32. doi:https://doi.org/10.1006/jmbi.2000.4158.
- Zhang H, Wang Y, Wu Y, Jiang X, Tao Y, Yao Y, Peng Y, Chen X, Fu Y, Yu L, et al. Therapeutic potential of an anti-HER2 single chain antibody-DM1 conjugates for the treatment of HER2-positive cancer. Signal Transduct Target Ther. 2017;2:17015. doi:https://doi.org/10.1038/sigtrans.2017.15.