ABSTRACT
Although therapeutically efficacious, ipilimumab can exhibit dose-limiting toxicity that prevents maximal efficacious clinical outcomes and can lead to discontinuation of treatment. We hypothesized that an acidic pH-selective ipilimumab (pH Ipi), which preferentially and reversibly targets the acidic tumor microenvironment over the neutral periphery, may have a more favorable therapeutic index. While ipilimumab has pH-independent CTLA-4 affinity, pH Ipi variants have been engineered to have up to 50-fold enhanced affinity to CTLA-4 at pH 6.0 compared to pH 7.4. In hCTLA-4 knock-in mice, these variants have maintained anti-tumor activity and reduced peripheral activation, a surrogate marker for toxicity. pH-sensitive therapeutic antibodies may be a differentiating paradigm and a novel modality for enhanced tumor targeting and improved safety profiles.
Introduction
CTLA-4 is a critical negative regulator of T cellsCitation1,Citation2 and its blockade with ipilimumab (Ipi) is an effective treatment for a wide range of cancers. In the tumor microenvironment, inhibition of CTLA-4 may restore antitumor immune responses by two separate but complementary actions: the activation and proliferation of T cells including tumor-infiltrating T effector cells,Citation3 and the reduction of immunosuppressive T regulatory cell (Treg) function.Citation4,Citation5 Despite the successes of Ipi in the clinic, including longer overall survival with higher doses, patients have experienced increased treatment-related adverse events particularly at the higher doses.Citation6 The dose-limiting toxicity of Ipi prevents its maximal anti-tumor activity potential, but has presented an opportunity for the discovery and development of next-generation molecules with enhanced therapeutic index against CTLA-4.
Additional forms of Ipi now in clinical development may increase anti-tumor efficacy, reduce side-effects for improved safety, or both. While Ipi uses an IgG1 isotype, a non-fucosylated (NF) form has been engineered to increase its binding affinity for FcγRIIIaCitation7 and result in enhanced antibody-dependent cellular cytotoxicity (ADCC) effector responses and depletion of intratumoral Tregs.Citation8,Citation9 Furthermore, to broaden the therapeutic window of CTLA-4 blockade for localized activity in the tumor versus normal tissue, a Probody® therapeutic technology platformCitation10 has been used to generate a peptide-masked Ipi, designed to remain inert in the periphery, but have activity restored when unmasked by tumor-associated proteases. Additional anti-CTLA-4 antibodies with similar enhanced Fc domains or masked forms are included in the clinical pipeline.
Other features of the tumor microenvironment may be leveraged to create the next generation of therapeutics with increased preferential activity in the tumor compared to peripheral tissues. For instance, acidic extracellular pH is a hallmark of cancer that contributes to tumor progression.Citation11,Citation12 Acidic pH-selective antibodies may have reversible activity that is dependent upon the local pH environment, i.e., binding may be enhanced at low pH to increase activity within acidic tumors, while binding and activity are reduced in neutral peripheral tissues. This modality is distinct from masking technologies, which result in molecules that function as pro-drugs, as they have irreversible activation after cleavage and would remain active outside the tumor. Indeed, acidic pH-selective antibodies have been engineered against VISTA, Her2, and CTLA-4.Citation13–15 The pH-engineered VISTA and CTLA-4 antibodies have demonstrated preferential accumulation in tumors in mice, as well as reduced toxicity in nonhuman primates, respectively, and have improved anti-tumor efficacy in comparison to their pH-independent counterparts. Conversely, neutral pH-selective antibodies have also been created, which have enhanced antibody serum half-life and favorable antibody pharmacokinetic properties.Citation16–20 While neutral pH-selective antibodies improve antibody recycling to overcome dosing limitations, the acidic pH-selective anti-VISTA antibody also has a longer mean residence time in mice and cynomolgus macaques (cyno) by reducing target-mediated drug disposition (TMDD) from peripheral sinks. As such, there is benefit of using pH-selective targeting approaches to augment the functionality of antibody therapeutics.
In this study, structure-guided designs combined with molecular evolution by phage display were used to select acidic pH-selective variants of ipilimumab (pH Ipi). These engineered antibodies had monovalent affinities that were matured at acidic pH, detuned at neutral pH, or both, and their pH-dependent activities were confirmed in cell binding and in vitro functional activity assays. When compared to Ipi using hIgG1-NF isotypes, the pH Ipi variants had similar anti-tumor efficacy and Treg depletion in hCTLA-4 knock-in mice and, importantly, had reduced peripheral activation and proliferation. These results suggest that the tumor microenvironment may be exploited using an acidic pH-selective Ipi to improve the therapeutic index of anti-CTLA-4 therapy.
Results
Engineering of acidic pH-selective ipilimumab variants
Ipi has pH-independent affinity to CTLA-4, and so structure-guided designs combined with in vitro evolution were used to engineer Ipi to have pH-sensitive activity (). The molecular surface electrostatics of the binding interface of the reported complex crystal structure were analyzed and complementarity-determining region (CDR) residues at or around the CTLA-4 epitope were substituted with pH-sensitive amino acids ( and Table S1). No additional tags or masking peptides were incorporated into the design. An initial mini library was constructed consisting of one histidine substitution at each heavy chain CDR. After rounds of selections with alternating acidic washes and neutral elutions, histidine enrichment was observed at HCDR positions Thr33, Asn56, and Thr95 (Figure S1). The triple mutant as well as variants with individual histidine substitutions were evaluated for pH-selectivity. The mutations in HCDR1 and HCDR2 slightly enhanced binding affinity, whereas the mutation in HCDR3 significantly reduced binding, and the combination of all three mutations improved affinity 10-fold (Table S2). However, these variants have equivalent binding to CTLA-4 irrespective of pH.
Figure 1. Protein engineering of acidic pH-selective ipilimumab variants. (a) Schematic of workflow for structure-guided design and phage display selections for acidic pH-selective binding. (b) Overlay of the Ipi CDR loops on the electrostatic surface potential of CTLA-4 at pH 7.4 and pH 6.0. The CDRs for the heavy and light chains are colored green and yellow, respectively. The residues selected for pH engineering are shown as sticks. (c) Sequence alignment of acidic pH-selective Ipi variants and associated (d) SPR sensorgrams at pH 7.4 and pH 6.0. (e) SPR binding affinities of acidic pH-selective Ipi variants to human CTLA-4 at incremental pH values.
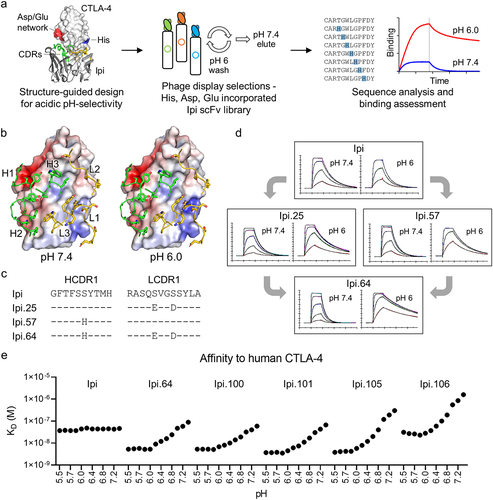
As the mini library had limited and restricted diversity, additional libraries were constructed consisting of up to three substitutions in either the heavy or light chain or up to six total mutations in the combinatorial library (Figure S2). Aspartic acid and glutamic acid residues were also incorporated in LCDR1, which is proximal to the only histidine of the CTLA-4 extracellular domain. Through rounds of phage selections by binding at pH 6.0 and eluting at pH 7.4, variants were enriched for the desired acidic pH-selective binding properties (Figure S3-5). Asp and Glu residues were observed at LCDR1 and their substitutions had acidic pH-selectivity (Table S3). While Thr33His variants were frequently seen at HCDR1, which has improved yet pH-independent affinity to CTLA-4, histidine substitutions were also enriched at HCDR1 Ser31 and had pH sensitivity. The individual LCDR1 Ser27aGlu Ser30Asp (Ipi.25) or HCDR1 Ser31His (Ipi.57) mutations had acidic-binding preferences and pH-selectivity was even further enhanced when they were combined to create Ipi.64. This variant has enhanced pH selectivity for binding CTLA-4 at pH 6.0 than at pH 7.4 compared to Ipi, as observed by faster association and slower dissociation binding rates ().
Additional variants with different levels of pH selectivity were engineered by incorporating other pH-sensitive or affinity altering mutations to Ipi.64. These mutations were distinct from Ipi.64 and were enriched after rounds of selection (Figure S3-5). Several antibodies exhibited enhanced preference for binding at pH 6.0 compared with pH 7.4 with HCDR2 Asn55His, HCDR3 Thr95His or LCDR1 Tyr32Glu substitutions, such as Ipi.92 and Ipi.95 (). However, they contain a “DS” peptide isomerization sequence liability in LCDR1 that was introduced by the Ser30Asp mutation, and isomerization was observed after 1 month in an accelerated stability study (Table S4). Switching the Ser30 position from Asp to Glu eliminated the isomerization sequence and preserved pH-selective binding properties (). Furthermore, surface plasmon resonance (SPR) kinetics of these variants were measured at incremental pH values between 5.5 and 7.4 and demonstrate that they are either affinity matured at acidic pH, affinity detuned at neutral pH, or both (). They all have similar association rates, which are faster in more acidic conditions, but their pH-dependent affinities are largely dictated by their off rates, as they dissociate quickly from CTLA-4 at neutral pH compared to acidic pH ( and Figure S6). Furthermore, the binding kinetics for each variant are constant below pH 6.0, indicating its maximal threshold of pH activation and may be a result of maximal protonation of the histidine side chains. Among these variants, Ipi.105 and Ipi.106 most notably have >50-fold affinity differences between pH 6.0 and pH 7.4 and have similar affinity and binding kinetics to Ipi at approximately pH 6.8 and pH 6.2, respectively. Although Ipi has weaker affinity to cyno CTLA-4 than to human CTLA-4, the pH-sensitive binding properties of these antibodies were preserved between the orthologs (Figure S7).
Table 1. Sequences and SPR kinetics at pH 7.4 and pH 6.0 of acidic pH-selective Ipi variants
In vitro characterization demonstrates acidic pH-selective activity
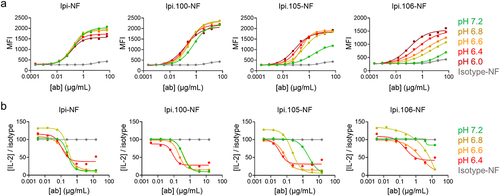
Cell binding experiments were performed to assess the in vitro pH selectivity of the Ipi variants. Binding of serially diluted fluorescent-labeled antibodies to BW cells expressing ~2,000 CTLA-4 receptor copies was measured in different pH buffer increments (pH 7.2, 6.8, 6.6, 6.4, and 6.0). Ipi has the same binding activity across the pH range and Ipi.100 has marginal pH-selectivity. At pH 7.2, Ipi.105 has weak cell binding and Ipi.106 is even more detuned, and they have improved cell binding with increasingly acidic pH conditions as observed by both an increase in maximum MFI and a shift in EC50 (). Similar binding results between these variants were observed with cell types expressing high copy numbers of CTLA-4 of up to ~100,000 receptor copies per cell (Figure S8a and S8b). Since bivalent IgGs show pH-selective binding despite different levels of CTLA-4 expression, these results demonstrate that avidity of the Ipi variants does not mask their pH-dependent properties in this assay. Importantly, both Ipi.105 and Ipi.106 show reduced binding relative to Ipi at neutral pH and equivalent binding at pH 6.8 and below 6.4, respectively, suggesting that these molecules have sufficiently different binding to CTLA-4 across pH to potentially result in functional differences.
Figure 2. pH-dependent cell binding and blocking of acidic pH-selective ipilimumab variants. One representative experiment of (a) binding and (b) blocking of Ipi, Ipi.100, Ipi.105, and Ipi.106 with BW cell lines at pH 7.2, 6.8, 6.6, 6.4, and 6.0. pH is indicated by color with isotype control in gray (values at all pH averaged). Blocking activity is normalized to isotype control.
CTLA-4 is estimated to be present at a copy number of 100–1,000 per cell on primary human T cells and human tumor infiltrating lymphocytes,Citation21 which is 20- to 1,000-fold less than the expression levels on the cell lines used to determine binding and functional blockade. In order to confirm that pH Ipi variants maintain pH-selective activity at physiological levels of CTLA-4 expression, staining was performed for total CTLA-4 on activated CD25+ cells at decreasing pH. As with the cell-line binding, Ipi.105 and Ipi.106 have reduced binding to CTLA-4 at neutral pH, and Ipi.105 and Ipi.106 have similar binding to Ipi at pH 6.8 and pH 6.4, respectively (Figure S9).
Blocking assays were performed to further investigate the functional activity of Ipi.100, Ipi.105 and Ipi.106. 58αβ-hCTLA-4/mCD3ζ reporter cells were stimulated with rhB7-1 for activation and then moved to pH-specific media containing the antibody variants. The isotype control showed decreasing signal in more acidic environments (Figure S8c), and so normalization to isotype control at each pH was performed to allow for a direct comparison of relative activity across pH. At neutral pH, the blocking activity of Ipi.105 was slightly reduced while Ipi.106 had minimal activity (). As the pH was lowered, Ipi.105 had increased blocking activity, matching Ipi at pH 6.8 and surpassing it at more acidic conditions. Ipi.106 had reduced blocking activity at neutral pH and similar blocking activity as Ipi at pH 6.4, consistent with the SPR and cell-binding results. Together these data are in agreement with cell binding where both Ipi.105 and Ipi.106 show clear changes in activity as pH changes, with Ipi.106 requiring a more acidic pH for equivalent activity to Ipi. Ipi.100, however, shows little to no pH-selectivity. Thus, Ipi.105 and Ipi.106 were selected for further evaluation.
Crystal structures of pH-selective Ipi-CTLA-4 complexes
To elucidate the molecular basis of pH-dependent recognition, the crystal structures of Ipi.105 and Ipi.106 were determined in complex with CTLA-4 at nominal resolutions of 2.73 Å and 2.75 Å, respectively (Table S5). The Ipi.105 complex crystallized at pH 5.2 while the Ipi.106 crystals grew at pH 8.3, and they may represent the high and low affinity pH-dependent binding states, respectively. Electron density at the interface between each antibody and CTLA-4 were well resolved for model building (Figure S10). Both pH-sensitive antibodies and Ipi recognize a similar epitope on CTLA-4 using a similar approach angle (within ~3°) and their Fv-CTLA-4 units have an all-atom RMSD of ~1–2 Å (). Each antibody uses all six CDRs to contact CTLA-4, where the heavy and light chains contribute to roughly 62% and 38% of the antigen-binding fragment (Fab) buried surface area, respectively (). While Ipi and Ipi.105 bury ~840 Å2 on CTLA-4, Ipi.106 binds a smaller footprint (~710 Å2).
Figure 3. Structural characterization of antibody recognition to CTLA-4 by pH-selective Ipi variants. (a) Overlay of the complex crystal structures of Ipi, Ipi.105, and Ipi.106 structurally aligned to CTLA-4. Ipi, Ipi.105, and Ipi.106 are colored gray, blue, and red, respectively. (b) Epitopes of Ipi, Ipi.105, and Ipi.106 colored onto the surface representation of CTLA-4. Regions contacted by the heavy chain or light chain are colored in a darker or lighter shade, respectively. (c) CDR loop comparisons of Ipi, Ipi.105, and Ipi.106 after alignment of the antibody variable domains. The pH-sensitive substitutions are shown as sticks and amino acids with different rotamer conformations compared to Ipi are shown as lines. (d) LCDR1 loop conformations of Ipi.105 and Ipi.106 compared to Ipi. The 2Fo–Fc electron density maps of the pH Ipi variants are represented as a gray mesh and are contoured at 1.5σ. (e) Heavy chain and (f) light chain CDR interactions to CTLA-4. The CTLA-4 surface is colored gray with its negatively charged and histidine residues colored red and blue, respectively.
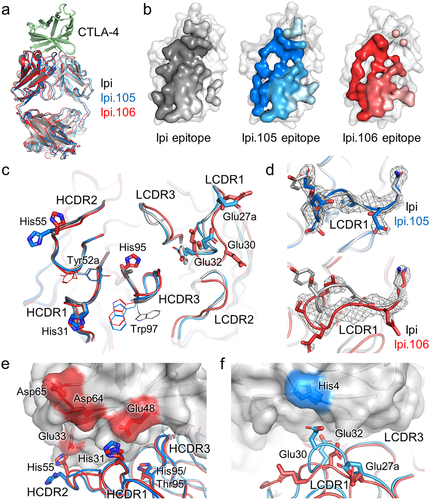
The variable domains of Ipi and Ipi.105 have good structural alignment, as their CDR loops have very similar backbone arrangements (). Furthermore, their amino acid side chains match well down to the level of rotamer conformations, where the pH-sensitive mutations of Ipi.105 are essentially substitutions of the Ipi residues on a fixed backbone with some differences (). The side chains of Trp97 in HCDR3 from Ipi.105 and Ipi.106 adopt an alternative conformation compared to Ipi. In addition, the rotamer of Tyr52a in HCDR2 of Ipi.105 aligns with Ipi, but is different for Ipi.106. These rotamer changes may be induced by the surrounding pH-sensitive substitutions. Furthermore, while the heavy chain CDRs of Ipi.106 also align well with Ipi, its LCDR1 and LCDR2 loops have different loop conformations (). Since the Ipi.105 and Ipi.106 complexes crystallized at acidic and basic conditions, these structural differences suggest that pH may influence the architecture of the CDR loops to confer high or low affinity pH-dependent interactions.
Ipi.105 and Ipi.106 use the same five acidic pH-selective substitutions and Ipi.106 has an additional HCDR3 Thr95His mutation. The HCDR1 Ser31His substitution, also used in Ipi.57 and Ipi.64, is directed near a cluster of negatively charged residues in CTLA-4 formed by Glu48, Asp64, and Asp65 (). While these residues are ordered in the Ipi.105 structure determined at low pH, they are disordered in the Ipi.106 complex, which may be a result of destabilization and poor binding at high pH state. Furthermore, the Asn55His mutation is proximal to the Glu33 of CTLA-4 (). Conversely, the LCDR1 Ser27aGlu, Ser30Glu, and Tyr32Glu residues form a cluster of negatively charged surface on the antibody that contacts the only histidine residue in CTLA-4 (). The HCDR3 Thr95His substitution of Ipi.106 is positioned near the center of the binding interface and the bulkier side chain may provide steric hindrance that results in detuned pH-independent binding activity. These molecular interactions provide a structural rationale of pH-dependency, where histidine residues, either in the Ipi variants or CTLA-4, become protonated in acidic conditions to form favorable electrostatic interactions with negatively charged residues.
pH-selective antibodies show pH-dependent uptake into CTLA-4+ T cells and enhancement of T cell function
For pH Ipi antibodies to have activity in acidic conditions, T cells must express CTLA-4 and be able to take up pH Ipi variants at low pH. Under neutral conditions, CTLA-4 is upregulated after T cell receptor (TCR) stimulation to negatively regulate T cell activation.Citation22 While it has been reported that T cell activation is reduced at low pH, CTLA-4 expression may be enhanced in acidic conditions.Citation23 To assess the impact of acidic pH on T cell activation, T cells were activated in different pH medias overnight and then stained for CTLA-4, as well as the canonical T cell activation marker CD25. It was observed that CD25 is upregulated after addition of TCR stimulation and is further enhanced by CD28 co-stimulation (, p = .0007, Two-way ANOVA). T cell activation is reduced as pH is lowered, and reduction of activation by low pH is somewhat mitigated by co-stimulation, as CD25 expression was observed down to pH 6.4 with CD28 present. However, no T cell activation was observed below pH 6.4, suggesting a physiological limit for T cell function at low pH. Interestingly, there was no evidence that acidic conditions enhance CTLA-4 expression. Conversely, CTLA-4 levels correlate closely with T cell activation and are thus reduced at low pH. Similar to CD25, some expression of CTLA-4 was observed down to pH 6.4, although CTLA-4 expression appeared less dependent on CD28 as a higher copy number of CTLA-4 was detected per cell when co-stimulation was present (, p = .0165, Two-way ANOVA). These results suggest that T cells can be activated under acidic conditions and that CTLA-4 is expressed at, but not enhanced by, low pH.
Figure 4. Acidic pH-selective ipilimumab variants enhance T cell activity at low pH. (a) T cell activation and CTLA-4 expression induced by anti-CD3 stimulation of human PBMCs, with and without CD28 co-stimulation, at pH 7.2, 6.8, 6.6, 6.4, and 6.0. (b) Uptake by activated human CD4+CD25+ cells of fluorescently labeled Ipi-NF, Ipi.105-NF and Ipi.106-NF as a function of pH. (c) Enhancement of SEA response by pH-selective Ipi variants at pH 7.2, 6.8 and 6.6. One representative donor is shown; percent change relative to Ipi-NF is summarized for n = 4 at 1.5 µg/mL. Statistical significance was determined by One-way ANOVA.
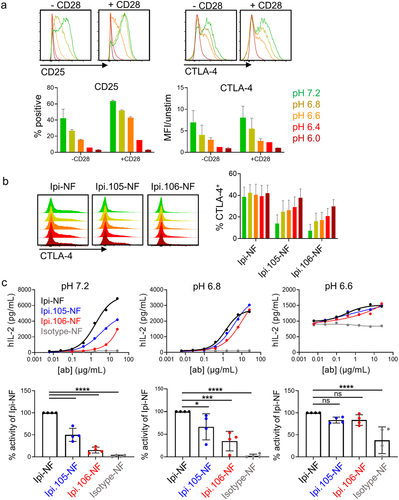
In addition to CTLA-4 expression, cycling of CTLA-4 must occur for binding of antibodies since CTLA-4 is stored in the endosomes of T cells and only exposed to the cell surface after stimulation.Citation23 To ensure that pH does not affect CTLA-4 cycling, CD4+CD25+ cells were activated in regular-pH media overnight to induce equivalent CTLA-4 expression and cycling, then cells were moved to media of varying pH containing fluorescently conjugated Ipi-NF, Ipi.105-NF, or Ipi.106-NF. Ipi-NF was found to have consistent uptake into T cells independent of pH, suggesting that activated T cells continue to cycle CTLA-4 regardless of pH changes (). As predicted by the cell-line binding data, the pH Ipi variants have acidic pH-dependent uptake by activated T cells, with Ipi.106-NF having greater loss of binding at neutral pH compared to Ipi.105-NF, confirming the acidic-selective CTLA-4 binding properties of these antibodies.
To assess functional activity on primary cells, the pH Ipi antibodies were tested in a superantigen assay. Staphylococcal enterotoxin A (SEA) crosslinks MHC class II and TCR and leads to T cell activation and interleukin-2 (IL-2) release, which can be enhanced by CTLA-4 blocking antibodies.Citation24 Because the release of cytokines is affected by low pH, cells were initially activated in regular media for 24 hours with SEA and then transferred to pH-specific medias containing antibodies for 72 hours. Although T cell activation was reduced by low pH in this assay, functional data could be obtained for T cells down to pH 6.6 before IL-2 production was no longer detected above background. Furthermore, background levels of IL-2 observed in the isotype control condition likely increased with decreasing pH due to reduced CD25 expression and IL-2 consumption in acidic conditions. Nonetheless, pH-dependent functional activity was indeed observed for the two pH Ipi variants (). To assess significance, four donors were compared at a single concentration (1.5 µg/mL) and the data were converted to percent activity relative to Ipi-NF to correct for variability in responsiveness between donors and across pH values. As observed with representative curves, Ipi.105-NF and Ipi.106-NF had significantly reduced activity at neutral pH (p < .0001) relative to Ipi-NF, while differences were not significant at pH 6.6. These results are consistent with the acidic pH-selective binding and blocking observed with CTLA-4 expressing cell lines, and enhanced binding and uptake observed in primary CD4+CD25+ cells. Importantly, they confirm a functional role for these antibodies in blocking CTLA-4 at low pH.
Improved therapeutic index in human CTLA-4 knock-in mice
To measure the activity of the pH-selective variants without the use of murine surrogate antibodies, Ipi-NF, Ipi.105-NF, and Ipi.106-NF were evaluated in human CTLA-4 knock-in (hCTLA-4-KI) mice bearing MC38 tumors. Expression of human CTLA-4 and not mouse CTLA-4 in these transgenic mice was validated by flow cytometry (Figure S11a). The KI mice had no overt phenotype and had similar levels of naïve cells as wild-type mice, supporting that human CTLA-4 is functional in mice and does not lead to inflammation or T cell activation (Figure S11b). To confirm that human CTLA-4 expression does not change responses to checkpoint blockade, mice were implanted with MC38 tumors and then treated with Ipi or an Fc-inert Ipi, as the Fc-inert Ipi should not result in tumor regression unless human CTLA-4 had reduced function in these mice.Citation4 Tumors in the wild-type mice and hCTLA-4-KI mice grew at similar rates, and only Ipi with an active Fc had activity (Figure S11 c). In addition to hCTLA-4 expression, the pH of MC38 tumors were evaluated. While tumors in general are accepted to be acidic, the pH may vary between cell lines. MC38 tumors respond to treatment with anti-CTLA-4 antibodies and have been shown to have a pH of ~6.7.Citation25 To confirm that our MC38 line results in acidic tumors, mice were subcutaneously implanted with MC38 cells and pH was measured with a probe. The average pH of MC38 tumors was observed to be 6.5 (Figure S12), supporting the use of this cell line for assessing pH-selective Ipi. Altogether, these data confirm that the hCTLA-4-KI mice are an appropriate model for measuring Ipi activity in vivo.
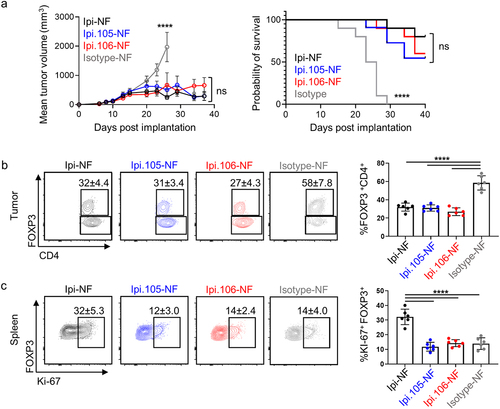
To test pH Ipi lead molecules in vivo, hCTLA-4-KI were implanted with MC38 tumor cells, then treated with Ipi-NF, Ipi.105-NF, Ipi.106-NF, or isotype control. Microbleeds were performed to confirm that there were no differences in exposure between Ipi-NF, Ipi.105-NF and Ipi.106-NF (p = ns, Figure S13). Mice were monitored for tumor growth and Ipi-NF, as expected, was able to significantly (p < .0001) reduce tumor volume relative to isotype (). Treatment with Ipi.105-NF and Ipi.106-NF also resulted in similar anti-tumor efficacy and survival (). Immune-monitoring was performed 4 days after treatment to look for pharmacodynamics (PD) markers of CTLA-4 engagement by the Ipi antibodies. Consistent with previous studies,Citation4 Treg depletion in tumors is highly correlated with efficacy in mice and Treg depletion was observed with all antibody treatments (). Importantly, no difference in Treg depletion was observed between Ipi-NF, Ipi,105-NF, and Ipi.106-NF, supporting the notion that the pH Ipi antibodies are targeting CTLA-4 in the tumor microenvironment. To determine CTLA-4 binding at neutral pH, spleens were also harvested from the tumored mice as Treg expansion, rather than depletion, has been shown with CTLA-4 blockade outside of the tumor microenvironment. Ipi-NF drove significant (p < .0001) proliferation of Tregs in the spleen as measured by the proliferation marker Ki-67 while both Ipi.105 and Ipi.106 had Treg Ki-67 levels similar to isotype control (p = ns) (). Similar to the spleen, T-cell activation in the tumor draining lymph nodes was observed with Ipi-NF but not by Ipi.105-NF or Ipi.106-NF (Figure S14). These results suggest that attenuated binding at neutral pH of these molecules resulted in less systemic effects in comparison to Ipi-NF.
Figure 5. Anti-tumor activity and Treg depletion are maintained by pH-sensitive ipilimumab variants while peripheral blockade is reduced. (a) Mean MC38 tumor volume (n = 10) and survival of human CTLA-4 knock-in mice treated with Ipi-NF, Ipi.105-NF, Ipi.106-NF, and isotype control. Statistical significance was determined by Two-way ANOVA (tumor volume) and Mantel-Cox test (survival). (b) Treg depletion as measured by FOXP3% in tumor CD4+ cells 5 days after treatment with pH-selective Ipi variants. (c) Expression of Ki-67 by Tregs in the spleen of mice treated with pH-selective Ipi variants. The statistical significance in panels (b) and (c) were determined by One-way ANOVA.
Discussion
Immune checkpoint blockade using monoclonal antibodies against CTLA-4 and programmed cell death protein-1 (PD-1) pathways has been a successful approach to achieve anti-tumor responses.Citation26 While CTLA-4 blockade has shown dose-dependent activity in the clinic, it comes at the cost of dose-dependent toxicity. Other second-generation anti-CTLA-4 antibodies have been developed using cleavable masks or Fc engineering approaches to overcome these toxicity issues. In this study, we describe an alternative strategy where acidic pH-selective Ipi variants were engineered that may preferentially target CTLA-4 in the tumor microenvironment, but remain inert in the periphery.
Engineered antibodies that exploit the tumor microenvironment using pH-selective properties are becoming an emerging modality of biotherapeutics that may alleviate undesirable properties or bolster specific activities. For instance, pH-selective antibodies have been engineered to modulate exposure and overcome short antibody half-lives caused by TMDD or sinks caused by high peripheral expression.Citation13,Citation16–18,Citation20 To build pH sensitivity into antibodies, incorporation of amino acids such as histidine and glutamic acid at antibody–antigen interfaces can create favorable electrostatic charge interactions in acidic pH environments. In addition, careful library designs and selection strategies can enrich clones with desired pH-dependent and affinity-tuned properties.Citation19 However, while histidine amino acids are considered to have a pKa ~6.5, the local molecular environmental features of its side chain within a protein, such as surface accessibility and the number of polar contacts, can result in a variable pKa.Citation27 The abundance of these pH-sensitive residues on a target protein can also affect the capacity for pH engineering. For example, VISTA is unusually rich with histidine residues and incorporation of negatively charged amino acids into antibody CDRs conferred pH selectivity.Citation13 However, pH engineering of Ipi in this study required incorporation of both negatively charged amino acids and histidine substitutions proximal to complementary charged residues or interfaces in CTLA-4. Altogether, since protein–protein interfaces are unique and the abundance and positions of pH-sensitive residues are variable, pH engineering may require a tailored approach through molecular modeling and in vitro evolution.
Other acidic as well as neutral pH-selective antibodies against CTLA-4 have been described.Citation15,Citation28,Citation29 The acidic pH-selective antibodies from others have shown enhanced anti-tumor activity and an increased CD8+ T cell to Treg ratio. These antibodies also had reduced toxicity in nonhuman primates in combination with an anti-PD-1 antibody. In contrast, a neutral pH-selective variant of tremelimumab (Treme) was reported to have alternative lysosomal degradation resulting in reduced CTLA-4 downregulation and increased antibody bioavailability.Citation28 These Treme variants, which were formatted in the IgG1 backbone, had increased depletion of intratumoral Tregs in comparison to Ipi. While these antibodies have opposing pH-selectivity profiles, they converge on improvement of anti-CTLA-4 therapy. The acidic pH-selective anti-CTLA-4 antibodies may be able to better target the tumor microenvironment and also prime T cell activation in lymph nodes, which have acidic niches produced by T cells.Citation30 On the other hand, neutral pH-selective antibodies may act to increase antibody recycling and prevent CTLA-4 downregulation. While both modalities appear to provide benefit for targeting CTLA-4, further investigation is needed to evaluate the distinct modes of action between these pH-dependent antibodies.
Application of pH-sensitive biologics requires a firm understanding of pH biology. Reliable, safe, and noninvasive ways to measure the pH of tumors is important to better define the ideal characteristics of pH-selective therapeutic antibodies. It is generally accepted that tumors have low pH due to oncogene expression, high rates of glycolysis, and accumulation of acidic waste due to poor vascularization and large tumor size.Citation31 Data to support the acidification of tumors has historically been generated with the use of physical probes to directly measure pH, but they are invasive and may not be universally amenable for clinical applications depending on tumor accessibility. pH-sensitive peptides such as pHLIP (pH-low insertion peptide) technologies can be used for diagnostic nuclear or fluorescent imaging and have also been used to deliver pH-sensitive peptide-drug anti-tumor conjugates.Citation32–34 Other noninvasive imaging techniques have been applied in preclinical mouse models as well as human tumors, such as photoacoustic imaging and MRI-CEST.Citation35–37 However, since the pH can be heterogenous in tumors or in peripheral tissues, it is critical that these methods are reliable and accurate to map the pH landscape. These promising technological advancements will better inform understanding of pH in patients to usher pH-selective antibodies into the clinic. In particular for pH Ipi, it would be desirable for clinical testing of pH to occur in indications where Ipi has shown activity as a single agent or in combination with anti-PD-1.
In addition to understanding pH gradients across different tissues, the expression of CTLA-4 and the consequences of its blockade are important to consider when designing an acidic-selective variant of ipilimumab. While lymph nodes have been recently described as having acidic niches that limit T cell activation under homeostatic conditions,Citation30 the role of CTLA-4 in these acidic niches has not been explored. Our results suggest that T cells activated at low pH can upregulate expression and cycling of CTLA-4, which can then be engaged by Ipi or the acidic-selective variants to enhance T cell function. Our results show no evidence of activity in lymph nodes, since the PD markers for CTLA-4 binding were not different between the pH Ipi variants and isotype control but significantly reduced compared to Ipi-NF, despite comparable activity in the tumor. However, a role for CTLA-4 blockade in tumor draining lymph nodes cannot be ruled out. Further studies to determine the relationship between CTLA-4 expression and regions of low pH in the lymph node, such as by imaging techniques or additional models of inflammation, are needed to understand how much CTLA-4 blockade is desirable in the lymph node and which pH Ipi variant may have the most appropriate pH-selective properties.
Context-dependent activation is an emerging modality for the next generation of therapeutic antibodies. In addition to the pH-dependent antibodies, other special attributes of the tumor microenvironment may be harnessed, such as elevated ATP levels.Citation38,Citation39 Furthermore, bispecific antibodies with appropriately adjusted affinities that account for differential receptor density between tumor and peripheral tissues can also fine-tune therapeutic activity under certain conditions.Citation40,Citation41 Altogether, combining protein engineering approaches toward disease biology may provide creative solutions to tackle challenging and unmet medical needs.
Materials and methods
pH Ipi design and phage library construction
The ipilimumab-CTLA-4 epitope and paratope were identified from the available complex crystal structure (PDB ID 5TRU) using CCP4 programsCitation42 Contact and AreaiMol and were then manually inspected. An initial mini library was built by splicing by overlap extension polymerase-chain reaction (SOE-PCR). One histidine substitution introduced into each heavy chain CDR was enforced at positions 28, 30, 31, 32, 33, 52, 52a, 53, 54, 55, 56, 95, 97, and 98 (Kabat numbering) for a total of three histidine mutations per variant, resulting in a theoretical diversity of 90 members. The single-chain variable fragment (scFv) library was inserted by Gibson Assembly (NEB) into the phagemid vector, which contains a phoA promoter, pelB leader sequence, and a C-terminal FLAG tag followed by a fragment of the phage coat protein pIII. The phagemid DNA was electroporated into SS320 cells (Lucigen) and the phage library was propagated and purified.
For the full pH Ipi library, 27 CDR residues across the heavy and light chains near or at the CTLA-4 interface were selected for substitution with histidine, aspartic acid, and glutamatic acid and 321 oligos from Integrated DNA Technologies were synthesized (24, 89, 8, 184, 8, and 8 oligos in CDRs H1, H2, H3, L1, L2, and L3, respectively). 19 VH and 19 VL sub-libraries were individually constructed in-house by SOE-PCR with no more than three substitutions per chain. The VH and VL libraries have a theoretical diversity of 690 and 1,079 members, respectively, and the combinatorial VH-VL library has a theoretical diversity of 744,510 members. The scFv libraries were inserted by Gibson Assembly (NEB) into the phagemid vector. The phagemid DNA was electroporated into SS320 cells (Lucigen) and the phage library was propagated and purified. Phage titers for each of the libraries (VH only, VL only, and combinatorial VH-VL) exceeded 3 × 1012 cfu/mL.
Phage selections
Positive and negative selections (i.e., binding at pH 6.0 and eluting at pH 7.4) were performed to select Ipi variants with pH-dependent binding properties. Each streptavidin pre-cleared phage library (1x1013 particles) was incubated with biotinylated human CTLA-4 at pH 6.0 (1x phosphate-buffered saline (PBS) pH 6.0, 0.05% Tween 20, 0.5% bovine serum albumin (BSA)). Rounds of phage selections were performed manually for the mini library. Biotinylated CTLA-4 were captured onto streptavidin-adsorbed plates. The phage libraries were then added to the plates and washed at pH 6.0 (1x PBS pH 6.0, 0.05% Tween 20, 0.5% BSA). Phage were eluted at pH 7.4 (1x PBS pH 7.4, 0.05% Tween 20, 0.5% BSA) and amplified overnight in XL1-blue cells (Agilent). Four rounds of panning were performed. Single colonies were plated from the eluted phage pools and the scFv insert was amplified by PCR for Sanger sequencing. The DNA sequences were inspected manually or analyzed with in-house software to track sequence trends in the CDRs.
Rounds of automated phage selections using the full pH Ipi library were performed using a KingFisher Flex Purification System (Thermo Fisher Scientific). Streptavidin-coated magnetic beads (Promega) were used to capture the CTLA-4-scFv-phage complexes and were washed at pH 6.0. Phage were eluted at pH 7.4 and amplified overnight in XL1-blue cells (Agilent). Four rounds of panning were performed. Single colonies were plated from the eluted phage pools and the scFv insert was amplified by PCR for Sanger sequencing. The DNA sequences were inspected manually or analyzed with in-house software to track sequence trends in the CDRs.
Protein expression and purification
The Fab and IgG1 constructs were individually cloned into the pTT5 expression plasmids (GenScript) with N-terminal osteonectin signal peptides. The Fabs have a poly-histidine tag fused to the C-terminus of the heavy chain. Human CTLA-4 was similarly cloned into the pTT5 expression plasmid with a C-terminal Myc, poly-histidine, and BirA biotinylation tag, as well as another plasmid with a C-terminal poly-histidine tag for crystallography studies. The proteins were expressed by transient transfection using the Expi293 Expression System (Thermo Fisher Scientific). For NF antibody production, FUT8-knockout Expi293 lines were transfected. The Fab and CTLA-4 supernatants were purified using Ni Sepharose excel (GE Healthcare Life Sciences) and buffer exchanged into 1x PBS. The IgG1 and IgG1-NF supernatants were purified using MAbSelect SuRe (GE Healthcare Life Sciences) and buffer exchanged into 1x PBS.
Biolayer interferometry binding measurements
Binding affinity of recombinant pH Ipi Fab variants to CTLA-4 was screened using an Octet HTX at neutral and acidic conditions using that buffers that contain 150 mM NaCl, 0.05% Tween 20, 0.5% BSA and either 50 mM Tris pH 7.5 or 50 mM MES pH 6.0 at 30°C. Enzymatically biotinylated human CTLA-4 (2 µg/mL) was loaded onto streptavidin biosensors (180 sec) and 50 nM Fab association (180–300 sec) and dissociation (300–1200 sec) was measured at each pH. Representative binding traces are shown in Figure S15.
Surface plasmon resonance spectroscopic determination of binding parameters
SPR was used to determine binding parameters for various Ipi variants to human and cyno CTLA-4 with a BIACORE® T200 SPR spectrometer (Biacore AB, Uppsala, Sweden) at 37°C. Fabs of Ipi variants were captured on a BIACORE® CM4 chip with immobilized anti-human kappa polyclonal capture antibody. Monomeric human or cyno CTLA-4 was flowed as analyte with up to 2 μM top concentration. Between cycles, the capture surface was regenerated with 75 mM phosphoric acid. Running buffers were HEPES buffered saline for pH 7.4 experiments and Bis-Tris buffered saline for experiments with lower pH. All buffers were supplemented with 0.05% Tween-20 and 1 g/L BSA. Double-referenced sensorgrams were fitted to a 1:1 Langmuir binding model with mass transport to determine equilibrium dissociation constants (KD), as well as association (ka) and dissociation (kd) rate constants where appropriate.
X-ray crystallography
CTLA-4 was expressed in Expi293 cells treated with kifunensine and purified protein was treated with EndoH. For complex formation, CTLA-4 was added to either Ipi.105 Fab or Ipi.106 Fab in a 1.1:1 molar excess at pH ≤ 6.0 and incubated at room temperature for 1 hr. The Ipi.105 or Ipi.106 complexes were then purified from unbound CTLA-4 by gel filtration using 50 mM NaCl, 25 mM MES pH 5.8 or 50 mM NaCl, 10 mM Bis-Tris pH 5.6 as the running buffers, respectively. Ipi.105-CTLA-4 crystals were grown by sitting drop vapor diffusion at 20°C by mixing 0.2 µL of concentrated protein sample (10.8 mg/mL) with 0.2 µL of mother liquor (0.2 M ammonium citrate dibasic, 20% w/v PEG 1,000, pH 5.2) and crystals appeared in 7 days. Ipi.106-CTLA-4 crystals were similarly grown at by mixing 0.2 µL of concentrated protein sample (10.8 mg/mL) with 0.2 µL mother liquor (0.2 M sodium citrate tribasic dehydrate, 22% PEG 3,350). Crystals were cryo-protected with 15% ethylene glycol and flash frozen in liquid nitrogen. X-ray diffraction data were collected at beamline 17ID (wavelength 1.0 Å) at the Advanced Photon Source (Argonne National Laboratory, Lemont, IL) under cryo conditions using a DECTRIS Eiger2 X 9 M detector. Diffraction data were processed with the autoPROCCitation43 toolbox that made use of external programs XDS/SCALE,Citation44 POINTLESS,Citation45 CCP4,Citation42 and STARANISO (Global Phasing Limited) for ellipsoidal truncation and anisotropic scaling. The structures were determined by molecular replacement with Phaser using Fab variable and constant domains (PDB IDs 5TRU, 4NM4, respectively) and CTLA-4 (PDB ID 3OSK). Two Ipi.105-CTLA-4 complexes and one Ipi.106-CTLA-4 complex was found in the asymmetric unit. The models were iteratively built using CootCitation46 and refined in Phoenix.Citation47 In the final structure, 94.7% and 95.8% of the residues are in favored regions of the Ramachandran plot with 0.3% and 0.4% outliers as calculated by MolProbityCitation48 for the Ipi.105 and Ipi.106 complexes, respectively. X-ray diffraction data collection and refinement statistics are reported in Table S5.
Receptor quantification
The Quantum Alexa Fluor 647 MESF kit (Bangs Laboratories) was used for the quantitation of AF647 fluorescence intensity in Molecules of Equivalent Soluble Fluorophore (MESF). The beads were analyzed on the Cytoflex-S and a standard curve was established. The MESF value at saturating concentrations of the Alexa Fluor 647 labeled Ipi variants were calculated from the QuickCal template provided. The number of surface receptors per cell was determined by dividing that MESF value by its degree of labeling.
In vitro cell binding assays
Ipi IgG-NF variants were labeled using the Alexa Fluor 647 labeling kit (Invitrogen) according to manufacturer instructions. The degree of labeling of each IgG was determined by a ND-ONE-C-W spectrophotometer (NanoDrop). Approximately 1.5 × 105 cells were added to each well in 96-well plates and incubated with the labeled antibodies in HBSS, 2% heat-inactivated fetal bovine serum (HI-FBS), 0.02% sodium azide, 2 mM EDTA, and titrated to their specific pH using sodium phosphate pH 5.4 for 30 minutes at 4°C in the dark. The Fab and IgG variants were titrated from 200 µg/mL or 60 µg/mL, respectively, at a 1:4 dilution across 8 or 12 points in their respective pH buffer. Cells were washed three times with their respective pH buffer and binding of the antibodies were detected on a Cytoflex-S instrument (Beckman Coulter). The Fabs were detected using an in-house anti-hKappa monovalent secondary antibody conjugated to Alexa Fluor 647. Data were analyzed with the FlowJo v10.6.1 software and binding was determined by mean fluorescence intensity.
For primary cell binding, cells were isolated from leukopaks (Discovery Life Sciences) via EasySep Human CD4+ T cell Isolation Kit (Stemcell, 17952) followed by Human CD25 II MicroBeads (Miltenyi, 130–092-983). CD4+CD25+ cells were then stimulated overnight with 1:1 Human T-Activator CD3/CD28 DynabeadsTM (ThermoFisher Scientific, 11132D) in RPMI 1640 + 10% HI-FBS and activation was confirmed by CD25 stain. For assessment of total CTLA-4: cells were stained with surface markers then fixed in eBioscience FOXP3/transcription factor fix buffer for 1 hour at room temperature followed by block with isotype in perm buffer (1x PBS + 1% HI-FBS +0.1% saponin + phosphoric acid (or NaOH for pH 7.2) for 15 minutes at room temperature; and stain with labeled Ipi IgG-NF variants in pH perm buffer for 1 hour at room temperature. For cycling assays: Overnight-stimulated CD4+CD25+ cells were washed and plated in RPMI 1640 + 10% HI-FBS titrated to desired pH containing 1 µg/mL labeled Ipi IgG-NF variants for 4 hours at 37°C, 8% CO2. Cells were then washed and stained for surface markers. All samples were run on BD LSRFortessaTM. Data were analyzed using FlowJo V10.
In vitro functional assays
For T cell activation at low pH, human peripheral blood mononuclear cells (PBMCs) were stimulated overnight in pH-media: RPMI-1640, 10% HI-FBS, sodium bicarbonate and sodium phosphate buffer (pH 5.4). Sodium phosphate buffer was added until the desired pH was reached (e.g., pH 7.2, 7.0, 6.8), then sterile filtered with 5 µg/mL CD3 and 1 mg/mL CD28, then stained for Zombie Aqua (BioLegend 771430), CD4 (BioLegend 317442), CD25 (Miltenyi 130–115-534), and CTLA-4 (BioLegend 369612 and 369626) per the manufacturer’s instructions.
Reporter cell line blocking: One day prior to running the assay, rhB7-1 was diluted in 1x PBS and plated in 100 µL/well at 1.0 µg/mL in 96-well flat-bottom plates and incubated at 4°C overnight. Ipi IgG-NF variants tested for blockade of 58αβ-hCTLA-4/mCD3ζ cells and plate-bound rhB7-1. 58αβ-hCTLA-4/mCD3ζ cells were collected and diluted to 1 × 106 cells/mL in RPMI + 10% HI-FBS. Coated plates were washed once with 150 µL/well of 1x PBS. Diluted cells were then added to the plates at 100 µL/well for a final count of 1 × 105 cells/well and incubated at 37°C, 8% CO2, for 6 hours. After incubation, plates were centrifuged at 500xg for 1 minute and supernatant was aspirated. pH Ipi variants were titrated for a final top concentration of 15 µg/mL at a 1:4 dilution across 8 points in triplicate in pH-specific media and 150 µL/well of antibody titrations were added to the plates containing pre-incubated cells. Plates were incubated at 37°C, 8% CO2, for 18 hours and supernatant was removed and tested for mouse IL-2 production by ELISA (BD).
Primary human cells: Frozen PBMCs were thawed and rested overnight at 37°C in RPMI 1640 + 10% HI-FBS. PBMCs were then plated at 5 × 105 cells/mL with 20 ng/mL SEA (Toxin Technology, AT101red) for 18 hours. Media was aspirated and replaced with pH-media containing SEA and Ipi IgG-NF variants titrated 1:4 from 25 mg/mL for 72 hours. Supernatants were collected and IL-2 was assessed by Human IL-2 ELISA Set (BD, BDB555190). Samples were read on MD SpectraMax 340PC at 450 nm and 570 nm.
In vivo studies
8–12 week old male and female huCTLA-4-KIflox:iTL (B6NJ) mice obtained from Taconic were subcutaneously injected with 1 × 106 MC38. After 7 days, tumor volumes were measured and mice were randomized into treatment groups of 50/50 male/female with comparable mean tumor volumes (80–120 mm3). 200 μg of Ipi IgG-NF variants or keyhole limpet hemocyanin isotype control antibodies formulated in PBS were administered intravenously (i.v.) on day 9. Tumor volumes were recorded 3 times weekly for efficacy assessment. For pharmacokinetics assessment, microbleeds were performed at 0.25, 2, 48, and 120 hours post treatment. Blood was diluted 10x in Rexxip Buffer and antibody concentrations were detected by Ligand binding assay on Gyrolab®. For pharmacodynamic assessment, mice were sacrificed and tumor, spleen, and draining lymph node were harvested for analysis on day 13 after tumor implantation. Single cell suspensions were prepared by dissociating tissue with gentleMACSTM C Tubes (Miltenyi) followed by enzymatic digestion of tumors (3 mL) and lymph nodes (0.5 mL) in digest mix containing 250 U/mL Collagenase IV, 100 µg/mL DNAse1, 5 mM CaCl2, and 5% FBS in HBSS for 30 mins at 37°C then quenched with 7 mL cold RPMI, 10% FBS, 2 mM EDTA. 1 × 106 cells per sample were stained with immune monitoring panel (Figure S16) and run on BD LSRFortessaTM. Data were analyzed using FlowJo V10.
Tumor pH was measured in C57Bl/6 N mice inoculated with 1 × 106 MC38 cells grown to 100–200 mm3, then measured with Presens pH sensor probe.
Accelerated stability studies
Stability samples: The mAbs were formulated in an optimized buffer at pH 6.0. All antibody solutions were incubated at 50 mg/mL in a refrigerator (4–5°C) and in specialized constant temperature ovens (25°C and 40°C) to monitor physical and chemical stability for 1 month.
Mass spectrometry-based peptide mapping: Proteins were denatured in the presence of 0.2% Rapigest surfactant (Waters Corp.) at 80°C for 30 min, reduced by dithiothreitol at 56°C for 30 min, alkylated by iodoacetamide at room temperature for 30 min in the dark, and digested by trypsin at 37°C for 4 hrs in Tris pH 7.4 followed by acidic quench. Peptides were analyzed on an ACQUITY UPLC system (Waters Manchester, U.K.) coupled to a Q-Exactive™ Plus mass spectrometer (Thermo Scientific, San Jose, CA). Peptides were eluted from a Waters BEH C18 column (130 Å 1.7 µm 2.1 × 150 mm), heated at 50°C, using a 60 min LC gradient. The gradient setting was 0.2% to 40% solvent B in 45 min at 0.2 mL/min flow rate. Solvent A was 0.1% formic acid (FA) in water and solvent B was 0.1% FA in acetonitrile. The mass spectrometer was operated under positive ion mode with ESI voltage at 3.5 kV, capillary temperature: 250°C, scan range: 300–2000 m/z, sheath gas flow rate: 45.
Abbreviations
Supplemental Material
Download Zip (7.9 MB)Acknowledgments
The authors thank Meaghan Happer, Andy Deng, Sean West, SJ Diong, Amanda Rhea, Chris Kimberlin, Felix Findeisen, Feng Wang, Srikanth Kotapati, Rahima Akter, and Mike Quigley for reagents, technical advice, and scientific discussions. The authors thank Steven Sheriff, Jodi Muckelbauer, and the staff at the IMCA-CAT at the Argonne National Laboratory for assistance in x-ray data collection and beamline support.
Disclosure statement
All authors are current or former employees of Bristol Myers Squibb. Bristol Myers Squibb has filed a patent application related to the subject matter of this paper; WO 2020/214748, Ipilimumab variants with enhanced specificity for binding at low pH.
Supplementary material
Supplemental data for this article can be accessed on the publisher’s website.
Additional information
Funding
References
- Krummel MF, Allison JP. CD28 and CTLA-4 have opposing effects on the response of T cells to stimulation. J Exp Med. 1995;182:459–14. doi:10.1084/jem.182.2.459.
- Fife BT, Bluestone JA. Control of peripheral T-cell tolerance and autoimmunity via the CTLA-4 and PD-1 pathways. Immunol Rev. 2008;224:166–82. doi:10.1111/j.1600-065X.2008.00662.x.
- Walunas TL, Lenschow DJ, Bakker CY, Linsley PS, Freeman GJ, Green JM, Thompson CB, Bluestone JA. CTLA-4 can function as a negative regulator of T cell activation. Immunity. 1994;1:405–13. doi:10.1016/1074-7613(94)90071-X.
- Selby MJ, Engelhardt JJ, Quigley M, Henning KA, Chen T, Srinivasan M, Korman AJ. Anti-CTLA-4 antibodies of IgG2a isotype enhance antitumor activity through reduction of intratumoral regulatory T cells. Cancer Immunol Res. 2013;1:32–42. doi:10.1158/2326-6066.CIR-13-0013.
- Arce Vargas F, Furness AJS, Litchfield K, Joshi K, Rosenthal R, Ghorani E, Solomon I, Lesko MH, Ruef N, Roddie C, et al. Fc effector function contributes to the activity of human anti-CTLA-4 antibodies. Cancer Cell. 2018;33:649–63 e4. doi:10.1016/j.ccell.2018.02.010.
- Ascierto PA, Del Vecchio M, Robert C, Mackiewicz A, Chiarion-Sileni V, Arance A, Lebbé C, Bastholt L, Hamid O, Rutkowski P, et al. Ipilimumab 10 mg/kg versus ipilimumab 3 mg/kg in patients with unresectable or metastatic melanoma: a randomised, double-blind, multicentre, phase 3 trial. Lancet Oncol. 2017;18:611–22. doi:10.1016/S1470-2045(17)30231-0.
- Okazaki A, Shoji-Hosaka E, Nakamura K, Wakitani M, Uchida K, Kakita S, Tsumoto K, Kumagai I, Shitara K. Fucose depletion from human IgG1 oligosaccharide enhances binding enthalpy and association rate between IgG1 and FcgammaRIIIa. J Mol Biol. 2004;336:1239–49. doi:10.1016/j.jmb.2004.01.007.
- Shields RL, Lai J, Keck R, O’Connell LY, Hong K, Meng YG, Weikert SHA, Presta LG. Lack of fucose on human IgG1 N-linked oligosaccharide improves binding to human Fcgamma RIII and antibody-dependent cellular toxicity. J Biol Chem. 2002;277:26733–40. doi:10.1074/jbc.M202069200.
- Wang W, Erbe AK, Hank JA, Morris ZS, Sondel PM. NK cell-mediated antibody-dependent cellular cytotoxicity in cancer immunotherapy. Front Immunol. 2015;6:368. doi:10.3389/fimmu.2015.00368.
- Desnoyers LR, Vasiljeva O, Richardson JH, Yang A, Menendez EE, Liang TW, Wong C, Bessette PH, Kamath K, Moore SJ, et al. Tumor-specific activation of an EGFR-targeting probody enhances therapeutic index. Sci Transl Med. 2013;5:207ra144. doi:10.1126/scitranslmed.3006682.
- Rohani N, Hao L, Alexis MS, Joughin BA, Krismer K, Moufarrej MN, Soltis AR, Lauffenburger DA, Yaffe MB, Burge CB, et al. Acidification of tumor at stromal boundaries drives transcriptome alterations associated with aggressive phenotypes. Cancer Res. 2019;79:1952–66. doi:10.1158/0008-5472.CAN-18-1604.
- El-Kenawi A, Gatenbee C, Robertson-Tessi M, Bravo R, Dhillon J, Balagurunathan Y, Berglund A, Vishvakarma N, Ibrahim-Hashim A, Choi J, et al. Acidity promotes tumour progression by altering macrophage phenotype in prostate cancer. Br J Cancer. 2019;121(7):556–66. doi:10.1038/s41416-019-0542-2.
- Johnston RJ, Su LJ, Pinckney J, Critton D, Boyer E, Krishnakumar A, Corbett M, Rankin AL, Dibella R, Campbell L, et al. Vista is an acidic pH-selective ligand for PSGL-1. Nature. 2019;574:565–70. doi:10.1038/s41586-019-1674-5.
- Sulea T, Rohani N, Baardsnes J, Corbeil CR, Deprez C, Cepero-Donates Y, Robert A, Schrag JD, Parat M, Duchesne M, et al. Structure-based engineering of pH-dependent antibody binding for selective targeting of solid-tumor microenvironment. MAbs. 2020;12:1682866. doi:10.1080/19420862.2019.1682866.
- Chang HW, Frey G, Liu H, Xing C, Steinman L, Boyle WJ, Short JM. Generating tumor-selective conditionally active biologic anti-CTLA4 antibodies via protein-associated chemical switches. Proc Natl Acad Sci U S A. 2021;118. doi:10.1073/pnas.2020606118.
- Igawa T, Ishii S, Tachibana T, Maeda A, Higuchi Y, Shimaoka S, Moriyama C, Watanabe T, Takubo R, Doi Y, et al. Antibody recycling by engineered pH-dependent antigen binding improves the duration of antigen neutralization. Nat Biotechnol. 2010;28(11):1203–07. doi:10.1038/nbt.1691.
- Chaparro-Riggers J, Liang H, DeVay RM, Bai L, Sutton JE, Chen W, Geng T, Lindquist K, Casas MG, Boustany LM, et al. Increasing serum half-life and extending cholesterol lowering in vivo by engineering antibody with pH-sensitive binding to PCSK9. J Biol Chem. 2012;287:11090–97. doi:10.1074/jbc.M111.319764.
- Devanaboyina SC, Lynch SM, Ober RJ, Ram S, Kim D, Puig-Canto A, Breen S, Kasturirangan S, Fowler S, Peng L, et al. The effect of pH dependence of antibody-antigen interactions on subcellular trafficking dynamics. MAbs. 2013;5:851–59. doi:10.4161/mabs.26389.
- Schroter C, Gunther R, Rhiel L, Becker S, Toleikis L, Doerner A, Becker J, Schönemann A, Nasu D, Neuteboom B, et al. A generic approach to engineer antibody pH-switches using combinatorial histidine scanning libraries and yeast display. MAbs. 2015;7:138–51. doi:10.4161/19420862.2014.985993.
- Fukuzawa T, Sampei Z, Haraya K, Ruike Y, Shida-Kawazoe M, Shimizu Y, Gan SW, Irie M, Tsuboi Y, Tai H, et al. Long lasting neutralization of C5 by SKY59, a novel recycling antibody, is a potential therapy for complement-mediated diseases. Sci Rep. 2017;7:1080. doi:10.1038/s41598-017-01087-7.
- Dovedi SJ, Elder MJ, Yang C, Sitnikova SI, Irving L, Hansen A, Hair J, Jones DC, Hasani S, Wang B, et al. Design and efficacy of a monovalent bispecific PD-1/CTLA4 antibody that enhances CTLA4 blockade on PD-1(+)PD-1+ activated T cells. Cancer Discov. 2021;11:1100–17. doi:10.1158/2159-8290.CD-20-1445.
- Alegre ML, Frauwirth KA, Thompson CB. T-cell regulation by CD28 and CTLA-4. Nat Rev Immunol. 2001;1:220–28. doi:10.1038/35105024.
- Bosticardo M, Ariotti S, Losana G, Bernabei P, Forni G, Novelli F. Biased activation of human T lymphocytes due to low extracellular pH is antagonized by B7/CD28 costimulation. Eur J Immunol. 2001;31:2829–38. doi:10.1002/1521-4141(200109)31:9<2829::AID-IMMU2829>3.0.CO;2-U.
- Krummel MF, Sullivan TJ, Allison JP. Superantigen responses and co-stimulation: CD28 and CTLA-4 have opposing effects on T cell expansion in vitro and in vivo. Int Immunol. 1996;8:519–23. doi:10.1093/intimm/8.4.519.
- Jin HS, Choi DS, Ko M, Kim D, Lee DH, Lee S, Lee AY, Kang SG, Kim SH, Jung Y, et al. Extracellular pH modulating injectable gel for enhancing immune checkpoint inhibitor therapy. J Control Release. 2019;315:65–75. doi:10.1016/j.jconrel.2019.10.041.
- Callahan MK, Postow MA, Wolchok JD. Targeting T Cell Co-receptors for Cancer Therapy. Immunity. 2016;44:1069–78. doi:10.1016/j.immuni.2016.04.023.
- Edgcomb SP, Murphy KP. Variability in the pKa of histidine side-chains correlates with burial within proteins. Proteins. 2002;49:1–6. doi:10.1002/prot.10177.
- Zhang Y, Du X, Liu M, Tang F, Zhang P, Ai C, Fields JK, Sundberg EJ, Latinovic OS, Devenport M, et al. Hijacking antibody-induced CTLA-4 lysosomal degradation for safer and more effective cancer immunotherapy. Cell Res. 2019;29:609–27. doi:10.1038/s41422-019-0184-1.
- Gao H, Cai H, Liu J, Wang X, Zheng P, Devenport M, Xu T, Dou F, Liu Y, Zhou A, et al. Structure of CTLA-4 complexed with a pH-sensitive cancer immunotherapeutic antibody. Cell Discov. 2020;6:79. doi:10.1038/s41421-020-00202-9.
- Wu H, Estrella V, Beatty M, Abrahams D, El-Kenawi A, Russell S, Ibrahim-Hashim A, Longo DL, Reshetnyak YK, Moshnikova A, et al. T-cells produce acidic niches in lymph nodes to suppress their own effector functions. Nat Commun. 2020;11:4113. doi:10.1038/s41467-020-17756-7.
- Huber V, Camisaschi C, Berzi A, Ferro S, Lugini L, Triulzi T, Tuccitto A, Tagliabue E, Castelli C, Rivoltini L., et al. Cancer acidity: an ultimate frontier of tumor immune escape and a novel target of immunomodulation. Semin Cancer Biol. 2017;43:74–89. doi:10.1016/j.semcancer.2017.03.001.
- Wyatt LC, Lewis JS, Andreev OA, Reshetnyak YK, Engelman DM. Applications of pHLIP Technology for Cancer Imaging and Therapy. Trends Biotechnol. 2017;35:653–64. doi:10.1016/j.tibtech.2017.03.014.
- Wyatt LC, Moshnikova A, Crawford T, Engelman DM, Andreev OA, Reshetnyak YK. Peptides of pHLIP family for targeted intracellular and extracellular delivery of cargo molecules to tumors. Proc Natl Acad Sci U S A. 2018;115:E2811–E8. doi:10.1073/pnas.1715350115.
- Gayle S, Aiello R, Leelatian N, Beckta JM, Bechtold J, Bourassa P, Csengery J, Maguire RJ, Marshall D, Sundaram RK, et al. Tumor-selective, antigen-independent delivery of a pH sensitive peptide-topoisomerase inhibitor conjugate suppresses tumor growth without systemic toxicity. NAR Cancer. 2021;3:zcab021. doi:10.1093/narcan/zcab021.
- Chen LQ, Howison CM, Jeffery JJ, Robey IF, Kuo PH, Pagel MD. Evaluations of extracellular pH within in vivo tumors using acidoCEST MRI. Magn Reson Med. 2014;72:1408–17. doi:10.1002/mrm.25053.
- Jo J, Lee CH, Kopelman R, Wang X. In vivo quantitative imaging of tumor pH by nanosonophore assisted multispectral photoacoustic imaging. Nat Commun. 2017;8:471. doi:10.1038/s41467-017-00598-1.
- Consolino L, Anemone A, Capozza M, Carella A, Irrera P, Corrado A, Dhakan C, Bracesco M, Longo DL. Non-invasive investigation of tumor metabolism and acidosis by MRI-CEST imaging. Front Oncol. 2020;10:161. doi:10.3389/fonc.2020.00161.
- Mimoto F, Tatsumi K, Shimizu S, Kadono S, Haraya K, Nagayasu M, Suzuki Y, Fujii E, Kamimura M, Hayasaka A, et al. Exploitation of elevated extracellular ATP to specifically direct antibody to tumor microenvironment. Cell Rep. 2020;33:108542. doi:10.1016/j.celrep.2020.108542.
- Kamata-Sakurai M, Narita Y, Hori Y, Nemoto T, Uchikawa R, Honda M, Hironiwa N, Taniguchi K, Shida-Kawazoe M, Metsugi S, et al. Antibody to CD137 activated by extracellular adenosine triphosphate is tumor selective and broadly effective in vivo without systemic immune activation. Cancer Discov. 2021;11:158–75. doi:10.1158/2159-8290.CD-20-0328.
- Piccione EC, Juarez S, Liu J, Tseng S, Ryan CE, Narayanan C, Wang L, Weiskopf K, Majeti R. A bispecific antibody targeting CD47 and CD20 selectively binds and eliminates dual antigen expressing lymphoma cells. MAbs. 2015;7:946–56. doi:10.1080/19420862.2015.1062192.
- Haber L, Olson K, Kelly MP, Crawford A, DiLillo DJ, Tavare R, Ullman E, Mao S, Canova L, Sineshchekova O, et al. Generation of T-cell-redirecting bispecific antibodies with differentiated profiles of cytokine release and biodistribution by CD3 affinity tuning. Sci Rep. 2021;11:14397. doi:10.1038/s41598-021-93842-0.
- Winn MD, Ballard CC, Cowtan KD, Dodson EJ, Emsley P, Evans PR, Keegan RM, Krissinel EB, Leslie AGW, McCoy A, et al. Overview of the CCP4 suite and current developments. Acta Crystallogr D Biol Crystallogr. 2011;67:235–42. doi:10.1107/S0907444910045749.
- Vonrhein C, Flensburg C, Keller P, Sharff A, Smart O, Paciorek W, Womack T, Bricogne G. Data processing and analysis with the autoPROC toolbox. Acta Crystallogr D Biol Crystallogr. 2011;67:293–302. doi:10.1107/S0907444911007773.
- Kabsch W. XDS. Acta Crystallogr D Biol Crystallogr. 2010;66:125–32. doi:10.1107/S0907444909047337.
- Evans P. Scaling and assessment of data quality. Acta Crystallogr D Biol Crystallogr. 2006;62:72–82. doi:10.1107/S0907444905036693.
- Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr D Biol Crystallogr. 2010;66:486–501. doi:10.1107/S0907444910007493.
- Adams PD, Afonine PV, Bunkoczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung L-W, Kapral GJ, Grosse-Kunstleve RW, et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66:213–21. doi:10.1107/S0907444909052925.
- Chen VB, Arendall WB 3rd, Headd JJ, Keedy DA, Immormino RM, Kapral GJ, Murray LW, Richardson JS, Richardson DC. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr D Biol Crystallogr. 2010;66:12–21. doi:10.1107/S0907444909042073.
