?Mathematical formulae have been encoded as MathML and are displayed in this HTML version using MathJax in order to improve their display. Uncheck the box to turn MathJax off. This feature requires Javascript. Click on a formula to zoom.
?Mathematical formulae have been encoded as MathML and are displayed in this HTML version using MathJax in order to improve their display. Uncheck the box to turn MathJax off. This feature requires Javascript. Click on a formula to zoom.ABSTRACT
Disulfide bonds play a crucial role in folding and structural stabilization of monoclonal antibodies (mAbs). Disulfide bond reduction may happen during the mAb manufacturing process, resulting in low molecular weight species and possible failure to meet product specifications. Although many mitigation strategies have been developed to prevent disulfide reduction, to the best of our knowledge, reforming disulfide bonds from the reduced antibody in manufacturing has not previously been reported. Here, we explored a novel rescue strategy in the downstream process to repair the broken disulfide bonds via in-vitro redox reactions on Protein A resin. Redox conditions including redox pair (cysteine/cystine ratio), pH, temperature, and reaction time were examined to achieve high antibody purity and a high reaction rate. Under the optimal redox condition, >90% reduced antibody could be reoxidized to form an intact antibody on Protein A resin in an hour. In addition, this study showed high flexibility on the range of the intact mAb fraction in the initial reduced mAb sample (the lower limit of intact mAb faction could be 14% based on the data reported in this study). Furthermore, a kinetic model based on elementary oxidative reactions was constructed to help optimize the reoxidation conditions and to predict product purity. Together, the deep understanding of interchain disulfide bond reoxidation, combined with the predictive kinetic model, provided a good foundation to implement a rescue strategy to generate high-purity antibodies with substantial cost savings in manufacturing processes.
Introduction
Recombinant monoclonal antibodies (mAbs) constitute a prominent class of therapeutic proteins.Citation1,Citation2 A typical mAb has a molecular weight of approximately 150 kilodalton (kDa) and consists of two identical light chains and two identical heavy chains, linked by inter-heavy-heavy (HH) and inter heavy-light (HL) disulfide bonds.Citation3–Citation5 Disulfide bonds that connect two heavy chains or connect a light chain and a heavy chain are known as interchain disulfide bonds. Disulfide bonds that connect the two β-sheets in a single domain (constant domain or variable domain) are known as intrachain disulfide bonds. In a typical mAb, there are 12 intrachain disulfide bonds (one per each domain), two interchain disulfide bonds between light chain and heavy chain, and two to eleven interchain disulfide bonds between two heavy chains.Citation3,Citation6 Disulfide bonds stabilize proteins thermodynamically and mechanically. Improper disulfide bond formation and disulfide bond reduction can affect process performance, protein stability, and biological functionality.Citation4,Citation5,Citation7–Citation10 In recent years, the disulfide bond reduction after cell culture harvest has been observed more often for many large-scale mAb manufacturing processes, resulting in out-of-specification levels of low-molecular-weight (LMW) species and potential batch failure in manufacturing.Citation7,Citation11,Citation12 Studies focusing on preventing interchain disulfide bond breakage have been reported previously.Citation11–Citation17 Yet, mitigation methods do not always adequately prevent the disulfide reduction from occurring. In addition, these mitigation methods may require extra equipment and increase the manufacturing cost. Alternatively, reoxidizing the disulfide from the reduced antibody species to generate intact antibody products as a rescue strategy could be developed to address the disulfide bond reduction challenge. The rescue strategy can “save” the reduced mAb batches without sacrificing the manufacturing process flexibility or increasing the manufacturing cost. To our best knowledge, this is the first time that the approach of developing the rescue strategy in manufacturing process has been reported.Citation8,Citation16,Citation18–Citation21
As a post-translational modification, a disulfide bond is formed by reoxidizing two neighboring free cysteine residues.Citation22–Citation24 Though there is a vast body of knowledge of in-vitro disulfide reoxidation for antibodies, the majority of these studies focused on the solution environment with limited investigational conditions.Citation8,Citation18,Citation19,Citation25,Citation26 The existing studies cannot be directly and practically implemented in manufacturing process for three reasons. First, the reaction parameters were not optimized under typical manufacturing operation conditions to achieve high intact purity and fast reoxidation kinetics. Second, there is limited information on whether any downstream unit operation (such as Protein A chromatography, ion exchange chromatograph, and ultrafiltration/diafiltration) can be selected to implement the rescue strategies. Also, the manufacturability of using the rescue strategies at a large scale remains to be seen. Third, there is a lack of comprehensive characterizations to confirm the product comparability between the rescued mAb and the reference material. Thus, to develop practical industrial rescue strategies, the gap between the existing disulfide bond formation studies and the general mAb manufacturing process must be bridged.
In this series of studies, we systematically evaluated how to implement the reduced mAb rescue strategy into the mAb manufacturing process to bridge the aforementioned gap. Our studies are discussed in two papers: 1) the “proof-of-concept” study evaluated the possibilities of rescuing reduced mAb, selected the unit operation step (Protein A chromatography) to include the rescue strategy and developed the kinetics models to optimize reaction parameters under typical manufacturing operation conditions; and 2) the “developability and manufacturability” study implemented a potential rescue strategy during Protein A chromatography and performed comprehensive characterizations of the recovered mAb, showing the capabilities of generating high-purity antibody products from the reduced form in the manufacturing process. The “proof-of-concept” study results are reported here, and “developability and manufacturability” study results are discussed in a separate paper (Tan et al., mAbs, in press).
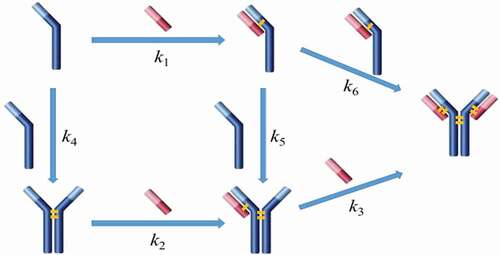
This paper focuses on the fundamental understanding of disulfide bond reoxidation in vitro, leading to a “proof-of-concept” implementation of this reoxidation-based rescue strategy in bioprocess with an expectation of eliminating LMW issue in manufacturing processes. This study had two main objectives: 1) to optimize process conditions to achieve high antibody purity and a higher reaction rate; and 2) to select the proper unit operation that can use the rescue strategy. We first examined factors (such as concentrations of redox components, pH, temperature, and reaction time) that potentially influenced the disulfide formation rate in order to define an optimal redox condition. Then, we laid out a rationale for using Protein A chromatography as a platform to implement the rescue strategy (details are included in Discussion section). We found that the disulfide reoxidation rate was considerably enhanced while immobilizing the reduced antibody on the Protein A resin in comparison to the one in free solution. Additionally, there were several practical considerations that justified the Protein A chromatography step as the most favorable step in the process to implement the rescue strategy. We also constructed a mathematical kinetic model of disulfide formation based on elementary oxidative reactions () using the Excel Visual Basic program ODExLims to determine rate constants, and hence, to choose optimal reoxidation conditions and help to predict product purity. In the end, an optimized condition was proposed to achieve high productivity and an acceptable product quality. Different mAbs were also tested to validate the optimized conditions, which demonstrated that the rescue strategy could have a potentially broad application in rescuing mAbs with severe disulfide reduction issues in mAb manufacturing process.
Results
Structure and conformation of reduced mAb
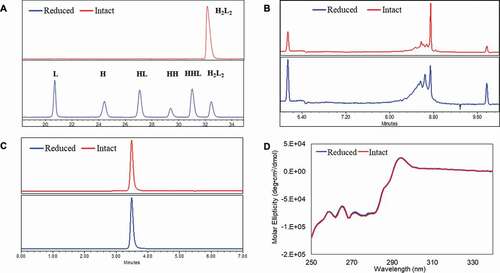
One concern in applying the reoxidation strategy to rescue reduced mAbs is that protein may undergo conformational changes while the interchain disulfide bonds are broken, which then affects the protein stability and biological properties. Thus, understanding the structure and conformation of the reduced mAb is essential to assess the feasibility of using reoxidation strategies in process development. In this study, two mAbs (IgG1 and IgG4) expressed using a Chinese Hamster Ovary (CHO) cell line were withdrawn from 500-L pilot-scale manufacturing (antibody sample details are available in Material and methods section). The IgG1 mAb (mAb-1) was used for most of the studies and the IgG4 mAb (mAb-2) was used for model comparison only. The structural profiles of the reduced mAb-1 and the intact mAb-1 were characterized using non-reduced capillary electrophoresis SDS (CE-SDS NR), size exclusion chromatography (SEC), imaged capillary electrophoresis focusing (icIEF), and circular dichroism (CD). ) showed that the majority of the mAb-1 was reduced into species heavy-heavy-light fragment (HHL), heavy-heavy fragment (HH), halfmer (HL), heavy chain (H), and light chain (L) during manufacturing. The icIEF analysis revealed a significant-skewed charge variant distribution for the reduced sample ()) in comparison to the intact sample. However, both reduced and intact samples showed an identical SEC profile ()) and near-UV CD profile ()). The results showed in were similar to previous reports that reduced mAb showed as one peak under native conditions, suggesting that the reduced mAb remained assembled through non-covalent interactions and retained the native tertiary structure although the interchain disulfide bonds had been largely broken.Citation3,Citation27
Figure 1. Conformations between intact and partially reduced monoclonal antibody (mAb), mAb-1 (IgG1). (a) Non-reduced CE-SDS analysis showed H2L2, HHL, HH, HL, H, and L for the reduced sample; (b) icIEF analysis showed different charge profiles between the reduced and intact mAb samples; (c) SEC analysis showed identical native sizes for the reduced and intact mAb samples; and (d) near-UV circular dichroism analysis showed identical high-order structures for the reduced and intact mAb samples. The intact mAb was defined with ≥ 90% purity based on non-reduced CE-SDS measurement and partially reduced mAbs was defined with < 90% purity. Analytical measurements details were documented in Material and Methods. H2L2: intact mAb, L: light chain, H: heavy chain, HH: heavy-heavy fragment, HL: halfmer, HHL: heavy-heavy-light fragment.
Factors that affect the in-vitro disulfide bond formation
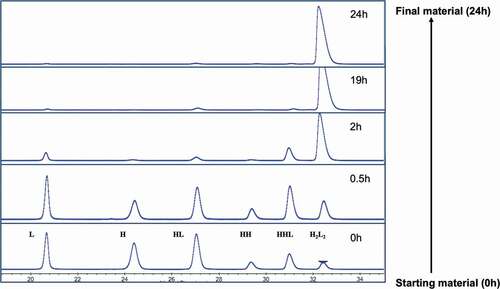
Glutathione (GSH)/glutathione disulfide (GSSG) is the most important redox pair in the endoplasm reticulum (ER), where antibody is synthesized, folded, and assembled.Citation28 Previous in-vitro experiments demonstrated that a GSH/GSSG ratio similar to that found in the ER could efficiently oxidize active-site cysteine in protein dimerization isomerase, which then could lead to the disulfide formation of substrate proteins.Citation28–Citation30 Since the intracellular disulfide formation is regulated by the redox system, we applied the same principle in an in-vitro environment. Cysteine (Cys) and cystine (Cys-Cys) have been reported as an effective combination to reoxidize reduced mAb fragments to form intact mAb.Citation30–32 In addition, the use of these two redox components is not expected to pose any product safety concerns as both are common upstream media components. Thus, we carried out in-solution studies to evaluate the possibilities of reoxidizing the reduced mAb-1 using a redox system containing varied amounts of cysteine and cystine. GSH was included in the studies due to its wide use in process development. Factors including pH and temperature were also evaluated. The starting material was a partially reduced mAb-1 molecule containing L, HL, HH, HHL, and H2L2 (intact mAb). showed a typical dynamic profile of mAb species overtime in an in-vitro redox system containing cysteine/cystine pair at pH 8. This allowed us to assess the possibility of adapting the principle of intracellular redox system into the in-vitro redox system without further optimizing the redox components and pH condition. The starting material, composed of less than 5% intact mAb-1 (presented as potential worst-case scenario), was recovered to reach the final product purity greater than 95% after 24 hours at room temperature.
Figure 2. Dynamic profile of the partially reduced mAb-1 species analyzed by non-reduced CE-SDS during the inter-chain disulfide bond reformation process. The starting material (< 5% intact mAb based on CE-SDS measurement) was incubated in the redox buffer system containing cysteine/cystine pair at pH 8 over a time course of 24 hours. H2L2: intact mAb, L: light chain, H: heavy chain, HH: heavy-heavy fragment, HL: halfmer, HHL: heavy-heavy-light fragment.
Composition of in-vitro redox system
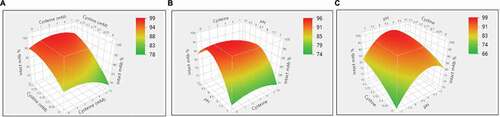
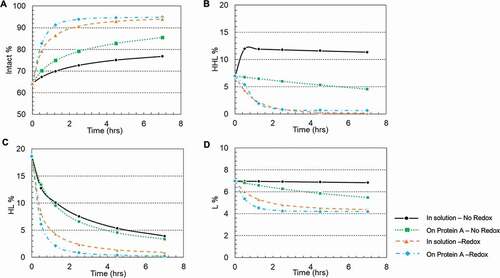
The study above showed it was possible to reoxidize the reduced mAb using the cysteine/cystine redox pair. A design-of-experiment (DoE) approach was used to further choose an optimal redox composition to achieve a kinetically higher disulfide formation rate. DoEs are widely applied in mAb downstream process development.Citation33–Citation35 Generally, their designs take into account the factors number and type, existing information, and reliability of the results to design the experiments. Thus, several factors can be changed in one set of the experiments, and the influences of these factors could be evaluated by a small number of experiments.Citation34 The design details, including concentrations of cysteine and cystine and pH, and resulting sample purities, are summarized in and Supporting Information Table S1. The JMP analysis based on Fit Least Square model showed that cystine concentration (p = .001) and pH (p = .03) were statistically significant factors that influenced the disulfide bond formation. While cysteine concentration (p = .55) did not show statistical significance, some interesting observations are worth mentioning. As shown in , at pH 10 condition, when there is only 0.3 mM cystine present (Run #3 in ), the intact mAb fraction increased by 7.7% compared with the negative control sample (Run #1 in ). Similarly, at pH 8 condition, when there is only 0.3 mM cystine present (Run #12 in ), the intact mAb fraction increased by 6.7% comparing with the negative control sample (Run #10 in ). However, at pH 7 condition, when there is both cysteine (1 mM cysteine for Run # 20, and 3 mM cysteine for Run #22) and 0.3 mM cystine present, the intact mAb fraction increased by 18.3% and 15.6% compared with the negative control sample (Run #19 in ), respectively. In brief, the presence of low-level of cysteine improved the mAb purity in general, but appeared more pronounced at the pH 7 condition than pH 8 or pH 10. Disulfide bond reoxidation was primarily dependent on pH and cystine concentration. This was consistent with those reported in literatures.Citation22,Citation30 Statistical analysis predicted that a redox composition of 1–3 mM cysteine, 0.3 mM cystine, and pH 8.5 could be an ideal redox condition for in-vitro disulfide formation ().
Table 1. Reoxidation results of the design-of-experiment investigation on cysteine, cystine, GSH, and pH.
Figure 3. Reoxidation results of the design of experiment (DoE) investigation on cysteine, cystine, and pH. (a) Surface plot of the resulted intact mAb% in response to the concentrations of cysteine and cystine; (b) Surface plot of the resulted intact mAb% in response to the concentrations of cysteine and pH; (c) Surface plot of the resulted intact mAb% in response to the concentrations of cystine and pH.
Disulfide formation on Protein A resin
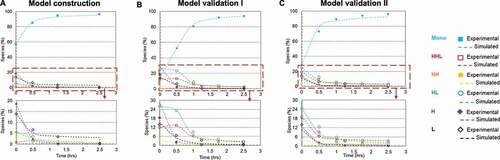
Redox systems containing cysteine and cystine have been used in studies for in-vitro oxidization of reduced mAb fragments in free solutions.Citation30–Citation32 However, there have been limited reports of disulfide bond formation on a solid surface, such as bound to a chromatographic resin.Citation36,Citation37 Here, we selected Protein A chromatography to evaluate the rescue strategy and studied the in-vitro disulfide formation on the Protein A resin surface; details regarding the unit operation step selection are included in the Discussion section below. We compared the disulfide formation rates in solution and on Protein A resin with and without the redox system (1 mM cysteine and 0.3 mM cystine at pH 8), respectively. Using the partially reduced mAb-1 sample (64% intact purity) in phosphate buffer matrix at pH 8, the experiment was performed at room temperature for over 7 hours in order to obtain a complete kinetic profile. ) show plots of all mAb species against reaction time based on experimental and kinetics model () simulated results for each studied condition. The kinetics model described the simplified reaction pathways as six elemental reactions, and the reaction rates fitted from the kinetics model were used for redox reaction rate comparisons in this study; kinetics model details are described in the Materials and Methods section. The kinetics model can also be applied as a predictive tool for selecting optimal redox condition and reoxidized product purity prediction (see section ‘Kinetics model construction and validation’ below for details).
Figure 4. Plots of disulfide formation for an IgG1 antibody (dots denoted the experimental data and the lines represented the kinetic model simulated results). (a) In free solution without redox; (b) On Protein A resin without redox; (c) In free solution with redox pair (1 mM cysteine and 0.3 mM cystine); (d) On Protein A resin with redox pair (1 mM cysteine and 0.3 mM cystine). Each plot included six kinetic profiles, representing all six elementary reactions in the kinetic model (). Mono: intact mAb, L: light chain, H: heavy chain, HH: heavy-heavy fragment, HL: halfmer, HHL: heavy-heavy-light fragment.
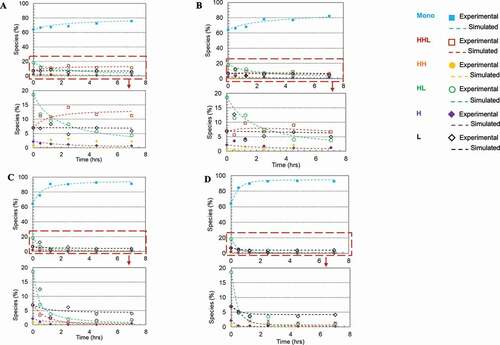
Each plot includes six kinetic profiles, representing all six elementary reactions. All four studied conditions showed that the simulated results were in good agreement with the experimental results. Additionally, plots () were also generated for each mAb species versus time to compare these four studied conditions and respective rate constants were determined by the kinetic modeling (kinetic model was illustrated in , and rate constants were summarized in ). In doing so, we observed a general trend that over time the overall intact mAb percentage increased and intermediate species decreased, but at different rates depending on the environmental conditions. Specifically, we first observed that the redox system significantly accelerated the disulfide formation rates in solution. All rate constants for the solution with redox system were increased at least 10-fold compared to the ones in solution redox-free system, as shown in . Secondly, in a redox-free system (labeled as “In solution without redox” and “Resin surface without redox” in ), we found that all the disulfide formation rates (k1-k6) were higher on the resin surface (“Resin surface without redox” sample in ) compared to the ones in solution (“In solution without redox” sample in ), suggesting that the disulfide formation kinetics were enhanced by binding the reduced mAb-1 onto Protein A resin. In contrast in a redox system (labeled as “In solution with redox” and “Resin surface with redox” in ), except for k6, all other elementary reaction rates on Protein A resin (“Resin surface with redox” in ) were noticeably different from those in a free solution (“In solution with redox” in ). Lastly, the compounding effect of using the redox condition on Protein A resin showed an overall improvement for the disulfide formation kinetics.
Table 2. The k1 to k6 values of mAb reoxidation in solution and on Protein A resin using sodium phosphate buffer, pH 8. The regression parameters are based on Equationequations 14(14)
(14) –Equation19
(19)
(19) .
Figure 5. Plots of intermediate specie fractions for an IgG1 antibody based on kinetic model simulated results. (a) Intact mAb; (b) HHL species; (c) HL species; (d) L species. Each plot included the result in four reaction environments: free solution without redox; On Protein A resin without redox; In free solution with redox pair cysteine and cystine; On Protein A resin with redox pair cysteine and cystine. Intact: intact mAb, L: light chain, HL: halfmer, HHL: heavy-heavy-light fragment.
pH impact on disulfide formation
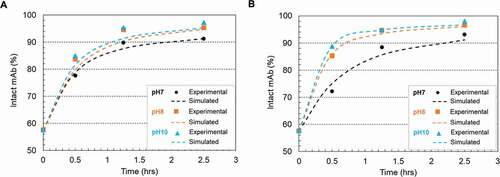
) depicted the disulfide formation kinetics under pH 7, 8 and 10 using two redox systems (0.5 mM cysteine/0.3 mM cystine and 1 mM cysteine/0.3 mM cystine), respectively. Cystine was controlled at a constant concentration of 0.3 mM for all conditions due to its limited solubility.Citation38 The experimental results showed a good agreement with the kinetic model simulated results. The calculated reaction rate constants, summarized in , showed a higher disulfide formation rate at higher pH conditions. We chose pH 8 as the condition for the following experiment to be consistent throughout the studies.
Table 3. The k3 and k6 values of mAb reoxidation in the presence of two redox buffer conditions (0.5 mM cysteine/0.3 mM cystine, 1 mM cysteine/0.3 mM cystine). The regression parameters are based on Equationequations 14(14)
(14) –Equation19
(19)
(19) .
Figure 6. Plots of disulfide formation represented as intact mAb % (mAb-1) change on the Protein A resin in 20 mM sodium phosphate buffer (pH 7 and pH 8) and 20 mM sodium carbonate buffer (pH10) and room temperature. (a) using a redox system containing 0.5 mM cysteine and 0.3 mM cystine; (b) using a redox system containing 1 mM cysteine and 0.3 mM cystine. The dots denoted the experimental data and the lines represented the simulation results. The kinetic model illustration was shown in scheme 1 and simulation parameters were listed in Table 3.
Temperature impact on disulfide formation
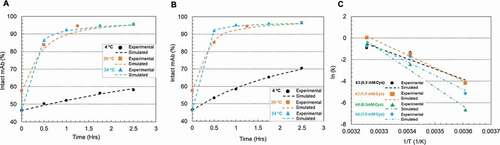
Temperature is known to affect reaction kinetics significantly. This study was conducted at three different temperatures (4°C, 20°C, and 34°C) using two redox systems at pH 8 (0.5 mM cysteine/0.3 mM cystine and 1 mM cysteine/0.3 mM cystine), respectively. In ), reaction rate decreases were observed with temperature decrease at both 0.5 and 1.0 mM cysteine levels. Selected kinetic parameters (k3 and k6 directly reflect disulfide formation rate of intact mAb) were listed in and their relations to temperature were simulated via Arrhenius Equation (EquationEquation (1))(1)
(1) .
Table 4. The k3 and k6 values at different cysteine concentrations and temperatures, and the activation energies calculated based on the Arrhenius EquationEquation (1)(1)
(1) .
Figure 7. Plots of mAb-1 disulfide reoxidation using 0.5 or 1 mM cysteine and 0.3 mM cystine in the sodium phosphate (pH 8) buffer on Protein A resin at different temperatures (4°C, 20°C and 34°C). (a) 0.5 mM cysteine, 0.3 mM cystine; (b) 1 mM cysteine, 0.3 mM cystine. The dots denoted the experimental data and the lines represented the simulation results; (c) Arrhenius Equation simulation of the selected kinetic parameters k3 and k6 at different cysteine concentrations. The kinetic model illustration was shown in Scheme 1. The kinetic parameters, the activation energy fitted from Arrhenius and R-squared value of the fitting were listed in Table 4.
where k0 is a constant for each reaction; Ea is the activation energy; R is the universal gas constant; T is the absolute temperature.
) showed that the temperature impact on k3 and k6 can be well illustrated using Arrhenius Equation. summarized the activation energy of two redox systems at different cysteine levels. A higher cysteine concentration appears to result in an increase of Ea for k3 but lowered Ea for k6, suggesting that the primary function of cysteine is to accelerate the reaction k6 instead of k3 by decreasing the Ea of k6, especially at room or lower temperatures. All the fittings in had R-square (R2) values close to 0.95 or higher than 0.95.
Kinetics model construction and validation
The studies on the factors affecting the in-vitro disulfide bond formation provided a general guidance to implement the rescue strategy into Protein A chromatography. To get a better understanding of the reaction mechanism, a model was built upon experimental results and the respective reaction rate constants were calculated using the Excel Visual Basic program ODExLims.Citation39 In our study, despite the fact that different types of fragments may exist in the antibody sample solution, free thiols of the reduced species can be reoxidized to form disulfide bonds, resulting in larger intermediate species and subsequently an intact mAb molecule.Citation18,Citation19,Citation25 Although the reoxidation kinetics depended on multiple factors, including redox pair, temperature, pH, the reaction pathways could be simplified to be expressed using the six elementary reactions () and further illustrated using a kinetic model described in Materials and Methods section.
Our kinetic study showed that a higher reoxidation rate and high intact antibody purity were achieved by carrying out the disulfide reoxidation on Protein A resin using the following optimal redox condition: 1 mM cysteine, 0.3 mM cystine, pH 8. In doing so, the purity of the initial mAb-1 sample (IgG1 containing 57.6% intact purity) was improved to 93.5% after a 1.25-h treatment (details shown in ). The kinetics data were simulated ()) to generate rate constants for each elementary reaction (). The kinetics of the same mAb under the above mentioned optimal redox condition were predicted based on the initial mAb composition. As shown in ), the disulfide formation kinetics of two batches of starting mAb-1 materials with intact purities of 14.0% and 29.0% were computed based on EquationEquations (14)(14)
(14) –(19) (dash line). The predicted results showed a high agreement with the experimental results for both intact and intermediate species ( and Supporting Information Table S2). For example, the intact purities of sample 1 and sample 2 reached 80.7% and 89.1%, respectively, after 1-h treatment versus predicted purities of 83.3% and 86.8%. The HL reached experimental values of 9.1% and 5.8% after 1-h treatment versus predicted values of 7.1% and 4.8% for sample 1 and sample 2, respectively. However, sample 1 showed less divergence between the experimental and predicted values than sample2, possibly due to the analytical assay variability. Overall, these results validated the kinetic modeling mechanism and confirmed the applicability of using this modeling method to predict the kinetic performance of antibody interchain disulfide formation.
Table 5. Validation of the kinetic model.
Figure 8. Model validation of mAb-1 disulfide bond formation kinetics using two starting materials with different purities. (a) Kinetic model was constructed based on data using the redox system containing 1 mM cysteine, 0.5 mM cystine, pH8 at 20°C on Protein A resin; (b) Model validation using starting material with 14% purity; (c) Model validation using starting material with 29% purity. The dots denote the experimental data and the lines represent the computational prediction results.. Each plot included six kinetic profiles, representing all six elementary reactions in the kinetic model (Scheme 1). Mono: intact mAb, L: light chain, H: heavy chain, HH: heavy-heavy fragment, HL: halfmer, HHL: heavy-heavy-light fragment.
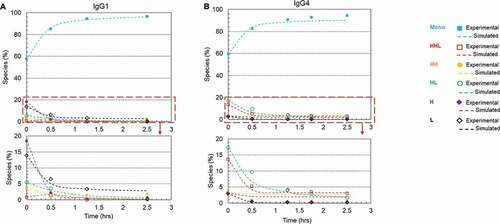
Disulfide formation for different IgG subclasses
Four major types of IgGs naturally exist in humans. Different IgG types may possess different disulfide reoxidation kinetics due to the difference of their interchain disulfide linkages.Citation3 To evaluate whether the rescue method can be applied to all IgG subclasses in general, we also studied the disulfide formation for an IgG4 (mAb-2) antibody. ) showed kinetics of disulfide formation on Protein A resin in the presence of 1 mM cysteine, 0.3 mM cystine, pH 8, for mAb-1 (IgG1) and mAb-2 (IgG4), respectively. The initial compositions of both mAbs and their calculated rate constants were summarized in . Under the redox condition 1 mM cysteine, 0.3 mM cystine, pH 8 for 1.25 hours, the intact purities of mAb-1 and mAb-2 were improved from 57.6% to 93.5% and 59.5% to 90.8%, respectively. This result showed that the rescue strategy, which was developed using mAb-1 (an IgG1 antibody), could be used as a general approach for reduced mAbs rescue, but the reaction rate varied for different mAbs.
Figure 9. Plots of disulfide formation for an IgG1 antibody and an IgG4 antibody on Protein A resin using a redox system containing 1 mM cysteine and 0.3 mM cystine at pH 8 and room temperature. (a) IgG1 mAb (mAb-1) with a starting purity of 57.6%; (b) IgG4 mAb (mAb-2) with a starting purity of 59.5%.. Each plot included six kinetic profiles, representing all six elementary reactions in the kinetic model (Scheme 1). Mono: intact mAb, L: light chain, H: heavy chain, HH: heavy-heavy fragment, HL: halfmer, HHL: heavy-heavy-light fragment.
Discussion
As the main goal of this study was to demonstrate a proof of concept of applying disulfide reoxidation as a rescue strategy in mAb manufacturing process, the method must achieve high antibody purity at a fast disulfide formation rate to meet manufacturing process requirements (e.g., cadence and facility fit). The chemistry of disulfide bond reduction and formation essentially is the chemistry of reduction-oxidation reactions. The redox environment and the physical proximity of the pairing cysteine residues are the key factors to drive the disulfide formation rate.Citation18,Citation40,Citation41
It has been reported that oxygen can function as an oxidant and be used to form disulfide bonds.Citation22 In fact, we observed an increase of product purity through downstream steps for several antibody programs with disulfide reduction issue. This spontaneous and often random air-oxidation reaction is thermodynamically favorable but kinetically slow, resulting in undesirable product quality and potential facility fit challenges.Citation30 Active aeration has been applied to increase dissolved oxygen levels to enhance cellular growth and promote the formation of disulfide bonds.Citation12,Citation42–Citation44 However, the active aeration alone cannot completely solve the issue.
Numerous studies have been conducted to understand in-vitro disulfide reoxidation kinetics fundamentally.Citation41,Citation45–Citation48 The rate of in-vitro disulfide formation was significantly affected by the pH and composition of the redox system. All biologically significant reactions of thiols involve the ionized thiolate form (S−).Citation18,Citation41,Citation49 Thiol-disulfide exchange is dependent on the concentration of the reactive thiolates anion (S−) relative to that of the unreactive thiol groups (SH), both of which are strongly dependent on solution pH. The concentration of reactive thiolate ion (S−) decreases while solution pH decreases. For disulfide formation, the starting species is the thiol group found on the cysteine residues in proteins. These fundamentally important studies informed the basics of our initial redox condition design. The pH value of ≥8 for our optimal redox condition is in agreement with the literature that alkaline condition is favorable for disulfide reoxidation.Citation41,Citation50
In the disulfide reoxidation process, another factor driving disulfide formation is the proximity of the pairing cysteine residues of the antibody molecule.Citation41 Here, the in-vitro system containing cysteine/cystine pair at pH ≥8 was optimized to enhance the interchain disulfide formation of an antibody expressed from a CHO cell line. Extensive characterization () showed that the reduced antibody, despite the interchain disulfide breakage, retained its native and high-order structures, likely being held together through non-covalent interactions.Citation3,Citation24 The conformation of the chains and the relative spatial relationships between chains, which provide evidence of the intact high-order structure, is essential to maintain the physical proximity of the partnering cysteine residues to enable the disulfide formation.Citation19
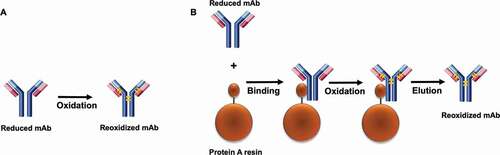
A distinctive aspect of this study was our examination of the kinetics of disulfide formation on the Protein A resin. Here, we selected Protein A chromatography as the unit operation to implement the rescue strategy based on the following three factors. First, the affinity between mAb and Protein A ligand is through Fc region of the antibody. Since the high-order structure is intact, the affinity between mAb and Protein A would remain unchanged. Thus, reduced mAb still binds to Protein A resin. As our dynamic binding capacity study (Tan et al, mAbs, in press) showed, at 10% breakthrough and 4-min residence time, mAb material containing three different LMW levels (90%, 50%, and 1%) achieved DBCs of 58.6, 58.6, and 58.5 g/Lresin, respectively. Second, Protein A chromatography is a general unit operation to purify mAb, and implementing the rescue strategy in Protein A chromatography could be a mAb platform process operation.Citation51,Citation52 Third, compared with ion-exchange chromatography, Protein A chromatography is less sensitive to operation buffer composition change and easier to include the redox pair into Protein A chromatography operation buffer.Citation53
Interestingly, we observed that the reoxidation kinetics was enhanced by binding the reduced antibody on the Protein A resin compared to the resin-free condition, all of which demonstrated that the Protein A chromatography step is a favorable unit operation for rescue strategy implementation. A possible reason of Protein A-enabled disulfide reoxidation enhancement was that the immobilization and concentration of the reduced antibody on the resin surface increased the molecular rigidity and created a closer proximity between the neighboring free sulfhydryl groups of the reduced molecule, resulting in a lower reaction activation energy and enabling easier disulfide formation. However, further analysis revealed that the rate constant k6 (hinge disulfide bond formation rate) did not show much difference between the redox reaction on the Protein A resin and the in-solution redox reaction, while all other reaction rates (k1 – k5) were clearly distinct between the two conditions. Such divergence of rate constants of these redox reactions in response to the Protein A resin or in solution could be related to the difference of the interchain solvent accessibility. It has been reported that the disulfide bonds in the hinge region (HH) are more solvent accessible,Citation3,Citation54 resulting in a higher rate of the HH disulfide formation than other interchains. Because of this, the HH disulfide bonds formation rate would appear to be less affected by whether the reduced mAb was bound onto the Protein A resin or in free solution. illustrates a schematic model of disulfide bond formation on the resin surface. The finding that the interchain disulfide bonds can be reformed and further enhanced on Protein A resin surface not only provides a novel way to probe in-vitro disulfide formation, which may help gain insight into the intracellular IgG assembly, but more importantly, further drives us to seek practical applications in the manufacturing process.
Figure 10. A model for the disulfide bond formation of mAb. (a) in solution. (b) on Protein A resin. A faster disulfide formation rate was observed on the solid surface compared to the reaction in solution. It was presumed that association of the reduced mAb and the solid surface led to an increase of rigidity of the molecule and a closer proximity of the free cysteines on the respective chains. The long blue bar represents the heavy chain, the short red bar represents the light chain, the orange bar represents the disulfide bond.
Besides the investigation on the reaction condition of the rescue strategy, a kinetic model was developed as a useful tool in selecting optimal redox conditions and provides an accurate prediction of the reoxidized product purity (). Our model was built based on all six elementary reactions. The percentage of each reduced species for the starting material is the only input that is needed for mAb purity prediction. It is important to note that the overall composition of the starting material is necessary to warrant an accurate model prediction. A given starting material containing a composition of mAb species will give a prediction of a mAb purity and all intermediate species at a given time. As such, a reaction time can be determined to achieve a predefined acceptable intact mAb purity, which is a critical quality attribute. Additionally, in order to implement the reoxidation strategy in manufacturing, the reaction time is a critical process parameter that needs further fine-tuning to meet the facility fit requirements.
Moreover, it is worth noting that our kinetic modeling, by determining six rate constants, provides more insightful information at a molecular level that relates to various disulfide formation pathways. In this work, we studied two model antibodies (IgG1 and IgG4) containing different H-L chain disulfide linkage,Citation3,Citation55–Citation57 inferring different disulfide bond reoxidation kinetics. The k3 value of the IgG4 was twofold larger than that of IgG1, while the k6 value of the IgG1 was 6-fold that of IgG4, suggesting that combination of HHL and L is the preferable assembly pathway for the IgG4 antibody and HL-HL assembly pathway for IgG1 antibody. The apparent divergence of these rate constants between IgG1 and IgG4 antibodies further confirms a previous report that different IgG subclasses may be involved in different preferable disulfide reoxidation pathways.Citation18
Protein purity is a critical quality attribute that must be controlled for consistent product quality. Disulfide bond reduction in mAbs leads to lower protein purity in manufacturing. It has drawn great attention across industry, and companies have since taken tremendous efforts to understand root causes and develop preventive mitigation strategies. Nevertheless, we propose a rescue strategy, in which reduced disulfide bonds are repaired via reoxidation, is also a viable option to contribute to the overall control strategy in mitigating disulfide bond reduction during the manufacturing process. Through in-vitro experiments, we developed an optimal reoxidation condition to achieve >90% intact purity from the starting material containing <5% intact purity within a 1-hr reaction time. A mathematic model based on all six elementary oxidative reactions was built to provide an accurate prediction of not only final intact mAb purity, but also compositions of each intermediate species. This model can be applied in a manufacturing setting to help achieve high product purity. Furthermore, this model may help us further understand the reduction/reoxidation pathways at a molecular level in a dynamic redox environment. This study provides proof-of-concept that reduced antibody can be reoxidized to form high-purity intact antibody at a higher reaction rate by using an optimal redox system on Protein A resin. Nevertheless, the feasibility and robustness of using this reoxidation strategy in manufacturing process still needs thorough evaluation. An extensive study focusing on developability and manufacturability in downstream process is described in our other report of results from this series of studies (Tan et al., mAbs, in press).
Materials and methods
Materials
The intact and partially reduced mAbs (IgG1 and IgG4) were generated in CHO cell culture at Bristol-Myers Squibb’s facility in Devens, MA. An intact mAb is defined with ≥90% purity and partially reduced mAbs is defined as <90% purity based on non-reduced CE-SDS measurement. Protein A resin (MabSelectTM SuReTM LX) was purchased from GE Healthcare. L-cysteine, L-cystine dihydrochloride, L-Glutathione reduced, Iodoacetamide were purchased from Sigma-Aldrich (Burlington, MA). Sodium phosphate, sodium carbonate, sodium chloride, hydrochloride acid, sodium acetate, acetate acid, and tris base were purchased from VWR (Radnor, PA).
Disulfide formation kinetic study
Study in solution
In 15 ml tubes, the partially reduced mAb sample was diluted to a target concentration of 5 mg/mL with 20 mM phosphate buffer (pH 7 and 8) or 20 mM carbonate buffer (pH 10) containing components of cysteine, cystine or glutathione. The tubes were then placed in water baths to maintain constant reaction temperatures. Samples were collected as a function of time for up to 7 hours. Samples were alkylated with iodoacetamide (IAM) and frozen prior to analysis.
Study on protein A resin
In 15 ml tubes, the partially reduced mAb sample was diluted to a target concentration of 5 mg/mL (~0.03 mM) with 20 mM phosphate buffer (pH 7 and 8) or 20 mM carbonate buffer (pH 10) containing redox components. The free thiol and disulfide molar concentration range in the reduced mAb were calculated to be 0.048 ~ 0.161 mM, 0.054 ~ 0.111 mM (Supporting Information Table S3). The immobilization of mAb to Protein A resin was carried out by immediately adding appropriate amounts of MabSelect SuRe LX resin (a target 30 g/Lresin binding) into the sample and mixing well for 3 minutes. Supernatants were assessed using UV-visible spectrophotometry measurement at 280 nm absorbance to ensure they were free of protein. The tubes were then placed in water baths to maintain constant reaction temperatures. Samples were collected as a function of time for up to 7 hours. The sample mixture was centrifuged for 1 minute at 1000 relative centrifugal force to remove the supernatant that was confirmed to be free of protein. The product was eluted with a low pH acetate buffer (pH3.5) and subsequently neutralized to pH 5.5 with Tris buffer. Finally, samples were alkylated with IAM and frozen prior to analysis.
DoE of redox reaction optimization
A custom DoE was used to evaluate factors that may affect the disulfide bond formation rate. Cysteine concentration (0–5 mM), cystine concentration (0–0.3 mM), and pH (7, 8, 10) were the main factors included in the experimental design. The experiment was carried out by mixing the starting material containing 57.6% H2L2 (intact mAb), 0.8% HHL, 3.7% HH, 5.5% HL, 18.4% H, and 13.9% L with redox components (). After incubating the mixtures at 20°C for 30 minutes, samples were alkylated with IAM. Samples were then tested for purity using non-reduced CE-SDS method. Statistical analysis was performed using JMP13.1.0 Statistical software from SAS.
Non-reduced CE-SDS analysis
SDS Microchip-based CE-SDS was performed on a LabChip GXII (Perkin Elmer) under non-reducing condition. Iodoacetamide was added into HT Protein Express Sample Buffer (Perkin Elmer) to a final IAM concentration of approximately 5 mM. A total of 5 μL antibody sample at approximately 1 mg/mL was mixed with 100 μL of the IAM containing sample buffer. The samples were incubated at 75°C for 10 min. The denatured proteins were analyzed with the “HT Protein Express 200” program.
Size exclusion chromatography
SEC was performed using a Waters BEH column (4.6 mm x 150 mm, 200 Å, 1.5 µm) with an isocratic gradient monitored at 280 nm on a Waters ACQUITY UPLC system (Milford, MA). The samples were injected onto the system at an isocratic flow rate of 0.4 mL/min using mobile phase of 25 mM phosphate, 300 mM arginine, pH 6.8.
Charge variants analysis
Charge variants were assayed by icIEF, which was performed on a Protein Simple iCE3 instrument with an Alcott 720 NV autosampler (San Jose, CA). Samples were mixed with appropriate pI markers, ampholytes, and urea and injected into a fluorocarbon-coated capillary cartridge. A high voltage was applied and the charged variants migrated to their respective pI values. A UV camera captured the image at 280 nm. The main peak was identified and the peaks that migrated into the acidic range and basic range were summed, quantitated, and reported as relative percent area.
Circular dichroism
CD spectra were measured using a Chirascan-auto CD spectropolarimeter (Applied Photophysics) fitted with a 0.5 mm pathlength quartz cuvette. Near-UV (CD) spectroscopy was used to monitor protein tertiary structure. Near-UV CD spectra were collected from 340 to 250 nm on solutions containing 10 mg/mL protein. All protein solutions were prepared and added to a 96-well plate temperature controlled at 10°C. Spectra were collected in triplicate, baseline subtracted, averaged, and converted from millidegrees to molar ellipticity using the theoretical molecular weight.
Kinetic modeling
Different types of fragments may exist in the mAb solution. Based on the primary CE-SDS NR analysis, the major contents in the initial solution are light chain (L), heavy chain (H), heavy-heavy fragment (HH), half-mer (HL), heavy-heavy-light fragment (HHL) and intact (H2L2 or Mono). The simplified reaction pathways can be illustrated as scheme 1and the reaction kinetics can be expressed as
Where ri (i = 1,.6) is the reaction rate for each elemental reaction, ki (i = 1,.6) is the rate constant for the corresponding reaction.
Based on EquationEquations (2)(2)
(2) –(Equation7
(7)
(7) ), mole balances of each fragment can be expressed as
Where t is reaction time.
By substituting EquationEquations (3)(3)
(3) –(8) into EquationEquations (8)
(8)
(8) –(13), the dynamic fragment concentrations at time t can be then integrated as:
Kinetic parameter calculation
The kinetic parameters were calculated based on EquationEquations (14)(14)
(14) –(Equation19
(19)
(19) ). The program OdexLims in Excel (2017) coded with Visual Basic for Applications was used to solve the equations.Citation39 The relationship of different impacting factors and the kinetic parameters were analyzed via the JMP13 software based on the DOE principle. Since EquationEquations (4)
(4)
(4) and (Equation7
(7)
(7) ) illustrate two major pathways to form intact mAb molecules, k3 and k6 were chosen as two key kinetic parameters to indicate the reoxidation properties for the intact mAb formation.
Abbreviations
| CD | = | circular dichroism |
| CE-SDS | = | capillary electrophoresis with sodium dodecyl sulfate |
| CHO | = | Chinese Hamster Ovary |
| DoE | = | design of experiment |
| ER | = | endoplasm reticulum |
| GSH | = | glutathione |
| GSSG | = | glutathione disulfide |
| HH | = | inter heavy-heavy |
| HL | = | inter heavy-light |
| IAM | = | iodoacetamide |
| icIEF | = | imaged capillary isoelectric focusing |
| LC | = | light chain |
| LMW | = | low molecular weight |
| mAb | = | monoclonal antibody |
| SEC | = | size exclusion chromatography |
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Supplemental Material
Download MS Word (22.8 KB)Acknowledgments
The authors would like to acknowledge Process Development Analytics for their contributions to this work. The authors also thanks Dr. Ronald Maurer for providing a thorough review and invaluable comments. All financial support was provided by BMS: Z.T., V.E., T.R., L.H., C.D., Y.S., L.T., A.L., S.G., and Z.L. are employees of BMS, and P.T. was supported by BMS during the preparation of this publication. The authors have no conflicts of interest to declare.
Supplementary material
Supplemental data for this article can be accessed on the publisher’s website.
References
- Ecker DM, Jones SD, Levine HL. The therapeutic monoclonal antibody market. MAbs. 2015;7:9–14. doi:https://doi.org/10.4161/19420862.2015.989042.
- Nelson AL, Dhimolea E, Reichert JM. Development trends for human monoclonal antibody therapeutics. Nat Rev Drug Discov. 2010;9:767–74. doi:https://doi.org/10.1038/nrd3229.
- Liu H, May K. Disulfide bond structures of IgG molecules: structural variations, chemical modifications and possible impacts to stability and biological function. mAbs. 2012;4:17–23. doi:https://doi.org/10.4161/mabs.4.1.18347.
- Fass D. Disulfide bonding in protein biophysics. Annu Rev Biophys. 2012;41:63–79. doi:https://doi.org/10.1146/annurev-biophys-050511-102321.
- Trivedi MV, Laurence JS, Siahaan TJ. The role of thiols and disulfides in protein chemical and physical stability. Curr Protein Pept Sci. 2009;10:614–25. doi:https://doi.org/10.2174/138920309789630534.
- Correia IR. Stability of IgG isotypes in serum. MAbs. 2010;2:221–32. doi:https://doi.org/10.4161/mabs.2.3.11788.
- Chung WK, Russell B, Yang Y, Handlogten M, Hudak S, Cao M, Wang J, Robbins D, Ahuja S, Zhu M, et al. Effects of antibody disulfide bond reduction on purification process performance and final drug substance stability. Biotechnol Bioeng. 2017;114(6):1264–74. doi:https://doi.org/10.1002/bit.26265.
- Wang T, Liu YD, Cai B, Huang G, Flynn GC. Investigation of antibody disulfide reduction and re-oxidation and impact to biological activities. J Pharm Biomed Anal. 2015;102:519–28. doi:https://doi.org/10.1016/j.jpba.2014.10.023.
- Manteca A, Alonso-Caballero Á, Fertin M, Poly S, De Sancho D, Perez-Jimenez R. The influence of disulfide bonds on the mechanical stability of proteins is context dependent. J Biol Chem. 2017;292:13374–80. doi:https://doi.org/10.1074/jbc.M117.784934.
- Liu H, Ponniah G, Zhang H-M, Nowak C, Neill A, Gonzalez-Lopez N, Patel R, Cheng G, Kita AZ, Andrien B, et al. In vitro and in vivo modifications of recombinant and human IgG antibodies. mAbs. 2014;6(5):1145–54. doi:https://doi.org/10.4161/mabs.29883.
- O’Mara B, Gao Z-H, Kuruganti M, Mallett R, Nayar G, Smith L, Meyer JD, Therriault J, Miller C, Cisney J, et al. Impact of depth filtration on disulfide bond reduction during downstream processing of monoclonal antibodies from CHO cell cultures. Biotechnol Bioeng. 2019;116(7):1669–83. doi:https://doi.org/10.1002/bit.26964.
- Mun M, Khoo S, Do Minh A, Dvornicky J, Trexler-Schmidt M, Kao YH, Laird MW. Air sparging for prevention of antibody disulfide bond reduction in harvested CHO cell culture fluid. Biotechnol Bioeng. 2015;112:734–42. doi:https://doi.org/10.1002/bit.25495.
- Handlogten MW, Zhu M, Ahuja S. Glutathione and thioredoxin systems contribute to recombinant monoclonal antibody interchain disulfide bond reduction during bioprocessing. Biotechnol Bioeng. 2017;114:1469–77. doi:https://doi.org/10.1002/bit.26278.
- Koterba KL, Borgschulte T, Laird MW. Thioredoxin 1 is responsible for antibody disulfide reduction in CHO cell culture. J Biotechnol. 2012;157:261–67. doi:https://doi.org/10.1016/j.jbiotec.2011.11.009.
- Du C, Huang Y, Borwankar A, Tan Z, Cura A, Yee JC, Singh N, Ludwig R, Borys M, Ghose S, et al. Using hydrogen peroxide to prevent antibody disulfide bond reduction during manufacturing process. MAbs. 2018;10(3):500–10. doi:https://doi.org/10.1080/19420862.2018.1424609.
- Trexler-Schmidt M, Sargis S, Chiu J, Sze-Khoo S, Mun M, Kao YH, Laird MW Identification and prevention of antibody disulfide bond reduction during cell culture manufacturing. Biotechnol Bioeng. 2010;106:452–61.
- Saccoccia F, Angelucci F, Boumis G, Carotti D, Desiato G, Miele AE, Bellelli A. Thioredoxin reductase and its inhibitors. Curr Protein Pept Sci. 2014;15:621–46. doi:https://doi.org/10.2174/1389203715666140530091910.
- Petersen JGL, Dorrington KJ. An in vitro system for studying the kinetics of interchain disulfide bond formation in immunoglobulin G. 1974.
- Sears DW, Mohrer J, Beychok S. A kinetic study in vitro of the reoxidation of interchain disulfide bonds in a human immunoglobulin IgGLk. Correlation between sulfhydryl disappearance and intermediates in covalent assembly of H2L2. Proc Natl Acad Sci U S A. 1975;72:353–57. doi:https://doi.org/10.1073/pnas.72.1.353.
- Kao YH, Hewitt DP, Trexler-Schmidt M, Laird MW. Mechanism of antibody reduction in cell culture production processes. Biotechnol Bioeng. 2010;107:622–32. doi:https://doi.org/10.1002/bit.22848.
- Hutterer KM, Hong RW, Lull J, Zhao X, Wang T, Pei R, Le ME, Borisov O, Piper R, Liu YD, et al. Monoclonal antibody disulfide reduction during manufacturing: untangling process effects from product effects. MAbs. 2013;5(4):608–13. doi:https://doi.org/10.4161/mabs.24725.
- Calce E, Vitale RM, Scaloni A, Amodeo P, De Luca S. Air oxidation method employed for the disulfide bond formation of natural and synthetic peptides. Amino Acids. 2015;47:1507–15. doi:https://doi.org/10.1007/s00726-015-1983-4.
- Reinwarth M, Glotzbach B, Tomaszowski M, Fabritz S, Avrutina O, Kolmar H. Oxidative folding of peptides with cystine-knot architectures: kinetic studies and optimization of folding conditions. Chembiochem. 2013;14:137–46. doi:https://doi.org/10.1002/cbic.201200604.
- Thies MJ, Talamo F, Mayer M, Bell S, Ruoppolo M, Marino G, Buchner J. Folding and oxidation of the antibody domain C(H)3. J Mol Biol. 2002;319:1267–77. doi:https://doi.org/10.1016/S0022-2836(02)00375-3.
- White FH. [30] Reduction and reoxidation at disulfide bonds. In: Kaplan N, Colowick N, Hirs C, Timasheff S. Methods in enzymology. Cambridge (MA): Academic Press; 1972. p. 387–92. doi:https://doi.org/10.1016/S0076-6879(72)25033-9.
- Ouellette D, Alessandri L, Chin A, Grinnell C, Tarcsa E, Radziejewski C, Correia I. Studies in serum support rapid formation of disulfide bond between unpaired cysteine residues in the VH domain of an immunoglobulin G1 molecule. Anal Biochem. 2010;397(1):37–47. doi:https://doi.org/10.1016/j.ab.2009.09.027.
- Zhang W, Czupryn MJ. Free sulfhydryl in recombinant monoclonal antibodies. Biotechnol Prog. 2002;18:509–13. doi:https://doi.org/10.1021/bp025511z.
- Bulleid NJ, Ellgaard L. Multiple ways to make disulfides. Trends Biochem Sci. 2011;36:485–92. doi:https://doi.org/10.1016/j.tibs.2011.05.004.
- Feige MJ, Hendershot LM, Buchner J. How antibodies fold. Trends Biochem Sci. 2010;35:189–98. doi:https://doi.org/10.1016/j.tibs.2009.11.005.
- Hatahet F, Ruddock LW. Protein disulfide isomerase: a critical evaluation of its function in disulfide bond formation. Antioxid Redox Signal. 2009;11:2807–50. doi:https://doi.org/10.1089/ars.2009.2466.
- Poole LB. The basics of thiols and cysteines in redox biology and chemistry. Free Radic Biol Med. 2015;80:148–57. doi:https://doi.org/10.1016/j.freeradbiomed.2014.11.013.
- Suzuki Y, Schwartz SL, Mueller NC, Schmitt MJ. Cysteine residues in a yeast viral A/B toxin crucially control host cell killing via pH-triggered disulfide rearrangements. Mol Biol Cell. 2017;28:1123–31. doi:https://doi.org/10.1091/mbc.e16-12-0842.
- Baumann P, Hubbuch J. Downstream process development strategies for effective bioprocesses: trends, progress, and combinatorial approaches. Eng Life Sci. 2017;17:1142–58. doi:https://doi.org/10.1002/elsc.201600033.
- Gronemeyer P, Ditz R, Strube J. Trends in upstream and downstream process development for antibody manufacturing. Bioengineering (Basel). 2014;1:188–212. doi:https://doi.org/10.3390/bioengineering1040188.
- Shukla AA, Hubbard B, Tressel T, Guhan S, Low D. Downstream processing of monoclonal antibodies–application of platform approaches. J Chromatogr B Analyt Technol Biomed Life Sci. 2007;848:28–39.
- Corma A, Ródenas T, Sabater MJ. Aerobic oxidation of thiols to disulfides by heterogeneous gold catalysts. Chem Sci. 2012;3:398–404. doi:https://doi.org/10.1039/C1SC00466B.
- Azuma T, Takeda J, Motoyama N, Okada H. Kinetics of inter-heavy chain disulfide bond formation of liganded and unliganded human immunoglobulin G by radioimmunoassay. Mol Immunol. 1992;29:37–44. doi:https://doi.org/10.1016/0161-5890(92)90154-P.
- Carta R, Tola G. Solubilities of l-Cystine, l-Tyrosine, l-Leucine, and Glycine in Aqueous Solutions at Various pHs and NaCl Concentrations. J Chem Eng Data. 1996;41:414–17. doi:https://doi.org/10.1021/je9501853.
- Liu S. Bioprocess engineering: kinetics, sustainability, and reactor design. 2nd ed. Amsterdam: Elsevier; 2017. doi:https://doi.org/10.1016/B978-0-444-63783-3.
- Rajpal G, Arvan P. Chapter 236 - disulfide bond formation. In: Kastin AJ. Handbook of biologically active peptides. 2nd ed. Boston (MA): Academic Press; 2013. p. 1721–29. doi:https://doi.org/10.1016/B978-0-12-385095-9.00236-0.
- Mamathambika BS, Bardwell JC. Disulfide-linked protein folding pathways. Annu Rev Cell Dev Biol. 2008;24:211–35. doi:https://doi.org/10.1146/annurev.cellbio.24.110707.175333.
- Fischer B, Sumner I, Goodenough P. Isolation, renaturation, and formation of disulfide bonds of eukaryotic proteins expressed in Escherichia coli as inclusion bodies. Biotechnol Bioeng. 1993;41:3–13. doi:https://doi.org/10.1002/bit.260410103.
- Menzella HG, Gramajo HC, Ceccarelli EA. High recovery of prochymosin from inclusion bodies using controlled air oxidation. Protein Expr Purif. 2002;25:248–55. doi:https://doi.org/10.1016/S1046-5928(02)00006-2.
- Naciri M, Kuystermans D, Al-Rubeai M. Monitoring pH and dissolved oxygen in mammalian cell culture using optical sensors. Cytotechnology. 2008;57:245–50. doi:https://doi.org/10.1007/s10616-008-9160-1.
- Kosuri P, Alegre-Cebollada J, Feng J, Kaplan A, Ingles-Prieto A, Badilla CL, Stockwell BR, Sanchez-Ruiz JM, Holmgren A, Fernández JM. Protein folding drives disulfide formation. Cell. 2012;151:794–806.
- Qin M, Wang W, Thirumalai D. Protein folding guides disulfide bond formation. Proc Natl Acad Sci U S A. 2015;112:11241–46. doi:https://doi.org/10.1073/pnas.1503909112.
- Liu M, Zhang Z, Zang T, Spahr C, Cheetham J, Ren D, Zhou ZS. Discovery of undefined protein cross-linking chemistry: a comprehensive methodology utilizing 18 O-labeling and mass spectrometry. Anal Chem. 2013;85:5900–08. doi:https://doi.org/10.1021/ac400666p.
- Wang Z, Rejtar T, Zhou ZS, Karger BL. Desulfurization of cysteine-containing peptides resulting from sample preparation for protein characterization by mass spectrometry. Rapid Commun Mass Spectrom. 2010;24:267–75. doi:https://doi.org/10.1002/rcm.4383.
- Jocelyn PC. Biochemistry of the SH group; the occurrence, chemical properties, metabolism and biological function of thiols and disulphides. London (NY): Academic Press; 1972.
- Arolas JL, Aviles FX, Chang JY, Ventura S. Folding of small disulfide-rich proteins: clarifying the puzzle. Trends Biochem Sci. 2006;31:292–301. doi:https://doi.org/10.1016/j.tibs.2006.03.005.
- Hahn R, Bauerhansl P, Shimahara K, Wizniewski C, Tscheliessnig A, Jungbauer A. Comparison of protein A affinity sorbents II. Mass Transfer Properties J Chromatogr A. 2005;1093:98–110.
- Hober S, Nord K, Linhult M. Protein A chromatography for antibody purification. J Chromatogr B Analyt Technol Biomed Life Sci. 2007;848:40–47. doi:https://doi.org/10.1016/j.jchromb.2006.09.030.
- Liu HF, Ma J, Winter C, Bayer R. Recovery and purification process development for monoclonal antibody production. MAbs. 2010;2:480–99. doi:https://doi.org/10.4161/mabs.2.5.12645.
- Liu H, Chumsae C, Gaza-Bulseco G, Hurkmans K, Radziejewski CH. Ranking the susceptibility of disulfide bonds in human IgG1 antibodies by reduction, differential alkylation, and LC-MS analysis. Anal Chem. 2010;82:5219–26. doi:https://doi.org/10.1021/ac100575n.
- Wypych J, Li M, Guo A, Zhang Z, Martinez T, Allen MJ, Fodor S, Kelner DN, Flynn GC, Liu YD, et al. Human IgG2 antibodies display disulfide-mediated structural isoforms. J Biol Chem. 2008;283:16194–205. doi:https://doi.org/10.1074/jbc.M709987200.
- Hess VL, Szabo A. Theoretical models for the covalent assembly of immunoglobulins. Biophys Chem. 1980;12:243–53. doi:https://doi.org/10.1016/0301-4622(80)80001-9.
- Magnusson CG, Bjornstedt M, Holmgren A. Human IgG is substrate for the thioredoxin system: differential cleavage pattern of interchain disulfide bridges in IgG subclasses. Mol Immunol. 1997;34:709–17. doi:https://doi.org/10.1016/S0161-5890(97)00092-8.