ABSTRACT
Antibodies facilitate targeted cell killing by engaging with immune cells such as natural killer cells through weak binding interactions with Fcγ receptors on the cell surface. Here, we evaluate the binding affinity of the receptor FcγRIIIa V158 (CD16a) for several therapeutic antibody classes, isoforms, and Fc-fusion proteins using an immobilized receptor affinity liquid chromatography (LC) approach coupled with online mass spectrometry (MS) detection. Aglycosylated FcγRIIIa was used in the affinity chromatography and compared with published affinities using glycosylated receptors. Affinity LC-MS differentiated the IgG1 antibodies primarily according to their Fc glycosylation patterns, with highly galactosylated species having greater affinity for the immobilized receptors and thus eluting later from the column (M5< G0F < G0 afucosylated ≅ G1F < G2F). Sialylated species bound weaker to their asialylated counterparts as reported previously. High mannose glycoforms bound weaker than G0F, contrary to previously published studies using glycosylated receptors. Also, increased receptor binding affinity associated with afucosylated antibodies was not observed with the aglycosylated FcγRIIIa. This apparent difference from previous findings highlighted the importance of the glycans on the receptors for mediating stronger binding interactions. Characterization of temperature-stressed samples by LC-MS peptide mapping revealed over 200 chemical and post-translational modifications, but only the Fc glycans, deamidation of EU N325, and an unknown modification to either proline or cysteine residues of the hinge region were found to have a statistically significant impact on binding.
Abbreviations: Antibody-dependent cell-mediated cytotoxicity (ADCC), chimeric antigen receptor (CAR), Chinese hamster ovary (CHO), dithiothreitol (DTT), electrospray ionization (ESI), hydrogen-deuterium exchange (HDX), filter aided-sample preparation (FASP), Fcγ receptor (FcγR), fragment crystallizable (Fc), high-pressure liquid chromatography (HPLC), immunoglobulin G (IgG), liquid chromatography (LC), monoclonal antibody (mAb), mass spectrometry (MS), natural killer (NK), N-glycolylneuraminic acid (NGNA), N-acetylneuraminic acid (NANA), principal component analysis (PCA), surface plasmon resonance (SPR), trifluoroacetic acid (TFA), and extracted mass chromatogram (XMC).
Introduction
Fcγ receptors (FcγR) are membrane-bound glycoproteins belonging to the immunoglobulin (Ig) superfamily that are found on the surfaces of many of the hematopoietic cells of the immune system. These receptors are responsible for the binding of IgG immune complexes and play an important role in modulating both adaptive and innate immune responses. Binding of FcγRs to IgG molecules plays an important role in activation and regulation of immune cells.1,Citation1–3 FcγRIIIa (CD16a) is a low affinity Fc receptor associated with the antibody-dependent cell-mediated cytotoxicity (ADCC) pathway. The FcγRIIIa-V158 allotype used in this study has a higher affinity for both monomeric and immune-complexed IgG1, IgG3, and IgG4 than IIIa-158 F.Citation4,Citation5,Citation6, ADCC results when antibodies recognize and bind to a cell-based target antigen and then recruit natural killer (NK) cells to actively lyse the antigen-expressing target cells. The ADCC response is often associated with NK cells, which express FcγRIIIa receptors on their cell surfaces. FcγRIIIa binds to the crystallizable fragment (Fc) region of an antibody and brings the effector (NK) cell into proximity of the antigen-expressing cell so that it may form a lytic synapse between the NK cell and the antigen-expressing cell.Citation1,Citation2,Citation3,Citation7
Effector function pathways such as ADCC are common secondary mechanism of action for oncology immunotherapies and have been shown to be effective at treating cancers with distinct overexpression of specific cell-based antigen markers using IgG1 based therapeutics.Citation8 Comparisons of in vitro binding assays such as surface plasmon resonance (SPR) with cell-based ADCC assays generally show good correlation between FcγRIIIa binding affinity and the ADCC potency of therapeutic monoclonal antibodies (mAbs).Citation9 The binding of the aFntibody Fc portion to FcγRIIIa is an enabling step in the ADCC pathway, and measuring the affinity of this interaction is an important part of the in vitro assessment of ADCC potency of mAb therapeutics.Citation8
Fc glycosylation affects receptor binding and effector response
The Fc glycan composition at the conserved N-glycosylation site (N297, EU numbering system) on the heavy chain plays an important and multifaceted role in regulating effector function and potency of mAb therapeutics. The Fc glycan composition of human IgG antibodies can be quite heterogeneous, owing to the hundreds of possible glycan combinations on the two heavy chains of the Fc.Citation10,Citation11 The glycan heterogeneity of the Fc region depends on several factors, including the host organism used for antibody expression and the types of glyco-processing machinery expressed by the host cell line.Citation10,Citation12,Citation13 Supplemental Figure S1 shows the nomenclature and structures of glycans commonly found on the Fc glycan sites. The nomenclature was adopted from Zhang and Shah,Citation14 but the antenna number upfront was omitted to save space in some figures and in the text. Biantennary glycans (A2) constituted the vast majority of detected glycans and were the only glycans quantitatively assessed in this study; therefore, the antenna number (A2) was omitted. These glycans are typically biantennary complex structures featuring two core N-acetylglucosamine (GlcNAc) residues and then branching at the third mannose residue into two arms via α-1,3- and α-1,6 linkages.
Structural studies of the Fc domain have shown that the glycans on N297 have a significant impact on the structure of the CH2 domain and affect the conformational dynamics and flexibility of this region.Citation15,Citation16,Citation17 The CH2 domain has a high degree of flexibility compared to the CH3 domain, which has a relatively static, rigid structure. Glycosylation of the CH2 domain acts to stabilize its structure and direct folding to specific conformations that permit binding to the various Fc receptors.Citation18,Citation19 Antibodies without Fc glycans show no affinity for Fcγ receptors, highlighting the importance of glycosylation for proper folding to yield functional mAbs capable of eliciting an immune response.Citation17,Citation18,Citation20,Citation21,Citation22,Citation23,Citation24
Afucosylation
Wild-type human IgGs are typically characterized by high levels of core fucosylation on the Fc glycans. Early work studying the potencies of therapeutic antibody glycoforms revealed that species lacking core fucose on the Fc glycan showed a significant increase in FcγRIIIa binding and ADCC potency (up to 50-fold increase).Citation25,Citation26,Citation27,Citation28 The enhanced affinity is attributed to a structural change in the afucosylated species that allows an additional favorable carbohydrate-carbohydrate interaction between the Fc and a glycan on FcγRIIIaCitation29,Citation30 as well as an additional sterically mediated H-bond between Tyr296 and FcγRIIIa. Increased ADCC potency is generally observed if either one or both Fc glycans is afucosylated with nearly identical enhancement.Citation31 Antibody molecules with high mannose glycans, also lacking the core fucose (Figure S1), also exhibited higher FcγRIIIa binding and ADCC activity.Citation32
Galactosylation
IgG1 antibodies having terminal galactose residues (e.g., G1F or G2F) show increased affinity for FcγRIIIa (CD16a), which are responsible for ADCC.Citation33,Citation34,Citation35 In the crystal structures of Fcs with larger biantennary glycans such as those with terminal galactose (e.g., G2F), a more open structure is observed as the CH2 region bends out from the CH3 domain and exposes a portion of the glycan where it may interact in receptor binding.Citation15 It is worth noting that there are differences observed in accessibility and flexibility of the α-1,3 and α-1,6 arms observed by both NMR and crystallography.Citation23 Shorter glycans (e.g., G0F) typically exhibit more “closed” conformations in the Fc region and also lower affinity for FcγRIIa and FcγRIIIa receptors in IgG1-based mAbs, indicating that these structural effects are important considerations for understanding the potency and efficacy of mAb based therapeutics.Citation14 It is worth noting that IgG glycans with α-1,3-galactose (as opposed to terminal) are not found in humans, and recombinant mAbs from mammalian cells such as murine Sp2/0 and NS0 lines that produce such glycoforms can trigger anaphylactic responses in some populations with allergies to α-1,3-galactose.Citation36,Citation37
Sialylation
Terminal sialic acid residues on Fc glycans act as a regulatory mechanism for controlling the pro- or anti-inflammatory functionality of IgGs.Citation38,Citation39,Citation40,Citation41 The effects of Fc sialylation on FcγRIIIa binding can vary according to the sialic acid linkage type (α-2,3- vs α-2,6-).41 Fc sialylation predominantly occurs after secretion from plasma B cells by exogenous post-translational modification machinery in circulation.Citation39 In this regard, sialylation can serve as a functional switch to change the behavior of antibodies from inflammatory to anti-inflammatory. Sialic acids are thus an important biomarker for understanding inflammatory diseasesCitation42,Citation43 as well as a critical attribute for tuning the function of therapeutic mAbs.
Sialic acid connectivity is different in human, hamster, and mouse cell lines, leading to differences in Fc stability and binding to FcγRIIIa V158, as follows. Terminal sialylation on N-glycans can take place through either an alpha 2,3-linkage or an alpha 2,6-linkage to galactose. Sialic acids on IgG Fc purified from human serum (made by human cells) are almost exclusively alpha 2,6-linked and on the 3-arm (Figure S5). Recombinant IgG molecules expressed in Chinese hamster ovary (CHO) cells, however, have sialic acids attached through alpha 2,3-linkages (because of the lack of the alpha 2,6-sialyltransferase gene in CHO) and on the 6-arm. Sialic acid-containing glycans were found by hydrogen-deuterium exchange (HDX) to destabilize the CH2 domain in CHO-expressed IgG, but not human-derived IgG.Citation16,Citation40 Only alpha 2,3-linked sialic acid on the 6-arm (the major sialylated glycans in CHO expressed IgG1) has been observed to destabilize the CH2 domain, presumably because of the steric effect that decreases the glycan-CH2 domain interaction. The alpha 2,6-linked sialic acid on the 3-arm (the major sialylated glycan in human-derived IgG), and the alpha 2,3-linked sialic acid on the 3-arm, do not have this destabilizing effect.Citation16 Sialic acid connectivity in IgG derived from mouse cell lines is similar to human, but N-glycolylneuraminic acid sialic acid residue (NGNA, Neu5Gc, mass 307 Da) is typically found instead of N-acetylneuraminic acid (NANA, mass 291 Da) for human.
IgG antibodies produced in CHO and containing terminal alpha 2,3-linked sialic acid on the 6-arm of their Fc N-glycans have been shown to decrease ADCC. IgG antibodies produced with the 2,6-linked sialic acid on the 3-arm – the major sialylated glycan produced by human cell lines – have been shown to increase ADCC.Citation40,Citation44,Citation45,Citation46 IgGs stably expressed in transfected mouse myeloma cells also showed decreased ADCC.Citation40
While glycosylation has an important role in defining the affinity of IgGs for Fc receptors, differences in IgG subclass (IgG1, IgG2, IgG3, and IgG4) also have a significant impact. The subclasses of IgG not only differ primarily in the size of the hinge region but also have subtle differences in the sequences of the CH2 portion of the conserved Fc region.45 Typically, IgG1 variants are associated with eliciting an ADCC response from immune cells, but other IgG subclasses can bind to FcγRIIIa.
Here, we describe an affinity chromatography approach using immobilized FcγRIIIa receptors with online mass spectrometry (MS) and offline fraction collection to investigate factors affecting FcγRIIIa affinity in a single non-targeted large-scale experiment. The high-throughput nature of this type of profiling can act as an alternative method for initial ADCC screening, which can be expensive and time consuming. Even though true cell-based potency measurements must still be performed at some point, higher throughput and decreased cost can be achieved with FcγRIIIa columns.
Results
Differentiation of antibody glycoforms based on Fcγ receptor affinity chromatography
The affinity of antibodies for FcγRIIIa is largely dependent on interactions of the antibody Fc glycan at position N297.Citation24 The glycan composition at N297 is heterogeneous, resulting in a mixture of glycoforms that have significant differences in binding affinity and ADCC potency. The selectivity of these receptors for differences in antibody glycoforms presents a unique approach for differentiation between mAb glycoforms according to their affinity. Recently, immobilization of glycosylated FcγRIIIa receptor into a chromatographic resin support and online MS of intact antibodies allowed for separation of specific mAb glycoforms and provided insight into their receptor affinity and potency.46 In this work, we used a similar approach using a commercially available affinity column containing immobilized aglycosylated FcγRIIIa receptors. A few glycosylated FcɣRIIIa columns, including the one described by Lippold et al.,46 were custom made and are not widely available to the research community. The commercial column we used is available to a large number of users who should benefit from the results of this study. shows the overall workflow of the native affinity LC-MS analyses described here. provides a list of all the therapeutic proteins used in these studies and a brief description of their molecular characteristics. shows the separation of individual glycoforms of mAb1 (rituximab) based on their affinity for the immobilized recombinant FcγRIIIa. The overall deconvoluted mass spectrum indicating the molecular weights of each observed glycoform is shown in . The UV absorbance trace at 280 nm for mAb1 is depicted in . The chromatogram has three well-defined peaks (labeled 2–4), a less resolved shoulder peak (1), and two low abundance pre-peaks (labeled as*). The most abundant glycoforms present are assigned based on their masses and consist primarily of biantennary complexes having core fucose and varying numbers of terminal galactose residues (+162 Da each). Each mAb Fc has one glycan on each heavy chain, as indicated in the cartoon structures and labeled accordingly (in the format glycan1/glycan2). shows the extracted mass chromatograms (XMC) of the most abundant glycoforms. The XMCs are generated by performing m/z deconvolution into mass and plotting the retention time and intensities of all ion charge states that correspond to a single deconvoluted mass. This extracted mass chromatogram feature is available in the ReSpect™ Intact Protein Deconvolution software package in Chromeleon 7 (See Materials and Methods section and Supplemental information for more details). These XMCs allow for differentiation of the individual glycoforms from the total chromatogram according to their masses and assessment of the affinity of each species based on the extracted retention time(s).
Table 1. Antibody samples used in this study
Figure 1. Overall analytical workflow of the native affinity LC-MS analyses and fraction collection for LC-MS peptide mapping
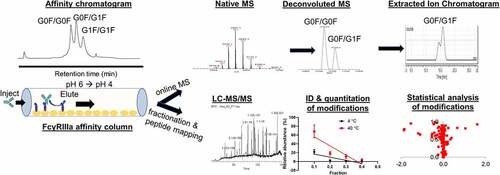
Figure 2. FcγRIIIa affinity LC-UV-MS characterization of rituximab. (a) Deconvoluted mass spectrum of entire sample; (b) LC chromatogram with UV detection at 280 nm. Asterisk indicates elution of truncated single antenna (A1G0F) species. (c) Extracted mass chromatograms (XMC) for individual intact antibody molecules with M5/M5, G0F/G0F, G0F/G1F, G1F/G1F, G1F/G2F, and G2F/G2F glycans
From the individual retention times, it is evident that galactosylation plays an important role in binding affinity to the receptor, with higher levels of galactose leading to increased affinity. This finding is consistent with previous studies and serves as a benchmark for the method.Citation32,Citation33,Citation34 Several of the glycoforms contain multiple peaks in the XMCs, for example, G1F/G1F. This is likely due to the possibility of positional isomers with identical masses (i.e., G1F/G1F and G0F/G2F contain the same number sugars but are arranged differently). Additionally, only one Fc glycan interacts with FcγRIIIa at a time,Citation47 such that asymmetrically glycosylated mAbs can have one side with higher affinity than the other, which can present as two different affinity peaks.
Two core afucosylated species are also detected (M5/M5 and G0F/G0). These types of afucosylated species are reported to have significantly increased affinity and ADCC potency,Citation24,Citation31 but this does not appear to be reflected in the retention times of these species. Most notably, the M5/M5 glycoform reported to have 4-6-fold increased affinity for FcγRIIIa,Citation31 but has the earliest elution time of all the glycoforms.
Only a slight increase in affinity of the afucosylated G0F/G0 glycoform relative to the fucosylated G0F/G0F counterpart was observed with the magnitude of the retention time shift comparable to the G1F/G1F glycoform, instead of the 10–50-fold increase that has been reported in literature.Citation24 We speculate that this deviation from the expected retention behavior may stem from the fact that the FcγRIIIa receptors on the column are aglycosylated, in contrast to the glycosylated FcγRs used in most published binding studies.
Antibody molecules with sialylation were observed with slightly earlier elution times relative to their asialylated counterparts. For example, in , the XMC for the glycoform G0F/S1G1F has the same elution time as G1F/G1F. The structure of G0F/S1G1F is analogous to G0F/G2F with a terminal sialic acid on one galactose, but its retention time is earlier (similar to G1F). This suggests that the sialic acid residue may block or inhibit the interaction of the second galactose with FcγRIIIa, making it more analogous to a G1F glycan. An isomeric form G1F/S1G0F is also possible according to intact mass, but this species (S1G0F) was not detected in the subsequent peptide mapping experiments described below. mAb1 (rituximab) is produced in CHO cells and therefore should mainly contain terminal alpha 2,3-linked sialic acids on the 6-arm. These types of sialic acid linkages have been observed in previous studies to bind weaker to FcγRIIIa V158 and decrease ADCC.Citation16,Citation40,Citation43
Assessment of Fc domain released by limited proteolysis
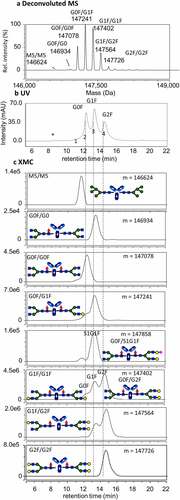
In IgG-based therapeutics, glycosylation most commonly occurs on N297 of the Fc region. However, additional glycosylation sometimes occurs in antigen-binding fragment (Fab) regions (e.g., cetuximab) or glycosylated receptors are attached to Fc to create Fc-fusion proteins (e.g., etanercept).Citation48 The additional glycosylation generally does not affect the interactions with Fcγ receptors because of their distance from the binding site, but it can lead to complications in interpreting the Fc glycan composition at the intact or released glycan levels due to increased heterogeneity and ambiguity in glycan location. Limited proteolysis can be used to address the issue of glycan ambiguity by cleaving the binding regions above the hinge region from Fc region.Citation49,Citation50,Citation51 shows the FcγRIIIa affinity chromatogram of a heavily glycosylated Fc-fusion protein before and after cleavage of the Fc region by limited proteolysis with Lys-C endoproteinase. A diagram of the Fc-fusion protein, which consists of two identical target binding domains fused to an IgG1 Fc domain, is inset in . The binding domains have several O- and N-linked consensus glycosylation sites, resulting in a highly heterogeneous distribution of total glycoforms. Performing the digestion reaction under native conditions and limiting the reaction time to 20 min resulted in a single cleavage at an exposed site between K223/T224. Cleavage of the binding domains above the hinge preserved the FcγRIIIa binding domain and allowed for affinity analysis of the Fc region by itself and with greater resolution between the different glycan peaks. shows the m/z and deconvoluted mass spectra of the main peak in the affinity chromatogram from the intact Fc fusion protein. Prior to cleavage of the binding domains, the m/z distribution is quite broad due to the presence of multiple heterogeneous N and O-glycosylation sites on the two binding domains. The average mass – 113 kDa – can be vaguely discerned, but the glycan heterogeneity results in many glycoforms with similar or identical masses that cannot be adequately resolved and assigned to individual glycoforms. Although information about the measured whole molecular mass may be useful for identification, it has a limited value for more detailed characterization. We also note a modest improvement in chromatographic resolution of the glycan peaks when measuring the cleaved Fc region compared to the intact protein. shows the mass spectra of the Fc portion alone following limited proteolysis with Lys-C. The mass spectra could be much more easily resolved and accurately deconvoluted, allowing for accurate assignment of the Fc glycans present that contribute to the peaks in the chromatograms shown in .
Figure 3. FcγRIIIa affinity chromatogram with (a) UV detection and (b) and (c) mass spectrometric detection of heavily glycosylated protein conjugated to IgG1 Fc (Fc-fusion protein 2). (a) Fc-fusion protein 2 analyzed as intact and cleaved by limited proteolysis with Lys-C endopeptidase. (b) Electrospray ionization (ESI) mass spectrum of intact Fc-fusion protein 2 (left) and deconvoluted mass spectrum (right). (c) ESI mass spectrum of Fc region generated by the limited proteolysis (left) and deconvoluted mass spectrum (right)
Correlation of the affinity chromatography results to AlphaLISA binding assay results
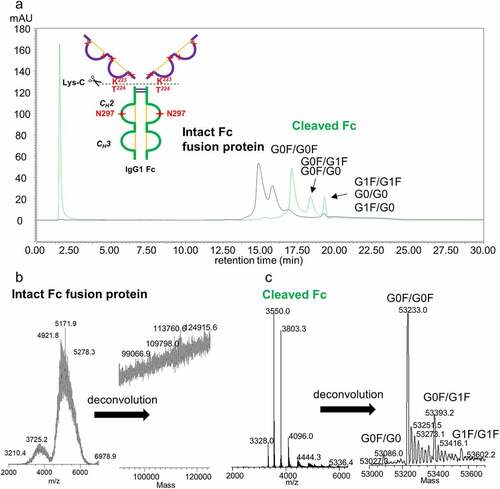
Measuring differences in FcγRIIIa binding affinity based on retention time is an attractive method for assessing different IgG subclasses, modalities and specific IgG1 glycan contributions, especially when glycoforms can be identified according to their unique masses. shows the affinity chromatograms of four therapeutic proteins ranked according to their average weighted retention times (weighted based on peak areas). A linear correlation of average retention time to % relative affinity from the AlphaLISA binding assay is shown in . Both assays report similar trends in which the aglycosylated Fc fusion protein shows no binding, mAb3 (IgG2) shows very weak binding, and increased binding for mAb2 (IgG1 with high levels of galactosylation) relative to mAb1. This provides validation for the use of the FcγRIIIa affinity chromatography method for characterizing the impact of product attributes that affect binding. It should be noted that two of the samples tested showed elevated activity in the AlphaLISA binding assay: a highly glycosylated Fc fusion protein, and an IgG1 species with increased levels of immature Fc glycans (e.g., high mannose, afucosylated). We speculate that this departure in strong correlation with the chromatography method may arise from the lack of glycosylation on the on-column immobilized FcγRIIIa receptor and presence of glycosylation on FcγRIIIa used for AlphaLISA binding assay.
Figure 4. (a) FcγRIIIa affinity LC-UV chromatograms of four representative therapeutic protein modalities: aglycosylated Fc fusion protein (purple), IgG2 mAb (red), and two IgG1 mAbs with different glycosylation patterns (blue and green). (b) Correlation plots showing relationship between relative binding affinity to average weighted retention time with outlier samples removed. (c) Correlation plots of relative binding affinity to weighted retention time including outlier samples
Differences in receptor affinities contribute to variation in effector functions for IgG subclasses
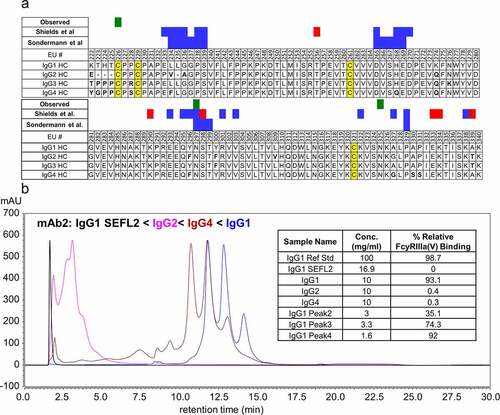
IgG subclasses (IgG1, IgG2, IgG3, and IgG4) have different roles in the immune system and elicit different effector functions based on their ability to bind to different Fc receptors and types of antigens.Citation4,Citation52 The Fc region among each subclass have high sequence homology but differ in a few key residues, which dictates their affinity for the various Fc receptors. The FcγRIIIa receptor binding sites are located on the lower hinge and CH2 domain, including AA positions 235–239, 265–269, 297–299, and 329 in EU numbering system.Citation47,Citation53 Another set of binding sites was identified by Shields et al.Citation54 One of them is a conserved N-glycosylation site at asparagine 297 (N297, EU numbering system). Aligned sequences of the hinge and CH2 regions of IgG subclasses 1–4 are shown in with the residues previously reported to be involved for binding to FcγRIIIa binding sites shown in blue and red shading (Sondermann et al.Citation47 and Shields et al.Citation54). Green shading marks residues that were identified in this study as impacting binding to FcγRIIIa when modified. Noteworthy differences include L234 (IgG2, IgG4) and H268 (IgG4), which have differences in residues directly involved in FcγRIIIa binding, and several differences in amino acids adjacent to the binding domains. To investigate the role of different Fc regions, variants of mAb2 with identical Fab regions, but the Fc region from IgG1, IgG2, IgG4, and an engineered stable effector functionless variant (SEFL2) were separated on the FcγRIIIa affinity column (). For reference, the SEFL2 IgG1 modality is an aglycosylated IgG1 variant (N297G) with an additional engineered hinge disulfide bond designed to have no ADCC function.Citation55,Citation56 Based on retention times, these data show that the engineered SEFL2 IgG1 modality has the lowest affinity, followed by IgG2, IgG4, and then IgG1. The overall elution order of these modalities IgG1 SEFL2 < IgG2 < IgG4< IgG1 is consistent with reported literature values for FcγRIIIa binding affinities.Citation4,Citation44,Citation57 However, the elution time for the IgG4 variant appears to overestimate the binding affinity, having an elution time only slightly earlier than the IgG1 variant. We hypothesize that this increased apparent affinity is related to the aglycosylated nature of the receptor on the column and should not be used to assess FcγRIIIa affinity for IgG4 subclass mAbs. Typically, IgG4 and IgG2 mAbs have only weak affinity for FcγRIIIa and do not lead to significant ADCC activity.Citation4
Figure 5. (a) Aligned sequences of CH2 region of human IgG heavy chains (EU numbering system). IgG Fc residues involved in FcγRIIIa binding are highlighted in green (measured experimentally in this study) and blue and red (reported in literature), and disulfide bonds are highlighted in yellow. Residues that differ from the IgG1 sequence are indicated in bold. (b) UV chromatogram (λ = 280 nm) of four variants of mAb2 having identical Fab regions and differing Fc regions corresponding to IgG1 (blue), IgG2 (magenta), IgG4 (maroon), and IgG1 SEFL2 with N297G mutation (black). The inset shows relative FcγRIIIa binding affinity values from AlphaLISA measurements. Affinity is measured relative to the IgG1 variant of mAb2 reference standard material
Disulfide isoform impact on FcγRIIIa binding in an IgG2 mAb
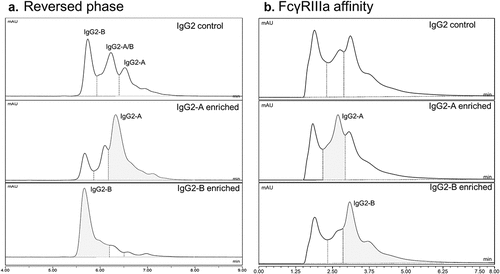
IgG2 mAbs do not produce significant ADCC activity through FcγRIIIa binding and commonly find use against targets when an immune response is not desired, e.g., blocking of targets.Citation52,Citation58 Given the heterogeneous nature of the IgG2 affinity chromatograms (see ), the IgG2 mab3 was selected for further characterization. Previous studies using reversed-phase LC-UV-MS analysis of this IgG2 mAb identified the presence of several naturally occurring disulfide bond structural isoforms, which can affect efficacy and Fc receptor binding behavior.Citation59,Citation60,Citation61 shows a side-by-side comparison of reversed-phase and FcγRIIIa affinity separations of mAb3 disulfide isoforms. It is interesting to note that the native-like conditions used in the affinity method are still capable of distinguishing between these structural isoforms, suggesting these isoforms have an effect on the structure of the hinge region. That these isoforms have different elution times highlights the importance of the hinge structure in mediating the binding interaction between the Fc and FcγRIIIa.
Figure 6. (a) Reversed-phase chromatograms with UV detection at 280 nm showing separation of a mixture of naturally occurring IgG2 disulfide variants and purified samples enriched for IgG2-A and IgG2-B. (b) FcγRIIIa affinity UV chromatograms of the same samples showing partial resolution of IgG2 disulfide isoforms. Mass spectra of the peak between 1–2 min contained relatively small abundance of protein ions, suggesting that the peak may be due to light scattering on the earlier eluting formulation excipients (sucrose, polysorbate) with different refractive indexes
Identification of modifications negatively impacting FcγRIIIa binding after stress conditions
Post-translational and chemical modifications can occur during all stages of production, storage, and administration of mAb therapeutics. Chemical modifications to residues located in or near receptor binding sites can affect the affinity of the antibody-receptor interactions, leading to changes in ADCC and efficacy. Such modifications can be induced by subjecting antibodies to typical conditions of production and storage and also to various stress conditions, such as elevated temperature, exposure to acidic or basic pH, and UV light exposure, imposed during elucidation of forced degradation pathways. In the case of the heat-stressed mAb1 sample (40°C for 6 weeks) used in this study, over 200 modifications were detected by LC-MS/MS peptide mapping with sufficient signal:noise and precision for confident identification. The majority of these modifications have little or no effect on binding and efficacy. To differentiate modifications having an effect on FcγRIIIa from those that do not, several statistical ranking approaches were used.
shows the affinity chromatograms of mAb1 with temperature stress (40°C for 6 weeks) compared to control sample that was maintained at 4°C. The main peaks from each sample were collected as four fractions (F1–F4) as indicated by dashed lines. Stress at 40°C led to a slight increase (2% → 4%) in the peak area of the lowest affinity fraction F1 with low FcγRIIIa binding. Peptide mapping was performed on each collected fraction to determine a trend (correlation) for relative percentages of modifications in fractions (). Enrichment of specific modifications in the low affinity fractions would indicate that these attributes have a negative impact on FcγRIIIa binding. Several examples of modifications with both negative and positive slopes are shown in . The fractions 1, 2, 3, and 4 were assigned values 0.1, 0.2, 0.3, and 0.4, respectively, so that the slope values could be easily comparable to R2, and the sum of R2 + absolute value of the slope could be used as a score to sort the modification list and for epitope mapping described later. Modifications with a positive slope (e.g., galactosylation and sialylation) indicates a favorable effect on receptor binding, while those with negative slopes (e.g., deamidation) indicate a negative effect on binding.
Figure 7. (a) Affinity UV chromatograms highlighting changes in peak intensities of mAb1 following heat stress at 40°C for 6 weeks (red trace) relative to 4°C control sample (black trace). (b–g) Changes in relative abundance of selected modifications in FcγRIII affinity fractions F1, F2, F3, and F4. (h, i) R squared versus slope of change for modifications percentages in the fractions for 4°C. R squared versus slope determined from linear regressions of all detected modifications in the fractions. In all panels, red color indicates heat-stressed samples (40°C, 6 weeks) and black indicates the 4°C control samples
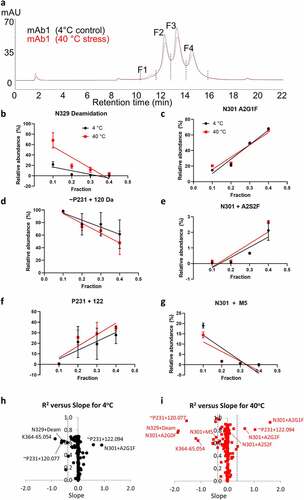
The most enriched modifications in F1 are deamidation of N329 (EU N325), an unknown modification attributed to P231 (EU P227) in the hinge motif EU C226PPC229, and several N-glycans on the conserved glycosylation site at N301 (EU N297). The N-glycans enriched in the low affinity fraction F1 are mostly immature sugars, having only a single antenna (A1G0F and A1G1F) and high mannose (M5) compositions. As noted before, the presence of M5 glycans in this early eluting fraction is surprising and may be a due to the aglycosylated FcγRIIIa receptor. Deamidation of asparagine is a common modification that occurs during production and storage of antibodies in conditions at or above neutral pH. Several deamidation modifications were detected, but only N329 was found to be preferentially enriched in F1, indicating that the other asparagine residues are not critical for FcγRIIIa binding. The modifications detected on the peptide containing P231 presented a challenge in the data interpretation. There are no common chemical modifications with a change in mass of 120 or 122 Da that could be attributed to proline. The +120 Da unknown modification assigned to P231 could also be attributed to cysteinylation (+119 Da) and a free thiol -SH group (+1 Da) on one of the adjacent cysteine residues, but the MS/MS data for this peptide cannot unambiguously determine its location (see supporting information Figure S6). As a result, we hypothesize that the unknown modifications may instead be on the adjacent C230, as cysteine is potentially a more reactive residue, and the difference in mass of 120 and 122 Da corresponds to an unpaired disulfide bond in this CPPC segment below the hinge in addition to the unknown 120 Da modification. If ~P231 + 122 Da represents an unpaired disulfide bond and ~P231 + 120 Da a fully paired structure, this suggests that the unpaired structure is actually more favorable for FcγRIIIa binding. This conclusion seems at first counterintuitive as unpaired disulfides are typically associated with misfolded or degraded structures, but structural studies have observed ambiguity in the pairing of the disulfides in this region,Citation62 and only one intact pair is required for ADCC.Citation63 This FcγRIIIa affinity approach provides a more refined view into the role of disulfide connectivity in receptor binding.
The slopes of chemical modifications (N329 deamidation) in the fractions were higher after stress (red lines in , d, f), while slopes for glycoforms before and after stress () remain similar, indicating that slopes are mainly defined by the effects of glycans on affinity to the receptor and presence in the fractions (F1, F2, F3, and F4). A slight increase in the levels of high affinity glycans, such as A2G0 (afucosylated A2G0, Figure S4) and A2S2F (highly galactosylated and sialylated, ) were also observed in F1 with heat stress (Figure S4). This suggests that antibody molecules with these glycoforms may be more susceptible to these chemical modifications. Hydrogen-deuterium exchange experiments have identified certain afucosylated and sialylated mAb species as having increased conformational flexibility and solvent exposure in the CH2 region,Citation16,Citation40,Citation49 which offers a possible explanation for their increased levels of chemical modification. A plot of slope versus R2 for all detected modifications is shown in for the 4°C and 40°C stressed samples, respectively. Modifications that negatively impact binding and have a negative slope appear in the left-hand corner, while modifications positively impacting binding appear in the right portion of the plot.
Bivariate statistical approaches such as volcano plots have previously been used for investigating critical attributes in systems with binary outcomes (e.g., treated vs control or bound vs unbound),Citation64 but the multiple affinity LC peaks present in the FcγRIIIa chromatograms make this type of analysis difficult for these data (Figure S3). In addition to the volcano plot, principal component analysis (PCA) was evaluated to consider multiple variables (i.e., fractions) to correlate which modifications were most associated with specific affinity-separated fractions. Figure S7 shows a PCA biplot of collected affinity fractions 1–4 and the modifications detected by peptide mapping of each fraction. Component 1 (x-axis) represents replicates and component 2 (y-axis) is defined by affinity (fraction numbers) as follows. The strength of the correlation with an individual modification to the affinity fraction is proportional to the distance between the two points. Replicate peptide mapping analyses from fraction F1 all cluster toward the top half of the plot, with many of the same negatively impacting modifications (e.g., EU N329 deamidation, M5 glycans, and truncated single antenna EU N297 glycans) found in this portion of the plot as well. Conversely, high affinity glycans on EU N297 such as A2G2F and A2G0 (afucosylated) are clustered toward the bottom of the plot nearest to the high affinity fractions F3 and F4, indicating their positive effect on FcγRIIIa affinity. Modifications on the left and right extremes of the plot are most strongly correlated with replicate number, indicating that these are likely due to sample preparation variability and can be treated as noise or digestion artifacts.
A summary of the identified critical attributes from all statistical analyses is shown in . Six metrics were chosen for evaluating the significance of the data to determine if the modifications were critical to FcγRIIIa binding. Three P-values were calculated for each modification comparing the modification abundance in the low affinity fraction (F1) to the high affinity fraction (F4), P-value comparing F1 modification abundance under heat stress (40°C 6 weeks) and control (4°C), and modification abundance in F2 vs F4 after heat stress. A P-value ≤ 0.05 was chosen as the threshold value for significance. In addition to P-values, several other metrics were also included based on the slope analysis shown in utilizing the magnitude of the slope across all four fractions, the R2 coefficient of the regression, and the relative standard deviation in the relative abundances in F1 from all replicate analyses. The evaluation revealed that the 11 modifications on three residues of mAb1 ~ P231 (EU P227), N301 (EU N297), and N329 (EU N325) affect binding with statistical significance ().
Table 2. Summary of statistical analyses to determine attribute criticality, statistically significant values are shown in green
Discussion
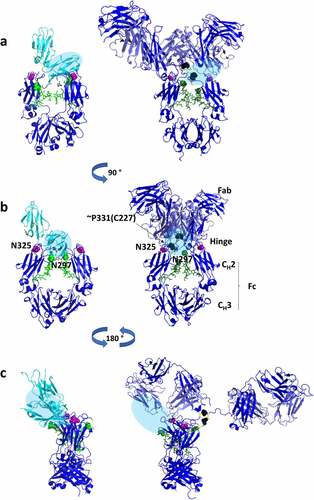
The residues that were identified as statistically significant to FcγRIIIa binding from the peptide mapping experiments are shown as a paratope map in , along with residues previously identified from crystallography and mutational studies as being involved in receptor binding.Citation46,Citation58 Each of the identified residues are in close proximity to the previously identified binding domains for IgG1 to FcγRIIIa. The proximity to the binding domains is further illustrated in , which shows the IgG1 Fc region bound to FcγRIIIa (PDB 5vu0) and an approximation of the binding domain superimposed on the intact antibody structure (PDB 1hzh). The proximity to the known binding domains of FcγRIIIa in both one-dimensional sequence space and three-dimensional conformational space provides additional confidence in the identification of these attributes as critical for binding. Out of 200+ modifications that were observed after temperature stress, modifications to only three residues were found to have a significant impact on FcγRIIIa binding. Other stresses not described here (UV light, oxidation, pH extremes) also showed very little decrease in binding, indicating that the Fc region is remarkably stable to typical (and atypical) stresses in biopharmaceutical processes. In some statistical analyses (Figure S3), a few chemical modifications (Y86, D166) and clipping (not shown here) appeared on residues in the Fab region as impacting binding to the receptor, but with low fold change and/or statistical significance. Those modifications suggested that Fabs may be also involved in binding to the FcγRIIIa, but more studies are needed to confirm those observations.
Figure 8. Paratope map of heavy chain on mAb1 for binding by the aglycosylated FcγRIIIa receptor immobilized in affinity column. R2 + slope of modification percentages versus fraction number in collected affinity chromatography fractions were used as y-axis, and amino acid residue numbers as x-numbers. The residue numbers are 4 higher relative to EU numbering. *Residues impacting binding according to Shields et al. 2001. Mutations at these residues decreased (blue) and increased (red) binding to FcγRIIIa. **Residues with modifications increasing and decreasing binding to FcγRIIIa as measured in this study
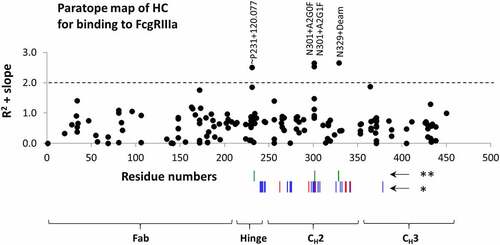
Figure 9. Three-dimensional structures of IgG1 Fc region bound to soluble FcγRIIIa (PDB 5vu0) (left) and the crystal structure of an intact IgG1 molecule (PDB 1hzh).Citation59 The location of the FcγRIIIa binding interaction is shown as a blue oval and mapped onto the intact IgG structure. Modifications on residues in EU numbering that were found to be critical in our study of mAb1 IgG1 antibody are also shown as colored spheres on the structures indicating their location relative to FcγRIIIa. Deamidation (N325) is shown in magenta, unknown modification (~P227) is shown in black, and glycosylation (N297) is shown in green
As mentioned above, carbohydrate-carbohydrate interactions between Fc-glycans and receptor glycans (particularly at N162) have been implicated in the increased affinity of afucosylated antibodies.Citation28,Citation29 Lack of the receptor N162 glycosylation appears to have reduced the affinity for afucosylated antibodies by over one order of magnitude, bringing the affinity down to the same level as for native IgG. Our measurements using the affinity chromatography with aglycosylated FcγRIIIa receptor agree with these earlier studies using SPR with aglycosylated FcγRIIIa.Citation28,Citation29 Additionally, in earlier studies, significant differences were revealed in rituximab binding to FcγRIIIa receptors expressed in different cell lines and with different glycosylation profiles, including CHO, human (HEK293) and mouse (NS0) cell lines, highlighting importance of receptor glycosylation profile.Citation65
Several observations from the studies using an aglycosylated FcγRIIIa affinity column appeared to deviate from more traditional binding assay results and published literature binding data using glycosylated FcγRIIIa. Notable examples of this being: (1) the lower than expected affinity for afucosylated species (e.g., high mannose, G0, G1 glycoforms), (2) no difference in affinity of sialylated and asialylated species, and (3) increased affinity for IgG4 mAbs. While this imposes a limit on the scope of use to species that do not appear to deviate from expected behavior (e.g., galactosylated IgG1 mAbs with core fucose), it also highlights the importance of receptor glycosylation in mediating binding interactions, and ultimately ADCC activity by immune cells. FcγRs have heterogeneous glycosylation patterns that can vary significantly based on their origin (e.g., isolated from different cell types or expression systems), which has been shown to have a significant impact on binding and effector function response.Citation66,Citation67,Citation68,Citation69,Citation70 Glyco-engineering of mAbs to tune the levels of effector response has become a widespread practice in the biopharmaceutical industry,Citation71 and more recently, similar methods have been applied to modulating the glycosylation of FcγRs.Citation72,Citation73 This approach of glyco-engineering FcγRs has emerged as a valuable tool for designing next generation cell-based immunotherapies such as chimeric antigen receptor (CAR) NK cells by providing a lever to improve their efficacy and predictability.Citation66 Such studies aimed at developing a better understanding of the impact of Fc receptor glycosylation on effector response will likely prove useful in the development of promising new targeted cellular therapeutics such as CAR NKs.Citation66
In conclusion, this analysis represents a powerful and systematic approach to identify attributes (known attributes or de novo) and gain a more comprehensive understanding of product quality attributes and the degradation pathways relevant to the development of biotherapeutics.
Materials and methods
Antibody samples
Rituximab samples were manufactured by Genentech/Roche Group (10 mg/mL in pH 6.5 formulation) and purchased from commercial sources. For additional drug information, see www.accessdata.fda.gov/drugsatfda_docs/label/2019/103705s5457lbl.pdf. For rituximab heat-stress studies, aliquots of the final drug product in formulation buffer were incubated at 40°C for up to six weeks. An equivalent aliquot was stored at 4°C to serve as a control. All other IgG samples were produced in-house at Amgen by recombinant expression technology in CHO cell lines, described in .
FcγRIIIa affinity LC-MS
A diagram of the experimental workflow is shown in . FcγRIIIa affinity chromatography separation was performed on an Agilent 1290 Infinity II HPLC (Agilent Technologies, Santa Clara, CA) using a TSKgel FcR-IIIA-NPR column, 4.6 mm ID x 7.5 cm L, (P/N 0023513, TOSOH Bioscience, King of Prussia, PA). HPLC mobile phases consisted of 50 mM ammonium acetate (Sigma Aldrich, St. Louis, MO) pH = 6 (phase A) and 50 mM ammonium acetate pH = 4 (phase B) in LC/MS grade water (Optima, Fisher Scientific, Waltham, MA). Substitution of ammonium acetate for the manufacturer recommended mobile phases (50 mM sodium citrate) resulted in a comparable peak resolution and identical elution order. For HPLC separation, 20 µg of protein was loaded onto the FcγRIIIa column and eluted by a 30-min pH gradient at a flow rate of 300 µL/min. The elution gradient is as follows: 0% B for 2 min followed by a linear gradient from 0–30% B over 18 min, then a step to 100% B for 5 min, and finally stepped to 0% B for 5 min to recondition the column.
On-line with the affinity chromatography, MS analysis () of intact proteins was performed using an Orbitrap Q Exactive Plus™ Biopharma (Thermo Scientific, San Jose, CA) equipped with a Thermo HESI-II ESI probe without the use of a flow splitting device. Online LC-MS analysis of intact proteins was performed in high mass range mode and mass resolution set to 17,500. Orbitrap maximum inject time was set to 1000 ms and averaged over 3 microscans. Electrospray capillary voltage was set to 3.8 kV and the transfer capillary temperature to 225°C.
Limited proteolysis with Endoproteinase Lys-C
Fc-fusion protein binding domains were cleaved with Lys-C endoproteinase (>200 units/mg, Roche, cat. no. 1 420 429 Basel, Switzerland) by performing the digestion reaction under non-denaturing conditions and limiting the reaction time to 20 min. Digestion was performed in 100 mM Tris buffer (pH = 8.0) at 37°C using a 1:400 enzyme:protein ratio.
Peptide mapping of FcγRIIIa affinity fractions
A larger semi-preparative scale FcR-IIIA-5PW 7.8 mm x 7.5 cm column (P/N 0023532, TOSOH Bioscience, King of Prussia, PA) was used to collect fractions of heat-stressed rituximab and control material (). The increased size and loading capacity of the semi-preparative column allowed for 1 mg or greater injections onto the column, requiring fewer collections to obtain a sufficient amount of material from each affinity fraction for peptide mapping (50–100 µg) analysis. The semipreparative column was operated with the same mobile phases and flow rate as the analytical version, but with a linear pH gradient from 0–100% B over 120 min followed by 100% B for 20 min to ensure complete elution. The column was equilibrated with 100% phase A for 10 min prior to starting the gradient and 10 additional minutes after completion.
Tryptic digestion
The peptide mapping procedure was adopted from Ren et al. with several modifications described below.73 Four fractions were collected for each sample as indicated by supplemental Figure S2. Approximate protein concentration for each fraction was determined by UV absorbance at 280 nm using a NanoDrop 2000 spectrophotometer (Thermo Fisher, Waltham, MA). The affinity fractions were concentrated using 0.5 mL Microcon centrifugal filter units with a 30 kDa molecular weight cutoff (Millipore Sigma, Burlington, MA, catalog # MRCF0R030) according to the manufacturer’s instructions. Fractions 2, 3, and 4 were concentrated to obtain 100 µg of mAb material in approximately 50 µL of buffer. Fraction 1 was concentrated to 50 µg of material in 50 µL of buffer due to sample limitations resulting from the low intensity of this peak. Each fraction was processed using a filter-aided sample preparation (FASP) style approach based on the protocol described by Wiśniewsk et al. with several modifications described below.Citation74 The fractions were buffer exchanged into denaturing buffer (8 M guanidine HCl, 50 mM Tris pH = 7.5) and centrifuged at 14,000 × g for 10 min to remove excess buffer. Disulfide reduction was performed by addition of 3 µL of 0.5 M dithiothreitol (DTT) in denaturing buffer added to each filter tube and incubated at 37°C for 30 min. The reduced samples were then alkylated by adding 7 µL of 0.5 M iodoacetic acid in denaturing buffer to each tube and incubating in the dark for 20 min at room temperature. The alkylation reaction was then quenched by addition of an additional 4 µL of 0.5 M DTT solution and centrifuged at 14,000 × g for 15 min to remove excess buffer. Each sample was buffer exchanged to remove the denaturing buffer by washing with 200 µL of digest buffer (50 mM Tris-HCl pH = 7.8, 20 mM L-methionine) and centrifuging at 14,000 × g for 15 min, three times each. Proteomics grade recombinant trypsin (Cat. No. 11 047 841 001, Roche Diagnostics GmbH Mannheim, Germany) was reconstituted in LC/MS grade water to a concentration of 1 mg/mL and added to each vial to give an enzyme:substrate ratio of 1:20. Filter tubes containing digestion reactions were incubated at 37°C for 90 min. Tryptic peptides were then collected by transferring the filter units to new collection tubes and centrifuging at 14,000 × g for 15 min followed rinsing and centrifuging the filters with 20 µL of digest buffer two times. The digests were then quenched by addition of 160 µL of quenching buffer (8 M guanidine-HCl, 250 mM sodium acetate pH = 4.7 to final 7 M guanidine) and transferred to autosampler vials for LC-MS analysis. All peptide mapping reagents were purchased from Sigma Aldrich (St Louis, MO).
LC-MS/MS peptide mapping
LC-MS/MS peptide mapping was performed using the same HPLC and MS instrumentation as the intact FcγRIIIa experiments described above. For each fraction, 40 µL of digest solution was injected and separated on a Polaris 180 Å C18-Ether 250 × 2.1 mm, 3-µm reversed phase HPLC column (PN A2021250X21, made in the Netherlands, Agilent Technologies). Tryptic peptides were eluted with a 190-min linear gradient from 0–50% B, followed by a 10-min wash step with 100% B, and 35 min of equilibration at 0% B. Mobile phase A is composed of LC-MS grade water and 0.1% trifluoroacetic acid (TFA) while phase B is 90% acetonitrile with 0.1% TFA. Flow rate was 200 µL/min. MS/MS analysis of the eluting tryptic peptides was performed on the same Orbitrap mass spectrometer as the intact analysis, using the full scan collected with a resolution of 70,000, and data dependent MSCitation1 (Top5) scans with resolution of 17,500 and normalized collision energy set to 26.
Data analysis
Raw MS data were processed using MassAnalyzer,Citation75,Citation76,Citation77 a software package (commercially available as Biopharma Finder from Thermo Fisher) for automated feature extraction, retention time alignment, and peptide identification/quantitation. Chromeleon 7 (Thermo Fisher, Waltham, MA) was used for peak integration of UV chromatograms as well as intact protein deconvolution using the ReSpect algorithm and Native Sliding Windows processing options. Additional details describing intact protein deconvolution parameters are described in Supplemental Table S1. SIMCA 15 software was used for PCA and data visualization of PCA results.
FcγRIIIa relative binding by AlphaLISA
Relative binding to FcγRIIIa (158 V) was measured using a competitive AlphaLISA® assay (Perkin Elmer) as described in detail previously.Citation78 Briefly, AlphaLISA™ assay is an Amplified Luminescent Proximity Homogeneous Assay (Alpha) designed to measure the level of FcγRIIIa binding to the Fc portion of IgG1 mAbs. When FcγRIIIa-GST and the biotinylated human IgG1 bind together, they also bring the acceptor and donor beads into close proximity. When laser light is applied to this complex, an energy transfer to the acceptor bead occurs, resulting in light production (luminescence), which is measured in a plate reader equipped for AlphaLISA™ signal detection. When a human IgG1 is present at sufficient concentrations to inhibit the binding of FcγRIIIa-GST to the biotinylated human IgG1, a dose-dependent decrease in emission at 520 to 620 nm is observed. Dose response curves were fit to a 4P logistical regression line and the relative binding was determined by comparing the IC50 of a sample to that of a reference standard.
Acknowledgments
The authors gratefully acknowledge Tosoh Corporation for providing a prototype semi-preparative scale affinity column for evaluation purposes. They would like to thank Gang Xiao, Zhongqi Zhang (ZZ), Scott Siera, Margaret Ricci, Chetan Goudar of Amgen; Ashli Simone, Philip Hoang, Atis Chakrabarti, Scott Melideo, and Heidi Vitrac from Tosoh Bioscience for fruitful discussions. Research funding was provided by Amgen, Inc.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Additional information
Funding
References
- Kim JM, Ashkenazi A. Fcγ receptors enable anticancer action of proapoptotic and immune-modulatory antibodies. J Exp Med. 2013;210(9):1647–17. doi:10.1084/jem.20131625.
- Nimmerjahn F, Ravetch JV. Fcγ receptors as regulators of immune responses. Nat Rev Immunol. 2008;8(1):34–47. doi:10.1038/nri2206. 2008/01/01.
- Nimmerjahn F, Gordan S, Lux A. FcγR dependent mechanisms of cytotoxic, agonistic, and neutralizing antibody activities. Trends Immunol. 2015 Jun;36(6):325–36. doi:10.1016/j.it.2015.04.005. PMID: 25981969.
- Bruhns P, Iannascoli B, England P, Mancardi DA, Fernandez N, Jorieux S, Daëron M. Specificity and affinity of human Fcγ receptors and their polymorphic variants for human IgG subclasses. Blood. 2009;113(16):3716–25. doi:10.1182/blood-2008-09-179754.
- Rascu A, Repp R, Westerdaal NA, Kalden, JR, and van de Winkel, JG, et al. Clinical relevance of Fc gamma receptor polymorphisms. AAnn NY Acad Sci. 1997 Apr 5;815:282–95. doi:10.1111/j.1749-6632.1997.tb52070.x. PMID: 9186665.
- Sugita N, Yamamoto K, Kobayashi T, Der Pol V, De Winkel V. Relevance of Fc gamma RIIIa-158V-F polymorphism to recurrence of adult periodontitis in Japanese patients. Clin.Exp.Immunol. 1999;117(2):350–54. doi:10.1046/j.1365-2249.1999.00984.x. PMID: 10444269.
- Orange JS. Formation and function of the lytic NK-cell immunological synapse. Nat Rev Immunol. 2008;8:713–25. doi:10.1038/nri2381.
- Kaplon H, Muralidharan M, Schneider Z, Reichert JM. Antibodies to watch in 2020. mAbs. 2020;12(1):1703531. doi:10.1080/19420862.2019.1703531. 2020/01/01.
- Kiyoshi M, Caaveiro JMM, Tada M, Tamura H, Tanaka T, Terao Y, Morante, K, Harazono, A, Hashii, N, Shibata, H, et al. Assessing the heterogeneity of the Fc-Glycan of a therapeutic antibody using an engineered FcγReceptor IIIa-immobilized column. Sci Rep. 2018;8:3955. doi:10.1038/s41598-018-22199-8.
- Jefferis R. Glycosylation of recombinant antibody therapeutics. Biotechnol Progr. 2005;21:11–16. doi:10.1021/bp040016j.
- Jefferis R. Recombinant antibody therapeutics: the impact of glycosylation on mechanisms of action. Trends Pharm Sci. 2009;30:356–62. doi:10.1016/j.tips.2009.04.007.
- Russell A, Adua E, Ugrina I, Laws S, Unravelling Immunoglobulin WW. G Fc N-glycosylation: a dynamic marker potentiating predictive, preventive and personalised medicine. Int J Mol Sci. 2018;19:390. doi:10.3390/ijms19020390.
- Liu L. Antibody glycosylation and its impact on the pharmacokinetics and pharmacodynamics of monoclonal antibodies and Fc-fusion proteins. J Pharm Sci. 2015;104:1866–84. doi:10.1002/jps.24444.
- Zhang Z, Shah B. Prediction of collision-induced dissociation spectra of common N-Glycopeptides for glycoform identification. Anal, Chem. 2010;82(24):10194–202. doi:10.1021/ac102359u. 2010/12/15.
- Houde D, Peng Y, Berkowitz SA, Engen JR. Post-translational modifications differentially affect IgG1 conformation and receptor binding. Mol Cell Proteomics. 2010;9:1716. doi:10.1074/mcp.M900540-MCP200.
- Krapp S, Mimura Y, Jefferis R, Huber R, Sondermann P. Structural analysis of human IgG-Fc glycoforms reveals a correlation between glycosylation and structural integrity. J Mol Bio. 2003;325:979–89. doi:10.1016/s0022-2836(02)01250-0.
- Zhang Z, Shah B, Richardson J. Impact of Fc N-glycan sialylation on IgG structure. mAbs. 2019 Nov-Dec;11(8):1381–90. doi:10.1080/19420862.2019.1655377. PMID: 31411531.
- Higel F, Seidl A, Sörgel F, Friess W. N-glycosylation heterogeneity and the influence on structure, function and pharmacokinetics of monoclonal antibodies and Fc fusion proteins. Eur J Pharm Biopharm. 2016;100:94–100. doi:10.1016/j.ejpb.2016.01.005.
- Zheng K, Bantog C, Bayer R. The impact of glycosylation on monoclonal antibody conformation and stability. mAbs. 2011;3:568–76. doi:10.4161/mabs.3.6.17922.
- Jefferis R, Lund J. Interaction sites on human IgG-Fc for FcgammaR: current models. Immunol Lett. 2002;82:57–65. doi:10.1016/s0165-2478(02)00019-6.
- Tao MH, Morrison SL. Studies of aglycosylated chimeric mouse-human IgG. Role of carbohydrate in the structure and effector functions mediated by the human IgG constant region. J Immunol. 1989;143(8):2595–601. PMID: 2507634.
- Jefferis R, Lund J, Pound JD. IgG-Fc-mediated effector functions: molecular definition of interaction sites for effector ligands and the role of glycosylation. Immunol Rev. 1998;163:59–76. doi:10.1111/j.1600-065x.1998.tb01188.x. PMID: 9700502.
- Arnold JN, Wormald MR, Sim RB, Rudd PM, Dwek RA. The impact of glycosylation on the biological function and structure of human immunoglobulins. Ann Rev Immunol. 2007;25:21–50. doi:10.1146/annurev.immunol.25.022106.141702.
- Barb AW, Prestegard JH. NMR analysis demonstrates immunoglobulin G N-glycans are accessible and dynamic. Nat Chem Bio. 2011;7(3):147–53. doi:10.1038/nchembio.511. PMID: 21258329.
- Shields RL, Lai J, Keck R, O’Connell LY, Hong K, Meng YG, Weikert SHA, Presta LG. Lack of Fucose on Human IgG1 N-Linked Oligosaccharide Improves Binding to Human FcγRIII and Antibody-dependent Cellular Toxicity. J Biol Chem. 2002;277:26733–40. doi:10.1074/jbc.M202069200. PMID: 11986321.
- Kanda Y, Yamada T, Mori K, Okazaki A, Inoue M, Kitajima-Miyama K, Kuni-Kamochi R, Nakano R, Yano K, Kakita S, et al. Comparison of biological activity among nonfucosylated therapeutic IgG1 antibodies with three different N-linked Fc oligosaccharides: the high-mannose, hybrid, and complex types. Glycobiology. 2007;17:104–18. doi:10.1093/glycob/cwl057.
- Junttila TT, Parsons K, Olsson C, Lu Y, Xin Y, Theriault J, Crocker L, Pabonan O, Baginski T, Meng G, et al. Superior In vivo efficacy of afucosylated trastuzumab in the treatment of HER2-amplified breast cancer. Cancer Res. 2010;70:4481. doi:10.1158/0008-5472.CAN-09-3704.
- Niwa R, Natsume A, Uehara A, Wakitani M, Iida S, Uchida K, Satoh M, Shitara K. IgG subclass-independent improvement of antibody-dependent cellular cytotoxicity by fucose removal from Asn297-linked oligosaccharides. J Immunol Meth. 2005;306:151–60. doi:10.1016/j.jim.2005.08.009.
- Ferrara C, Grau S, Jäger C, Sondermann P, Brünker P, Waldhauer I, Hennig M, Ruf A, Rufer AC, Stihle M, et al. Unique carbohydrate-carbohydrate interactions are required for high affinity binding between FcgammaRIII and antibodies lacking core fucose. Proc Nat Acad Sci. 2011;108:12669–74. doi:10.1073/pnas.1108455108.
- Mizushima T, Yagi H, Takemoto E, Shibata-Koyama M, Isoda Y, Iida S, Masuda K, Satoh M, Kato K. Structural basis for improved efficacy of therapeutic antibodies on defucosylation of their Fc glycans. Genes Cells. 2011;16:1071–80. doi:10.1111/j.1365-2443.2011.01552.x.
- Shatz W, Chung S, Li B, Marshall B, Tejada M, Phung W, Sandoval W, Kelley RF, Scheer JM. Knobs-into-holes antibody production in mammalian cell lines reveals that asymmetric afucosylation is sufficient for full antibody-dependent cellular cytotoxicity. mAbs. 2013;5:872–81. doi:10.4161/mabs.26307.
- Yu M, Brown D, Reed C, Chung S, Lutman J, Stefanich E, Wong A, Stephan JP, Bayer R. Production, characterization, and pharmacokinetic properties of antibodies with N-linked mannose-5 glycans. MAbs. 2012;4(4):475–87. doi:10.4161/mabs.20737. PMID: 22699308.
- Reusch D, Tejada ML. Fc glycans of therapeutic antibodies as critical quality attributes. Glycobiology. 2015;25:1325–34. doi:10.1093/glycob/cwv065.
- Dekkers G, Treffers L, Plomp R, Bentlage AEH, de Boer M, Koeleman CAM, Lissenberg-Thunnissen SN, Visser R, Brouwer M, Mok JY, et al. Decoding the human immunoglobulin G-glycan repertoire reveals a spectrum of Fc-Receptor- and complement-mediated-effector activities. Front Immunol. 2017;8:877. doi:10.3389/fimmu.2017.00877.
- Subedi GP, Barb AW. The immunoglobulin G1 N-glycan composition affects binding to each low affinity Fc γ receptor. mAbs. 2016;8:1512–24. doi:10.1080/19420862.2016.1218586.
- Chung CH, Mirakhur B, Chan E, Le Q-T, Berlin J, Morse M, Murphy BA, Satinover SM, Hosen J, Mauro D, et al. Cetuximab-induced anaphylaxis and IgE specific for galactose-α-1, 3-galactose. N Engl J Med. 2008;358:1109–17. doi:10.1056/NEJMoa074943.
- Chitnavis M, Stein DJ, Commins S, Schuyler AJ, Behm B. First-dose anaphylaxis to infliximab: a case of mammalian meat allergy. J Allerg Cl Imm-Prac. 2017;5:1425–26. doi:10.1016/j.jaip.2017.04.044.
- Kaneko Y, Nimmerjahn F, Ravetch JV. Anti-Inflammatory activity of immunoglobulin G resulting from Fc Sialylation. Science. 2006;313:670. doi:10.1126/science.1129594.
- Jones MB, Oswald DM, Joshi S, Whiteheart SW, Orlando R, Cobb BA. B-cell–independent sialylation of IgG. Proc Nat Acad of Sci. 2016;113:7207. doi:10.1073/pnas.1523968113.
- Washburn N, Schwab I, Ortiz D, Bhatnagar N, Lansing JC, Medeiros A, Tyler S, Mekala D, Cochran E, Sarvaiya H, et al. Controlled tetra-Fc sialylation of IVIg results in a drug candidate with consistent enhanced anti-inflammatory activity. Proc Nat Acad of Sci. 2015;112:E1297. doi:10.1073/pnas.1422481112.
- Kuhne F, Bonnington L, Malik S, Thomann M, Avenal C, Cymer F, Wegele H, Reusch D, Mormann M, Bulau P. The impact of immunoglobulin G1 Fc sialylation on backbone amide H/D exchange. Antibodies (Basel). 2019;8:49. doi:10.3390/antib8040049.
- Parekh RB, Dwek RA, Sutton BJ, Fernandes DL, Leung A, Stanworth D, Rademacher TW, Mizuochi T, Taniguchi T, Matsuta K, et al. Association of rheumatoid arthritis and primary osteoarthritis with changes in the glycosylation pattern of total serum IgG. Nature. 1985;316(6027):452–57. doi:10.1038/316452a0. 1985/08/01.
- Wong AHY, Fukami Y, Sudo M, Kokubun N, Hamada S, Yuki N. Sialylated IgG-Fc: a novel biomarker of chronic inflammatory demyelinating polyneuropathy. J Neurol Neurosur Ps. 2016;87:275. doi:10.1136/jnnp-2014-309964.
- Scallon BJ, Tam SH, McCarthy SG, Cai AN, Raju TS. Higher levels of sialylated Fc glycans in immunoglobulin G molecules can adversely impact functionality. Mol Immunol. 2007;44:1524–34. doi:10.1016/j.molimm.2006.09.005.
- Vidarsson G, Dekkers G, Rispens T. IgG subclasses and allotypes: from structure to effector functions. Front Immunol. 2014:5. doi:10.3389/fimmu.2014.00520.
- Lippold S, Nicolardi S, Domínguez-Vega E, Heidenreich A-K, Vidarsson G, Reusch D, Haberger M, Wuhrer M, Falck D. Glycoform-resolved FcɣRIIIa affinity chromatography–mass spectrometry. mAbs. 2019;11:1191–96. doi:10.1080/19420862.2019.1636602.
- Sondermann P, Huber R, Oosthuizen V, Jacob U. The 3.2-A crystal structure of the human IgG1 Fc fragment-Fc gammaRIII complex. Nature. 2000;406:267–73. doi:10.1038/35018508.
- Houel S, Hilliard M, Yu YQ, McLoughlin N, Martin SM, Rudd PM, Williams JP, N- CW. and O-glycosylation analysis of etanercept using liquid chromatography and quadrupole time-of-flight mass spectrometry equipped with electron-transfer dissociation functionality. Anal Chem. 2014;86:576–84. doi:10.1021/ac402726h.
- Kleemann GR, Beierle J, Nichols AC, Dillon TM, Pipes GD, Bondarenko PV. Characterization of IgG1 Immunoglobulins and Peptide−Fc fusion proteins by limited proteolysis in conjunction with LC−MS. Anal Chem. 2008;80:2001–09. doi:10.1021/ac701629v.
- Gadgil HS, Bondarenko PV, Pipes GD, Dillon TM, Banks D, Abel J, Kleemann GR, Treuheit MJ. Identification of cysteinylation of a free cysteine in the Fab region of a recombinant monoclonal IgG1 antibody using Lys-C limited proteolysis coupled with LC/MS analysis. Anal Biochem. 2006;355:165–74. doi:10.1016/j.ab.2006.05.037. PMID: 16828048.
- Fang J, Richardson J, Du Z, Zhang Z. Effect of Fc-Glycan structure on the conformational stability of IgG revealed by hydrogen/deuterium exchange and limited proteolysis. Biochemistry. 2016;55:860–68. doi:10.1021/acs.biochem.5b01323. PMID: 26812426.
- de Taeye SW, Bentlage AEH, Mebius MM, Meesters JI, Lissenberg-Thunnissen S, Falck D, Sénard T, Salehi N, Wuhrer M, and Schuurman J, et al. FcγR binding and ADCC activity of human IgG allotypes Front Immunol . 2020 2020-May-06;11(740). doi:10.3389/fimmu.2020.00740.
- Edelman GM, Cunningham BA, Gall WE, Gottlieb PD, Rutishauser U, Waxdal MJ. The covalent structure of an entire gammaG immunoglobulin molecule. Proc Natl Acad Sci. 1969;63(1):78–85. doi:10.1073/pnas.63.1.78. PMID: 5257969.
- Shields RL, Namenuk AK, Hong K, Meng YG, Rae J, Briggs J, Xie D, Lai J, Stadlen A, Li B, et al. High resolution mapping of the binding site on human IgG1 for Fc gamma RI, Fc gamma RII, Fc gamma RIII, and FcRn and design of IgG1 variants with improved binding to the Fc gamma R. J Biol Chem. 2001;276:6591–604. doi:10.1074/jbc.M009483200. PMID: 11096108.
- Jacobsen FW, Stevenson R, Li C, Salimi-Moosavi H, Liu L, Luo WJ, Quanzhou DK, Buck L, Miller S, Miller S, et al. Engineering an IgG scaffold lacking effector function with optimized developability. J Biol Chem. 2017;292:1865–75. doi:10.1074/jbc.M116.748525. PMID: 27994062.
- Liu L, Jacobsen FW, Everds N, Zhuang Y, Yu YB, Li N, Clark D, Nguyen MP, Fort M, Narayanan P, et al. Biological characterization of a stable effector functionless (SEFL) monoclonal antibody scaffold in vitro. J Biol Chem. 2017;292(5):1876–83. doi:10.1074/jbc.M116.748707. PMID: 27994063.
- Yu J, Song Y, Tian W. How to select IgG subclasses in developing anti-tumor therapeutic antibodies. J Hematol Oncol. 2020;13(1):45. doi:10.1186/s13045-020-00876-4. PMID: 32370812.
- Schneider-Merck T, Lammerts van Bueren JJ, Berger S, Rossen K, van Berkel PH, Derer S, Beyer T, Lohse S, Wk B, Peipp M, et al. Human IgG2 antibodies against epidermal growth factor receptor effectively trigger antibody-dependent cellular cytotoxicity but, in contrast to IgG1, only by cells of myeloid lineage. J Immunol. 2010;184:512–20. doi:10.4049/jimmunol.0900847. PMID: 19949082.
- Wypych J, Li M, Guo A, Zhang Z, Martinez T, Allen MJ, Fodor S, Kelner DN, Flynn GC, Liu YD, et al. Human IgG2 antibodies display disulfide-mediated structural isoforms. J Biol Chem. 2008;283:16194–205. doi:10.1074/jbc.M709987200. PMID: 18339624.
- Dillon TM, Ricci MS, Vezina C, Flynn GC, Liu YD, Rehder DS, Plant M, Henkle B, Li Y, Deechongkit S, et al. Structural and functional characterization of disulfide isoforms of the human IgG2 subclass. J Biol Chem. 2008;283:16206–15. doi:10.1074/jbc.M709988200. PMID: 18339626.
- Zhang A, Fang J, Chou RYT, Bondarenko PV, Zhang Z. Conformational difference in human IgG2 disulfide isoforms revealed by hydrogen/deuterium exchange mass spectrometry. Biochemistry. 2015;54:1956–62. doi:10.1021/bi5015216. PMID: 25730439.
- Saphire EO, Parren PW, Pantophlet R, Zwick MB, Morris GM, Rudd PM, Dwek RA, Stanfield RL, Burton DR, Wilson IA. Crystal structure of a neutralizing human IgG against HIV-1: a template for vaccine design. Science. 2001;293:1155. doi:10.1126/science.1061692. PMID: 11498595.
- Michaelsen TE, Brekke OH, Aase A, Sandin RH, Bremnes B, Sandlie I. One disulfide bond in front of the second heavy chain constant region is necessary and sufficient for effector functions of human IgG3 without a genetic hinge. Proc Nat Acad Sci. 1994;91:9243. doi:10.1073/pnas.91.20.9243. PMID: 7937748.
- Shi RL, Xiao G, Dillon TM, McAuley A, Ricci MS, Bondarenko PV. Identification of critical chemical modifications by size exclusion chromatography of stressed antibody-target complexes with competitive binding. mAbs. 2021;13:1887612. doi:10.1080/19420862.2021.1887612. PMID: 33616001.
- Hayes JM, Frostell A, Karlsson R, Müller S, Martín SM, Pauers M, Reuss F, Cosgrave EF, Anneren C, Davey GP, et al. Identification of Fc gamma receptor glycoforms that produce differential binding kinetics for rituximab. Mol Cell Proteomics. 2017;16:1770–88. doi:10.1074/mcp.M117.066944. PMID: 28576848.
- Barb AW. Fc γ receptor compositional heterogeneity: considerations for immunotherapy development. J Biol Chem. 2021;296:100057. doi:10.1074/jbc.REV120.013168. PMID: 33172893.
- Patel KR, Roberts JT, Subedi GP, Barb AW. Restricted processing of CD16a/Fc γ receptor IIIa N-glycans from primary human NK cells impacts structure and function. J Biol Chem. 2018;293:3477–89. doi:10.1074/jbc.RA117.001207. PMID: 29330305.
- Washburn N, Meccariello R, Duffner J, Getchell K, Holte K, Prod’homme T, Srinivasan K, Prenovitz R, Lansing J, Capila I, et al. Characterization of endogenous human FcγRIII by mass spectrometry reveals site, allele and sequence specific glycosylation. Mol Cell Proteomics. 2019;18:534–45. doi:10.1074/mcp.RA118.001142. PMID: 30559323.
- Hayes JM, Frostell A, Cosgrave EF, Struwe WB, Potter O, Davey GP, Karlsson R, Anneren C, Rudd PM. Fc gamma receptor glycosylation modulates the binding of IgG glycoforms: a requirement for stable antibody interactions. J Proteome Res. 2014;13:5471–85. doi:10.1021/pr500414q. PMID: 25345863.
- Wojcik I, Sénard T, de Graaf EL, Janssen GMC, de Ru AH, Mohammed Y, van Veelen PA, Vidarsson G, Wuhrer M, Falck D. Site-Specific Glycosylation Mapping of Fc gamma receptor IIIb from neutrophils of individual healthy donors. Anal Chem. 2020 Oct 6;92(19):13172–81. doi:10.1021/acs.analchem.0c02342. PMID: 32886488.
- Yamaguchi Y, Barb AW. A synopsis of recent developments defining how N-glycosylation impacts immunoglobulin G structure and function. Glycobiology. 2020 Mar 20;30(4):214–25. doi:10.1093/glycob/cwz068. PMID: 31822882.
- Cambay F, Forest-Nault C, Dumoulin L, Seguin A, Henry O, Durocher Y, De Crescenzo G. Glycosylation of Fcγ receptors influences their interaction with various IgG1 glycoforms. Mol Immunol. 2020;121:144–58. doi:10.1016/j.molimm.2020.03.010. 2020/05/01/.
- Ren D, Pipes GD, Liu D, Shih LY, Nichols AC, Treuheit MJ, Brems DN, Bondarenko PV. An improved trypsin digestion method minimizes digestion-induced modifications on proteins. Anal Biochem. 2009;392:12–21. doi:10.1016/j.ab.2009.05.018. PMID: 19457431.
- Wiśniewski JR, Zougman A, Nagaraj N, Mann M. Universal sample preparation method for proteome analysis. Nat Methods. 2009;6:359–62. doi:10.1038/nmeth.1322. PMID: 19377485.
- Zhang Z. Large-scale identification and quantification of covalent modifications in therapeutic proteins. Anal Chem. 2009;81:8354–64. doi:10.1021/ac901193n.
- Zhang Z. Prediction of low-energy collision-induced dissociation spectra of peptides. Anal Chem. 2004;76:3908–22. doi:10.1021/ac049951b. PMID: 15253624.
- Retention Time ZZ. Alignment of LC/MS data by a divide-and-conquer algorithm. J Am Soc Mass Spectrom. 2012;23:764–72. doi:10.1007/s13361-011-0334-2. PMID: 22298290.
- Seo N, Polozova A, Zhang M, Yates Z, Cao S, Li H, Kuhns S, Maher G, McBride HJ, Liu J. Analytical and functional similarity of Amgen biosimilar ABP 215 to bevacizumab. mAbs. 2018;10:678–91. doi:10.1080/19420862.2018.1452580. PMID: 29553864.
