ABSTRACT
Anti-SARS-CoV-2 monoclonal antibodies and vaccines have shown improvement in lowering viral burden and hospitalization. However, emerging SARS-CoV-2 variants contain neutralizing antibody-escape mutations. Therefore, several reports have suggested the administration of recombinant angiotensin-converting enzyme 2 (rACE2) as a soluble receptor trap to block SARS-CoV-2 infection and limit viral escape potential. Several strategies have been implemented to enhance the efficacy of rACE2 as a therapeutic agent. Fc fusions have been used to improve pharmacokinetics and boost the affinity and avidity of ACE2 decoys for the virus spike protein. Furthermore, the intrinsic catalytic activity of ACE2 can be eliminated by introducing point mutations on the catalytic site of ACE2 to obtain an exclusive antiviral activity. This review summarizes different evolution platforms that have been used to enhance ACE2-Fc (i.e., immunoadhesins) as potential therapeutics for the current pandemic or future outbreaks of SARS-associated betacoronaviruses.
Introduction
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) has been identified as the seventh coronavirus that is known to cause human respiratory disease.Citation1 This novel virus, which belongs to the genus betacoronavirus, has a single-stranded positive-sense RNA genome. It is the causative agent of the coronavirus disease 19 (COVID-19) that emerged in China and swiftly spread to the rest of the world starting in late 2019.Citation2 According to the World Health Organization, COVID-19 has affected nearly every healthcare system in the world, and infection with the virus has led to millions of deaths.
The 30-kb SARS-CoV-2 genome encodes four main structural proteins: the spike (S) glycoprotein, membrane (M) protein, an envelope (E) protein and a nucleocapsid (N) protein.Citation3 The S protein on the surface of the virus is known to play the most important roles in viral attachment, fusion and entry. Therefore, S protein has been exhaustively studied as a key target for vaccine and therapeutic development.Citation4
The entry of SARS-CoV-2 into the host cells is mediated by the binding of the S protein to the host cellular membrane-bound angiotensin-converting enzyme 2 (ACE2) receptors.Citation5–8 The S protein on the surface of the virus is composed of S1, which is responsible for binding, and S2, which is responsible for membrane fusion.Citation9 The S1 subunit contains an N-terminal domain (NTD) and a receptor-binding domain (RBD) at the C-terminal that contains the receptor-binding motif (RBM). However, the S2 subunit contains a fusion peptide (FP), heptad repeat 1 (HR1) and 2 (HR2) domains, a transmembrane (TM) and a cytoplasmic (CP) domain (). Upon binding the RBD on the S1 to the peptidase domain (PD) of ACE2, the FP on the S2 site is inserted into the cell membrane to promote fusion with the viral membrane.Citation5,Citation10 This process is primed by several host transmembrane protease serine proteases, including TMPRSS2 and TMPRSS4, to cleave S1 and S2 subunits.Citation11–13
Figure 1. SARS-CoV-2 genome and the encoded S protein. A. Schematic representation of the single-stranded positive-sense RNA (+ ssRNA) genome of SARS-CoV-2 (27–32kb in length). Different domains are shown by different colors. ORF, open reading frame. The spike (s) protein consists of secretion signal (SS); N-terminal domain (NTD); receptor-binding domain (RBD); subdomain 1(SD1); subdomain 2 (SD2); protease cleavage site (S1/S2); heptad repeat 1 (HR1); central helix (CH); connector domain. (CD); heptad repeat (HR); transmembrane domain. (TM); and cytoplasmic tail (CT). B. The crystallographic primary structure of the S protein.
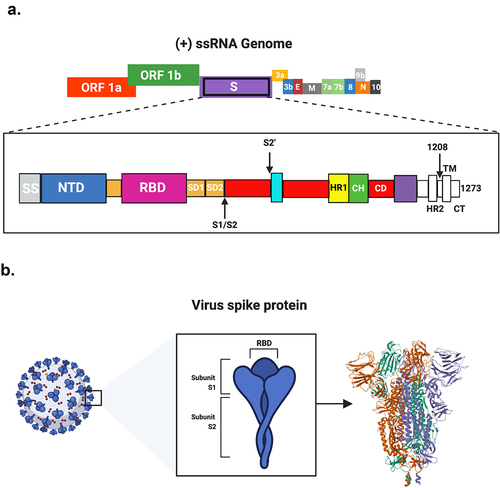
Given the important role of RBD in initiating the invasion of SARS-CoV-2 into host cells, it is reasonable to define the RBD as the most promising target for the development of virus attachment inhibitors, neutralizing antibodies and vaccines. Therefore, several studies have suggested the administration of human recombinant ACE2 (rACE2) protein to block the RBD, and thereby prevent COVID-19.
Recombinant human ACE2
Human ACE2 is a monocarboxylic peptidase that is widely expressed in several organs, including the brain, heart, lung, gut, kidneys and testis,Citation14 and circulates in the plasma as a soluble form.Citation15,Citation16 Physiologically, ACE2 converts angiotensin II into angiotensin 1–7, to counterbalance induction of the renin-angiotensin system (RAS) and thereby regulates hypertension, sodium-water retention and protects multiple organs, including the heart, kidneys and lung.Citation17 Therefore, rACE2 can be used to alleviate angiotensin II–induced diseases.
APN01 (alunacedase alfa, GSK-258688; Apeiron Biologics), a human soluble rACE2,Citation18–20 was evaluated in a Phase 2 clinical trial as a treatment for SARS-CoV-2 infection (NCT04335136).Citation21,Citation22 Due to its catalytic activity, APN01 is able to regulate the RAS to minimize organ injuries and improve the symptoms in patients with severe COVID-19.Citation22,Citation23
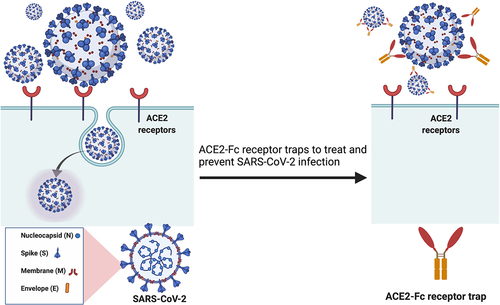
The ACE2 cell receptor is a common entry gateway of several human coronaviruses, including SARS-CoV-1, HCoV-NL63 and SARS-CoV-2, wherein the viral spike proteins are used for receptor binding.Citation9,Citation24,Citation25 Therefore, rACE2 has been evaluated as a potential antiviral therapy in which the protein acts as a decoy to facilitate immune clearance of such viruses.Citation19,Citation20,Citation26,Citation27 SARS-CoV-2 is an RNA virus that is expected to have a high mutation rate,Citation28 which might consequently enable the virus to acquire resistance against vaccines and antibodies.Citation29–34 Thus, using ACE2 as a decoy receptor for SARS-CoV-2 is particularly attractive as it aims to combat the interactions between SARS-CoV-2 S protein and cellular ACE2 receptor (). Moreover, ACE2 has the ability to neutralize emerging SARS-CoV-2 variants of concern that harbor antibody-escape mutations, as has been observed for vaccine escape mutations.Citation33,Citation35
Figure 2. Soluble recombinant human ACE2-Fc protein as a decoy receptor to SARS-CoV-2. Recombinant human ACE2-Fc protein generated by fusing the C-terminus of the human ACE2 extracellular domain to a human IgG Fc region could work as a potential SARS-CoV-2 inhibitor.
Several studies performed deep mutagenesis analysis to identify critical amino acid changes in ACE2 that could increase affinity for SARS-CoV-2 S protein. For example, the engineered decoy receptor ACE22.v2.4, showed high affinities to broadly diverse SARS-CoV-2 S proteins from humans and bats, despite the fact that the ACE2-binding surface region have high diversity. These results suggest that resistance to such engineered decoy receptors will most probably be rare and that they might be active against future emerging outbreaks of SARS-related betacoronaviruses.Citation36,Citation37
Other computational, site-directed mutagenesis and glycosidase treatment studies investigated S protein N-glycosylation involved in RBD-ACE2 interaction to engineer an ACE2 decoy receptor with enhanced S protein binding affinity and improved virus neutralization. Recent studies indicated that S N-glycans attached to N343, N165, N234, N90 and N322 positions of ACE2 play important role in determining spike binding, facilitating RBD opening and stabilizing RBD-ACE2 interaction.Citation38–41
As with any therapeutic, there are strengths and limitations associated with the rACE2-based treatment. For example, supplementation of exogenous rACE2 might be a double‐edged sword in COVID‐19 patients with underline cardiovascular disorders. It has been reported that there are a close association between increased levels of circulating soluble ACE2 (sACE2) and cardiovascular diseases, which are known to be COVID-19 risk factors.Citation42,Citation43 The elevated levels of sACE2 would initiate the formation of high numbers of circulating SARS-CoV2–sACE2 complex, which might be responsible for vascular occlusions, autoimmune inflammation, and organ ischemia. Therefore, further laboratory and clinical research are needed to assess the use of rACE2 to treat COVID‐19 patients with cardiovascular diseases.
Additionally, pharmacological studies showed that rACE2 as a decoy receptor exhibits a short half-life in both human and mice and is limited by its fast clearance.Citation18 Moreover, there have been opinions that rACE2 may unintentionally upregulate endogenous ACE2 expression, alter the balance of ACE2 hormonal substrates and worsen COVID-19 recovery.Citation44,Citation45 To overcome this, it has been suggested that fusion of enzymatically inactivated ACE2 to the Fc region of human immunoglobulin G offers superior pharmacokinetic and pharmacological benefits compared to rACE2-based therapy. Thus, this review is focused on different strategies used to engineer human rACE2 as Fc fusion proteins (i.e., immunoadhesins) to trap SARS-CoV-2.
ACE2-Fc immunoadhesin as a decoy receptor
ACE2-Fc immunoadhesins, as antibody-like molecules, offer substantial advantages over other traditional antiviral treatments. The effector functions of the Fc domain allow the recruitment of several phagocytic immune cells, including dendritic cells, macrophages and natural killer cells, and facilitate the activation of the host antiviral immune response against the virus.Citation46,Citation47 Additionally, fusing ACE2 with Fc would improve the recombinant protein half-life, binding affinity, long-acting time, serum stability, antiviral specificity, neutralization efficacy and transport into the lung.Citation48–51 For example, neonatal Fc receptors (FcRn) are widely expressed by a variety of cell types, including endothelium and pulmonary epithelial cells, and thus these cells are capable of transporting IgG and Fc fusion molecules into the respiratory tract through the mucosal barriers.Citation52–54
Immunoadhesin-based drugs have been widely used in modern biopharmaceuticals. Around 13 immunoadhesins have been approved in the United States or Europe and are currently marketed as treatments for different disease conditions, including systemic lupus erythematosus, multiple sclerosis and rheumatoid arthritis, and many more are in different phases of clinical trials.Citation55–58 However, no antiviral immunoadhesins have been approved, though at least one has entered clinical trials for human immunodeficiency virus (HIV) treatment.Citation59 While HIV is not kinetically and clinically similar to the SARS-CoV-2, previous research on HIV suggests that decoy receptor could be a potential therapeutic strategy against SARS-CoV-2.
Recent research showed that making a cocktail therapeutic by mixing neutralizing antibodies that do not bind to the RBD with RBD-targeting antibodies and engineered ACE2-Fc would generate synergistic inhibitory effect against viral infections.Citation60 Furthermore, it has been found that a ACE2-Fc fusion protein induced a broad neutralization capacity against many SARS-CoV-2 variants, including D614G, B.1.1.7 (Alpha), B.1.351 (Beta), B.1.617.1 (Kappa) and B.1.617.2 (Delta).Citation61 Thus, engineering immunoadhesins as receptor traps and stockpiling them would be critical for a fast, preemptive approach against emerging and reemerging betacoronaviruses to help control future outbreaks.
Inactivating ACE2 intrinsic catalytic activity
ACE2 at optimum concentration can lower virus infectivity in vitro, in a similar pattern as neutralizing antibodies.Citation21,Citation62–64 Moreover, it has an intrinsic catalytic activity that regulates cardiovascular functions and fluid balance.Citation65 However, the role of ACE2 receptors for viral entry is not directly linked with their enzymatic and nonenzymatic actions. Thus, it is critical to introduce individual point mutations on the catalytic site of ACE2 to inactivate the unnecessary carboxypeptidase activity, and to achieve an exclusive antiviral effect.
The ACE2 metallopeptidase activity requires a divalent cation such as Zn2+, wherein the zinc ion binding site is buried in the catalytic cleft within the proximal lobe, which is close to the viral binding site on ACE2. Three proximal lobe residues, His374, His378 and Glu402, which control the Zn+Citation2 binding with their side chains, are the apparent candidates to be mutagenized to generate enzymatically inactive ACE2 mutants. For example, Lei et al. constructed an ACE2 immunoadhesin by fusing the extracellular domain of a modified version of the human rACE2 (1–740) to the Fc region of the human immunoglobulin IgG1.Citation63 The modified ACE2-Fc variant includes two specific mutations for histidine residues 374 and 378 to asparagine (H374N and H378N) to reduce the intrinsic ACE2 catalytic activity. The fusion protein has shown high binding affinity toward the RBDs of SARS-CoV-1 (170 nM) and SARS-CoV-2 (11 nM), which was determined with BIAcore-binding assay. The ACE2-Fc receptor trap was also able to neutralize and inhibit S protein-mediated fusion of both SARS-CoV-1 and SARS-CoV-2, despite the H374N and H378N mutation, suggesting that fusing recombinant sACE2 to the Fc region of the human IgG1 can remarkably improve the neutralization potency and pharmacokinetics of the receptor traps.Citation63,Citation66
Although mutations at the Zn+2 ion binding sites H374 and H378 can produce enzymatically inactive mutants, mutations within zinc-binding pocket may cause protein instability because the divalent cation is essential for metallopeptidase protein structure and stability.Citation67–69 Therefore, mutations within the ACE2 substrate-binding pocket are less likely to cause construct instability or affect the binding affinity. A study performed by Liu et al. investigated individual point mutations using alanine within the catalytic site of ACE2 to inactivate the unnecessary carboxypeptidase activity and to achieve stable and effective version of the receptor trap.Citation62 They were able to engineer enzymatically inactive ACE2-Fc immunoadhesin variants that can block and neutralize the virus without the potential adverse side effects from its catalytic activities. Six other residues located in the proximal and distal lobe (Glu145, Arg273, His345, Pro346, Asp368 and His505) were also mutagenized to produce enzymatically inactive ACE2 mutants because their side chains formed direct interactions with hormonal substrates.Citation62 Among all enzymatically inactive ACE2-Fc variants, the Arg273Ala mutant, which is a substrate-binding residue, showed complete inhibition of the peptidase activity and is expected to maintain protein stability and binding affinity to the viral RBD. There are, however, compelling arguments that ACE2 catalytic activity can help alleviate COVID-19 symptoms through blocking receptor-binding sites on the S protein to neutralize SARS-CoV-2 infection or promoting angiotensin II breakdown and angiotensin-1-7 production.Citation22
Engineering ACE2-Fc immunoadhesin through guided evolution platforms
The main focus of engineering ACE2 as receptor traps is to enhance their binding affinity, avidity, and specificity, as well as pharmacokinetic and pharmacological efficacy.Citation36,Citation47,Citation63,Citation70 Wild-type ACE2 is not an optimal anti-SARS-CoV-2 biotherapeutic due to its modest affinity toward the virus RBD.Citation71–73 Therefore, it is essential to engineer ACE2 to have a binding affinity (KD) in the low- to sub-nanomolar range, which is comparable to SARS-CoV-2 S protein-specific monoclonal antibodies.Citation74–78
The crystal structure of the SARS-CoV-2/human ACE2 complex revealed that the interacting segments of ACE2 include the residues S19 to A614,Citation79 which indicates the amino acid side mutations that are crucial for enhancing the ACE2–RBD interaction. As a result, different directed evolution approaches were performed to enhance ACE2 properties as receptor traps such as using artificial intelligent (AI), mutagenesis and display technologies. For instance, Glasgow et al. described a novel approach to generate numerous affinity matured and enzymatically inactive recombinant ACE2 immunoadhesins that act as receptor traps to block and prevent SARS-CoV-2 entry into host cells.Citation66 In their two-phase stepwise approach, AI was first used to engineer ACE2 variants by introducing specific amino acid changes that have resulted in several-folds tighter binding to the RBD compared to the wild-type ACE2. Secondly, they used the re-engineered receptor traps as templates for affinity maturation using yeast surface display, which enabled the isolation of variants with binding affinities that have several folds higher to the RBD compared to the original templates. A human ACE2 that comprises residues 18 to 614 extracellular domain [ACE2(614)] was used as a template for the computational affinity optimization strategy.Citation66 A computationally guided selection followed by affinity maturation using yeast surface display were used to generate variant 313 [K31F, N33D, H34S, E35Q ACE2(614)], which was one of the best clones in terms of binding affinity and neutralization potency. The computational design methods based on AI bypass the bias that can arise from using experimental affinity maturation platforms alone, and remarkably accelerate the overall process. Yeast was used as a eukaryotic display system due to its effective surface display and its ability to produce glycosylated proteins.Citation80
The ACE2 ectodomain (18 to 740) contains an enzymatic domain (18–615) and a collectrin-like domain (CLD). Interestingly, Glasgow et al. also observed a significant enhancement for the ACE2-Fc decoy receptors affinity, avidity, and stability by including the natural ACE2 CLD [ACE2(740)-Fc]. This is consistent with another study that showed that ACE2(740)-Fc is more effective in blocking viral infection compared to ACE2(614)-Fc.Citation81 Also, to avoid the off-target effects that accompanied ACE2 catalytic enzyme activity without affecting the binding affinity toward RBD and ACE2(740)-Fc scaffold, they inactivated the peptidase activity by including an H345L mutation, which is important for substrate binding.Citation82 Noteworthy, replacing the leucine with alanine for His345 residue did not inhibit the ACE2-Fc catalytic activity toward angiotensin II, but, on the contrary, it enhanced the construct activity toward the substrate compared to the wild-type.Citation83,Citation84 Further investigations are required for better understanding of ACE2 mechanism and substrate specificity.
The computationally designed, affinity maturation, and enzymatic inactive variant 313 [K31F, N33D, H34S, E35Q, H345L ACE2(740)-Fc] was able to bind the RBD of SARS-CoV-2 a hundred times higher than wild-type ACE2-Fc and neutralize pseudoviruses and authentic SARS-CoV-2 virus with half-maximal inhibitory concentrations (IC50) of less than 100 ng/mL.
Higuchi et al. demonstrated another strategy to enhance the binding affinity of ACE2 to the SARS-CoV-2 S RBD. They performed a mammalian cell (HEK-293 T)-based guided evolution using surface display of mutagenized library in association with fluorescence-activated cell sorting (FACS).Citation85 Screening system based on mammalian cells were used instead of yeast display to isolate ACE2 variants with proper posttranslational modifications patterns and favorable biophysical and biological attributes.Citation86,Citation87
The viral S protein interface is found in the protease domain, which is located in the top-middle segment of the ACE2 ectodomain. Thus, Higuchi et al. mutagenized the protease domain using error-prone PCR. They generated small plasmid library (~105 mutants), which were transformed into competent cells and packaged into lentivirus before being expressed in human HEK-293 T cells. The library was then screened for 3 cycles and only the highest binding cells to SARS-CoV-2 RBD-GFP were collected by FACS. Mutant 3N39v2 (A25V, K31N, E35K, L79F) showed binding affinity at sub-nanomolar levels to SARS-CoV-2 RBD (KD ∼0.64 nM) due to its slow dissociation rate. This is because 3N39v2 saturate all three RBDs on the S protein trimer complex, while wild-type ACE2 binds to S protein mainly as 1:1.Citation85,Citation88 3N39v2 has also demonstrated potent neutralization of SARS-CoV-1 and SARS-CoV-2 pseudoviruses and authentic SARS-CoV-2 virus. In a COVID-19 hamster model, the 3N39v2 fusion protein has showed efficacy in mitigating lung abnormalities, viral RNA copies and cytokine expression, which are associated with COVID-19 severity.
Engineering ACE2-Fc immunoadhesin using mutations based on host-ortholog receptors
The binding domain of the viral cellular receptor may be used to render immunoadhesins. However, zoonotic viruses can bind to their animal-derived ortholog cellular receptors with higher affinities than human cell-surface receptors resulting from natural evolution.Citation89 Therefore, it was proposed that immunoadhesins built with mutations based on host-ortholog receptors can provide stronger antiviral therapeutics.
SARS-CoV-2 has a genome that is close to bat-derived SARS-like coronaviruses, which make them a probable origin of SARS-CoV-2.Citation90 Since ACE2 orthologs from different species may serve as SARS-CoV-2 entry receptors, these species may have served as intermediate reservoir hosts before virus spreading to humans.Citation90 For SARS-CoV-2, the human-ACE2 receptor has been shown to be a suboptimal receptor.Citation36 As a result, it was proposed that human-ACE2 can be re-designed to have a higher binding affinity for SARS-CoV-2 in order to establish an efficient immunotherapy that can effectively block and prevent virus infection.
According to Cohen-Dvashi et al., sequence alignment of many ACE2 orthologues derived from mammals revealed many non-conserved residues of the RBD-recognition site on ACE2 receptor,Citation91 suggesting that there are several potential sequence mutations in ACE2 that can be implemented to enhance the binding affinity to SARS-CoV-2. Cohen-Dvashi et al., therefore, subjected tens of orthologous ACE2 genes with at least 80% similarity to human-ACE2 to Rosetta atomistic modeling calculations to find beneficial ACE2 mutations that could maximize construct stability and binding affinity to the virus RBD. They generated potent, enzymatic inactivated, affinity-matured human ACE2-Fc immunoadhesin variant with eight incorporated mutations (T27L, D30E, Q42R, E75R, L79Y, N330F, T92R and E375L). For example, T92R mutation enhanced the affinity to SARS-CoV-2 RBD because of the elimination of N-linked glycan site, which imposes steric constraints for the binding to the RBD, while the arginine can form polar interactions with proximate glutamine. The modified ACE2-Fc variant demonstrated superior IC50 and KD values compared to the unmodified ACE2-Fc.
In another report, Mou et al. tested HEK-293 T cells expressing several ACE2 orthologs for their ability to bind to the recombinant SARS-CoV-2 RBD.Citation92 The study showed that human, pangolin, and horseshoe-bat ACE2 orthologs bind to SARS2-CoV-2 RBD more efficiently compared to other organisms. Bats have been suggested to be the species of origin of SARS-CoV-2 since the virus genome has more than 95% similarity with other bat coronaviruses,Citation25 while pangolin have been suggested to be the intermediate host of SARS-CoV-2 rather than a long-term reservoir.Citation93 Mou et al. also suggested that SARS-CoV-2 S protein is not completely adapted to human ACE2,Citation92 and, therefore, mutations derived from pangolin and horseshoe-bat ACE2 orthologs were implemented to augment binding affinity and neutralization potency of human ACE2-Fc to SARS-CoV-2. Their report showed that the five bat-derived mutations (Q24E, T27K, H34S, N49E and N90D) enhanced ACE2-Fc binding affinity toward SARS-CoV-2 RBD (5.454 nM) compared to the wild-type ACE2-Fc (11.64 nM). Furthermore, the capacity of the modified ACE2-Fc variant to neutralize SARS-CoV-2 pseudo- and live viruses was significantly more potent than wild-type ACE2-Fc.
It is well documented that SARS-CoV-2 uses the ACE2 receptor as a gateway to infect host cells.Citation94 However, it is not confirmed until now if domestic animals have a role in SARS-CoV-2 transmission.Citation95–97 Nonetheless, since wild animals such as bats have been shown to play an essential part in the transmission of notorious coronaviruses,Citation81 it is vital to study the virus’s distributions among different hosts. Thus, the capacity of SARS-CoV-2 to bind to ACE2 animal orthologs was performed to assist identifying potential animal hosts. Accordingly, Le et al. evaluated 16 ACE2 orthologs to determine if they support SARS-CoV-2 entry by using recombinant RBD-IgG, pseudovirures and live virus.Citation81 The purified SARS-CoV2 RBD-IgG was able to bind to human ACE2 and ACE2 orthologs from a variety of domestic mammals, including camels, horses, cats, and rabbits, suggesting that these animals can be infected and might act as intermediate hosts for SARS-CoV2 viruses. Moreover, they tested the neutralization efficiency of different forms of ACE2-IgG against SARS-CoV-2 pseudovirures and live virus.Citation81 ACE2(615)-IgG and ACE2(740)-IgG variants were designed to inactivate ACE2s’ intrinsic enzymatic activity and enhance the immunoadhesins affinity toward the SARS-CoV-2 RBD. These mutations were based on adding hydrophobic residues (Y83W, H34Y and M82K) or enhancing the salt-bridge interactions (D30E). The ACE2(740)-IgG variants were substantially more potent than ACE2(615)-IgG variants. Additionally, compared to all other ACE2-IgG variants, ACE2(740)-D30E-IgG mutant had the best neutralizing activity against SARS-CoV-2. Furthermore, they engineered a more potent ACE2(740)-DE30-IgG version that has four sACE2 domains in a single molecule instead of two. This quadruple ACE2(740)-DE30-IgG showed more than several-fold improvement in IC50 compared to the original dimer version in pseudovirus neutralization assay. Additionally, ACE2(740)-DE30-IgG with antibody-like configuration showed more potent neutralization of live virus infection at 0.16 μg/ml compared to bivalent ACE2(740)-DE30-IgG, which makes it a powerful entry inhibitor against SARS-CoV-2 virus.
Limitation of the ACE2-Fc immunoadhesin
One potential limitation of the ACE2-Fc immunoadhesin strategy is that it could inadvertently activate the immune system. ACE2 is secreted in all human tissues, including the heart, kidney, and gastrointestinal tissues. Therefore, the elevation of circulating extracellular ACE2 domain for a prolonged time via Fc domain extended half-life could have unknown long-term clinical outcomes.
Another potential concern is that the presence of Fc might increase complement and cytokine responses, which might exacerbate the inflammation, worsen the patient’s condition, leading to infection advancement.Citation98–100 Although the constant heavy-chain regions of the IgG isoforms have similar amino acid sequences, they exert different levels of effector functions. IgG1- and IgG3-Fc are known for strong complement-dependent cytotoxicity (CDC), antibody-dependent cellular phagocytosis (ADCP) and antibody-dependent cell-mediated cytotoxicity (ADCC), whereas, IgG4-Fc has lower Fc-mediated antibody effector functions.Citation101,Citation102 Therefore, Svilenov et al. engineered IgG4-Fc-based ACE2(732)-Fc fusion constructs to avoid Fc-mediated cytotoxic side effects.Citation103 Nonetheless, the importance and the potential roles of Fc-associated effector functions in virus elimination should be further studied, as they could contribute to ACE2-Fc inhibitory effects against SARS-CoV-2.
Moreover, naturally occurring IgG4 antibodies are less stable than IgG1 versions, which makes them less valuable as biopharmaceutical products.Citation104–107 Therefore, Svilenov et al. incorporated the immunoglobulin Fc region of an IgG4 isotype with an S228P sequence mutation in the hinge region to minimize Fab arm exchange and increase the stability of the ACE2-IgG4-Fc fusion protein.Citation108 The ACE2(732)-IgG4-FcS228P construct was successfully expressed and purified with high binding affinity (KD ~4 nM) to immobilized SARS-CoV-2 RBD and potent antiviral activity at nano- to picomolar levels.
Ferrari et al. demonstrated a catalytically inactive ACE2(740)-H374N:H378N-Fc decoy receptor with abrogated FcγR interaction to reduce the risk of antibody-dependent enhancement.Citation109 They engineered an Fc L234A/L235A/P329G mutant in the CH2 domain (ACE2(740)-H374N:H378N-FcLALA-PG), which showed complete abrogation of human FcγR engagement while preserving the FcRn interaction to provide extended half-life. The generated construct displayed a potent neutralization activity in vitro against four SARS-CoV-2 variants, including D614G, B.1.1.7, B.1.351 and P.1. Additionally, administration of ACE2(740)-H374N:H378N-FcLALA-PG in a SARS-CoV-2 hamster model showed significant reduction in viral RNA copy as well as lung damage.
Recombinant ACE2-Fc is expected to have an acceptable safety profile based on APN01 clinical trials, wherein no serious undesirable reactions were reported.Citation22,Citation110 However, similar to monoclonal antibodies, patients receiving ACE2-Fc immunoadhesins might produce anti-drug antibodies, which would negatively affect the pharmacokinetic, safety and efficacy of the therapeutic molecule.Citation111,Citation112 While acquiring immunological cross-reactivity against endogenous ACE2 after receiving recombinant ACE2-Fc would have a long-term devastating effect on patients, clinicians can use well-evaluated methods to delineate and reduce any undesirable immunological reactions of therapeutic biologics during the course of ACE2-Fc treatment.Citation113
Conclusion
ACE2-Fc immunoadhesins offer considerable advantages over other therapeutics that aim to neutralize SARS-related betacoronaviruses. Here, we discussed different optimization strategies that helped generate several modified ACE2-Fc versions as promising anti-SARS-CoV-2 candidates (summarized in ). Beyond coronaviruses, ACE2-Fc-based therapeutics could be used as a quick therapeutic option for future viral outbreaks that use the ACE2 receptor for entry without risk of virus mutational escape. Moreover, engineered ACE2-Fc as an antibody-like biomolecule would have the potential to be used as a prophylactic agent for those who are at high risk of COVID-19, such as healthcare workforces especially in the absence of an effective vaccine against new variants.
Table 1. Examples of modified ACE2-Fc as possible anti-COVID-19 agents
Acknowledgments
Figures were created by biorender.com
Author contributions
Conceptualization – MA; Original draft Preparation – MA, AZ, SA; Review and editing – MA, AZ, SA, AH; Figures and visualizations – MA. All authors have read and agreed to the published version of the manuscript.
Abbreviations
ACE2, Angiotensin-converting enzyme 2; KD, Binding affinity; CLD, Collectrin-like domain; IC50, Half-maximal inhibitory concentrations, RBD, Receptor-binding domain; rACE2, Recombinant ACE2, SARS-CoV-2, Severe acute respiratory syndrome coronavirus 2; S, Spike protein.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Additional information
Funding
References
- Zhu N, Zhang D, Wang W, Li X, Yang B, Song J, Zhao X, Huang B, Shi W, Lu R, et al. A novel coronavirus from patients with pneumonia in China, 2019. N Engl J Med. 2020;382(8):727–11. doi:10.1056/NEJMoa2001017.
- Wang C, Horby PW, Hayden FG, Gao GF. A novel coronavirus outbreak of global health concern. Lancet. 2020;395:470–73. doi:10.1016/S0140-6736(20)30185-9.
- Luan J, Lu Y, Jin X, Zhang L. Spike protein recognition of mammalian ACE2 predicts the host range and an optimized ACE2 for SARS-CoV-2 infection. Biochem Biophys Res Commun. 2020;526:165–69. doi:10.1016/j.bbrc.2020.03.047.
- Tai W, He L, Zhang X, Pu J, Voronin D, Jiang S, Zhou Y, Du L. Characterization of the receptor-binding domain (RBD) of 2019 novel coronavirus: implication for development of RBD protein as a viral attachment inhibitor and vaccine. Cell Mol Immunol. 2020;17:613–20. doi:10.1038/s41423-020-0400-4.
- Hoffmann M, Kleine-Weber H, Schroeder S, Kruger N, Herrler T, Erichsen S, Schiergens TS, Herrler G, Wu NH, Nitsche A, et al. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020;181:271–80 e8. doi:10.1016/j.cell.2020.02.052.
- Donoghue M, Hsieh F, Baronas E, Godbout K, Gosselin M, Stagliano N, Donovan M, Woolf B, Robison K, Jeyaseelan R, et al. A novel angiotensin-converting enzyme-related carboxypeptidase (ACE2) converts angiotensin I to angiotensin 1-9. Circ Res. 2000;87:E1–9. doi:10.1161/01.res.87.5.e1.
- Turner AJ, Tipnis SR, Guy JL, Rice G, Hooper NM. ACEH/ACE2 is a novel mammalian metallocarboxypeptidase and a homologue of angiotensin-converting enzyme insensitive to ACE inhibitors. Can J Physiol Pharmacol. 2002;80:346–53. doi:10.1139/y02-021.
- Li W, Moore MJ, Vasilieva N, Sui J, Wong SK, Berne MA, Somasundaran M, Sullivan JL, Luzuriaga K, Greenough TC, et al. Angiotensin-converting enzyme 2 is a functional receptor for the SARS coronavirus. Nature. 2003;426:450–54. doi:10.1038/nature02145.
- Wrapp D, Wang N, Corbett KS, Goldsmith JA, Hsieh CL, Abiona O, Graham BS, McLellan JS. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science. 2020;367:1260–63. doi:10.1126/science.abb2507.
- Hoffmann M, Hofmann-Winkler H, Pöhlmann S. Priming time: how cellular proteases arm coronavirus spike proteins. Activation of Viruses by Host Proteases. 2018:71–98. doi:10.1007/978-3-319-75474-1_4.
- Glowacka I, Bertram S, Muller MA, Allen P, Soilleux E, Pfefferle S, Steffen I, Tsegaye TS, He Y, Gnirss K, et al. Evidence that TMPRSS2 activates the severe acute respiratory syndrome coronavirus spike protein for membrane fusion and reduces viral control by the humoral immune response. J Virol. 2011;85:4122–34. doi:10.1128/JVI.02232-10.
- Matsuyama S, Nagata N, Shirato K, Kawase M, Takeda M, Taguchi F. Efficient activation of the severe acute respiratory syndrome coronavirus spike protein by the transmembrane protease TMPRSS2. J Virol. 2010;84:12658–64. doi:10.1128/JVI.01542-10.
- Shulla A, Heald-Sargent T, Subramanya G, Zhao J, Perlman S, Gallagher T. A transmembrane serine protease is linked to the severe acute respiratory syndrome coronavirus receptor and activates virus entry. J Virol. 2011;85:873–82. doi:10.1128/JVI.02062-10.
- Luft FC. ACE in the hole. J Mol Med (Berl). 2014;92:793–95. doi:10.1007/s00109-014-1172-z.
- Gheblawi M, Wang K, Viveiros A, Nguyen Q, Zhong JC, Turner AJ, Raizada MK, Grant MB, Oudit GY. Angiotensin-Converting enzyme 2: SARS-CoV-2 receptor and regulator of the renin-angiotensin system: celebrating the 20th anniversary of the discovery of ACE2. Circ Res. 2020;126:1456–74. doi:10.1161/CIRCRESAHA.120.317015.
- Sama IE, Ravera A, Santema BT, van Goor H, Ter Maaten JM, Cleland JGF, Rienstra M, Friedrich AW, Samani NJ, Ng LL, et al. Circulating plasma concentrations of angiotensin-converting enzyme 2 in men and women with heart failure and effects of renin-angiotensin-aldosterone inhibitors. Eur Heart J. 2020;41:1810–17. doi:10.1093/eurheartj/ehaa373.
- Santos RA, Simoes E, Silva AC, Maric C, Silva DM, Machado RP, de Buhr I, Heringer-Walther S, Pinheiro SV, Lopes MT, et al. Angiotensin-(1-7) is an endogenous ligand for the G protein-coupled receptor Mas. Proc Natl Acad Sci U S A. 2003;100:8258–63. doi:10.1073/pnas.1432869100.
- Haschke M, Schuster M, Poglitsch M, Loibner H, Salzberg M, Bruggisser M, Penninger J, Krahenbuhl S. Pharmacokinetics and pharmacodynamics of recombinant human angiotensin-converting enzyme 2 in healthy human subjects. Clin Pharmacokinet. 2013;52:783–92. doi:10.1007/s40262-013-0072-7.
- Kuba K, Imai Y, Rao S, Gao H, Guo F, Guan B, Huan Y, Yang P, Zhang Y, Deng W, et al. A crucial role of angiotensin converting enzyme 2 (ACE2) in SARS coronavirus-induced lung injury. Nat Med. 2005;11:875–79. doi:10.1038/nm1267.
- Imai Y, Kuba K, Rao S, Huan Y, Guo F, Guan B, Yang P, Sarao R, Wada T, Leong-Poi H, et al. Angiotensin-converting enzyme 2 protects from severe acute lung failure. Nature. 2005;436:112–16. doi:10.1038/nature03712.
- Monteil V, Kwon H, Prado P, Hagelkruys A, Wimmer RA, Stahl M, Leopoldi A, Garreta E, Hurtado Del Pozo C, Prosper F, et al. Inhibition of SARS-CoV-2 infections in engineered human tissues using clinical-grade soluble human ACE2. Cell. 2020;181:905–13 e7. doi:10.1016/j.cell.2020.04.004.
- Zoufaly A, Poglitsch M, Aberle JH, Hoepler W, Seitz T, Traugott M, Grieb A, Pawelka E, Laferl H, Wenisch C, et al. Human recombinant soluble ACE2 in severe COVID-19. Lancet Respir Med. 2020;8:1154–58. doi:10.1016/S2213-2600(20)30418-5.
- Khan A, Benthin C, Zeno B, Albertson TE, Boyd J, Christie JD, Hall R, Poirier G, Ronco JJ, Tidswell M, et al. A pilot clinical trial of recombinant human angiotensin-converting enzyme 2 in acute respiratory distress syndrome. Crit Care. 2017;21:234. doi:10.1186/s13054-017-1823-x.
- Yan R, Zhang Y, Li Y, Xia L, Guo Y, Zhou Q. Structural basis for the recognition of SARS-CoV-2 by full-length human ACE2. Science. 2020;367:1444–48. doi:10.1126/science.abb2762.
- Zhou P, Yang XL, Wang XG, Hu B, Zhang L, Zhang W, Si HR, Zhu Y, Li B, Huang CL, et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579:270–73. doi:10.1038/s41586-020-2012-7.
- Gu H, Xie Z, Li T, Zhang S, Lai C, Zhu P, Wang K, Han L, Duan Y, Zhao Z, et al. Angiotensin-converting enzyme 2 inhibits lung injury induced by respiratory syncytial virus. Sci Rep. 2016;6:19840. doi:10.1038/srep19840.
- Zou Z, Yan Y, Shu Y, Gao R, Sun Y, Li X, Ju X, Liang Z, Liu Q, Zhao Y, et al. Angiotensin-converting enzyme 2 protects from lethal avian influenza A H5N1 infections. Nat Commun. 2014;5:3594. doi:10.1038/ncomms4594.
- van Dorp L, Richard D, Tan CCS, Shaw LP, Acman M, Balloux F. No evidence for increased transmissibility from recurrent mutations in SARS-CoV-2. Nat Commun. 2020;11:5986. doi:10.1038/s41467-020-19818-2.
- Williams TC, Burgers WA. SARS-CoV-2 evolution and vaccines: cause for concern? Lancet Respir Med. 2021;9:333–35. doi:10.1016/S2213-2600(21)00075-8.
- Xie X, Liu Y, Liu J, Zhang X, Zou J, Fontes-Garfias CR, Xia H, Swanson KA, Cutler M, Cooper D, et al. Neutralization of SARS-CoV-2 spike 69/70 deletion, E484K and N501Y variants by BNT162b2 vaccine-elicited sera. Nat Med. 2021;27:620–21. doi:10.1038/s41591-021-01270-4.
- Chen RE, Zhang X, Case JB, Winkler ES, Liu Y, VanBlargan LA, Liu J, Errico JM, Xie X, Suryadevara N, et al. Resistance of SARS-CoV-2 variants to neutralization by monoclonal and serum-derived polyclonal antibodies. Nat Med. 2021;27:717–26. doi:10.1038/s41591-021-01294-w.
- Starr TN, Greaney AJ, Addetia A, Hannon WW, Choudhary MC, Dingens AS, Li JZ, Bloom JD. Prospective mapping of viral mutations that escape antibodies used to treat COVID-19. Science. 2021;371:850–54. doi:10.1126/science.abf9302.
- Weisblum Y, Schmidt F, Zhang F, DaSilva J, Poston D, Lorenzi JC, Muecksch F, Rutkowska M, Hoffmann HH, Michailidis E, et al. Escape from neutralizing antibodies by SARS-CoV-2 spike protein variants. Elife. 2020;9. doi:10.7554/eLife.61312.
- Kemp SA, Collier DA, Datir RP, Ferreira I, Gayed S, Jahun A, Hosmillo M, Rees-Spear C, Mlcochova P, Lumb IU, et al. SARS-CoV-2 evolution during treatment of chronic infection. Nature. 2021;592:277–82. doi:10.1038/s41586-021-03291-y.
- Yuan M, Huang D, Lee CD, Wu NC, Jackson AM, Zhu X, Liu H, Peng L, van Gils MJ, Sanders RW, et al. Structural and functional ramifications of antigenic drift in recent SARS-CoV-2 variants. Science. 2021;373:818–23. doi:10.1126/science.abh1139.
- Chan KK, Dorosky D, Sharma P, Abbasi SA, Dye JM, Kranz DM, Herbert AS, Procko E. Engineering human ACE2 to optimize binding to the spike protein of SARS coronavirus 2. Science. 2020;369:1261–65. doi:10.1126/science.abc0870.
- Chan KK, Tan TJC, Narayanan KK, Procko E. An engineered decoy receptor for SARS-CoV-2 broadly binds protein S sequence variants. bioRxiv. 2020. doi:10.1101/2020.10.18.344622.
- Mehdipour AR, Hummer G. Dual nature of human ACE2 glycosylation in binding to SARS-CoV-2 spike. Proc Natl Acad Sci U S A. 2021:118. doi:10.1073/pnas.2100425118.
- Casalino L, Gaieb Z, Goldsmith JA, Hjorth CK, Dommer AC, Harbison AM, Fogarty CA, Barros EP, Taylor BC, McLellan JS, et al. Beyond shielding: the roles of glycans in the SARS-CoV-2 spike protein. ACS Cent Sci. 2020;6:1722–34. doi:10.1021/acscentsci.0c01056.
- Sztain T, Ahn SH, Bogetti AT, Casalino L, Goldsmith JA, Seitz E, McCool RS, Kearns FL, Acosta-Reyes F, Maji S, et al. A glycan gate controls opening of the SARS-CoV-2 spike protein. bioRxiv. 2021. doi:10.1101/2021.02.15.431212.
- Capraz T, Kienzl NF, Laurent E, Perthold JW, Foderl-Hobenreich E, Grunwald-Gruber C, Maresch D, Monteil V, Niederhofer J, Wirnsberger G, et al. Structure-guided glyco-engineering of ACE2 for improved potency as soluble SARS-CoV-2 decoy receptor. Elife. 2021;10. doi:10.7554/eLife.73641.
- Wallentin L, Lindback J, Eriksson N, Hijazi Z, Eikelboom JW, Ezekowitz MD, Granger CB, Lopes RD, Yusuf S, Oldgren J, et al. Angiotensin-converting enzyme 2 (ACE2) levels in relation to risk factors for COVID-19 in two large cohorts of patients with atrial fibrillation. Eur Heart J. 2020;41:4037–46. doi:10.1093/eurheartj/ehaa697.
- Sama IE, Voors AA, van Veldhuisen DJ. New data on soluble ACE2 in patients with atrial fibrillation reveal potential value for treatment of patients with COVID-19 and cardiovascular disease. Eur Heart J. 2020;41:4047–49. doi:10.1093/eurheartj/ehaa761.
- Mehra MR, Desai SS, Kuy S, Henry TD, Patel AN. Cardiovascular disease, drug therapy, and Mortality in Covid-19. N Engl J Med. 2020;382:e102. doi:10.1056/NEJMoa2007621.
- Guo J, Huang Z, Lin L, Lv J. Coronavirus Disease 2019 (COVID-19) and cardiovascular disease: a viewpoint on the potential influence of angiotensin-converting enzyme inhibitors/angiotensin receptor blockers on onset and severity of severe acute respiratory syndrome coronavirus 2 Infection. J Am Heart Assoc. 2020;9:e016219. doi:10.1161/JAHA.120.016219.
- Yasui F, Kohara M, Kitabatake M, Nishiwaki T, Fujii H, Tateno C, Yoneda M, Morita K, Matsushima K, Koyasu S, et al. Phagocytic cells contribute to the antibody-mediated elimination of pulmonary-infected SARS coronavirus. Virology. 2014;454-455:157–68. doi:10.1016/j.virol.2014.02.005.
- Kruse RL. Therapeutic strategies in an outbreak scenario to treat the novel coronavirus originating in Wuhan, China. F1000Res. 2020;9:72. doi:10.12688/f1000research.22211.2.
- Bitonti AJ, Dumont JA, Low SC, Peters RT, Kropp KE, Palombella VJ, Stattel JM, Lu Y, Tan CA, Song JJ, et al. Pulmonary delivery of an erythropoietin Fc fusion protein in non-human primates through an immunoglobulin transport pathway. Proc Natl Acad Sci U S A. 2004;101:9763–68. doi:10.1073/pnas.0403235101.
- Strohl WR. Fusion proteins for half-life extension of biologics as a strategy to make biobetters. BioDrugs. 2015;29:215–39. doi:10.1007/s40259-015-0133-6.
- Unverdorben F, Richter F, Hutt M, Seifert O, Malinge P, Fischer N, Kontermann RE. Pharmacokinetic properties of IgG and various Fc fusion proteins in mice. MAbs. 2016;8:120–28. doi:10.1080/19420862.2015.1113360.
- Czajkowsky DM, Hu J, Shao Z, Pleass RJ. Fc-fusion proteins: new developments and future perspectives. EMBO Mol Med. 2012;4:1015–28. doi:10.1002/emmm.201201379.
- Sockolosky JT, Szoka FC. The neonatal Fc receptor, FcRn, as a target for drug delivery and therapy. Adv Drug Deliv Rev. 2015;91:109–24. doi:10.1016/j.addr.2015.02.005.
- Latvala S, Jacobsen B, Otteneder MB, Herrmann A, Kronenberg S. Distribution of FcRn Across Species and Tissues. J Histochem Cytochem. 2017;65:321–33. doi:10.1369/0022155417705095.
- Tzaban S, Massol RH, Yen E, Hamman W, Frank SR, Lapierre LA, Hansen SH, Goldenring JR, Blumberg RS, Lencer WI. The recycling and transcytotic pathways for IgG transport by FcRn are distinct and display an inherent polarity. J Cell Biol. 2009;185:673–84. doi:10.1083/jcb.200809122.
- Kariolis MS, Miao YR, Jones DS 2nd, Kapur S, Mathews II, Giaccia AJ, Cochran JR. An engineered Axl ‘decoy receptor’ effectively silences the Gas6-Axl signaling axis. Nat Chem Biol. 2014;10:977–83. doi:10.1038/nchembio.1636.
- Huang C. Receptor-Fc fusion therapeutics, traps, and MIMETIBODY technology. Curr Opin Biotechnol. 2009;20:692–99. doi:10.1016/j.copbio.2009.10.010.
- Wycoff K, Maclean J, Belle A, Yu L, Tran Y, Roy C, Hayden F. Anti-infective immunoadhesins from plants. Plant Biotechnol J. 2015;13:1078–93. doi:10.1111/pbi.12441.
- Duivelshof BL, Murisier A, Camperi J, Fekete S, Beck A, Guillarme D, D’Atri V. Therapeutic Fc-fusion proteins: current analytical strategies. J Sep Sci. 2021;44:35–62. doi:10.1002/jssc.202000765.
- Shearer WT, Israel RJ, Starr S, Fletcher CV, Wara D, Rathore M, Church J, DeVille J, Fenton T, Graham B, et al. Recombinant CD4-IgG2 in human immunodeficiency virus type 1-infected children: phase 1/2 study. The Pediatric AIDS Clinical Trials Group Protocol 351 Study Team. J Infect Dis. 2000;182:1774–79. doi:10.1086/317622.
- Chi X, Yan R, Zhang J, Zhang G, Zhang Y, Hao M, Zhang Z, Fan P, Dong Y, Yang Y, et al. A neutralizing human antibody binds to the N-terminal domain of the spike protein of SARS-CoV-2. Science. 2020;369:650–55. doi:10.1126/science.abc6952.
- Zhang Z, Zeng E, Zhang L, Wang W, Jin Y, Sun J, Huang S, Yin W, Dai J, Zhuang Z, et al. Potent prophylactic and therapeutic efficacy of recombinant human ACE2-Fc against SARS-CoV-2 infection in vivo. Cell Discov. 2021;7:65. doi:10.1038/s41421-021-00302-0.
- Liu P, Xie X, Gao L, Jin J. Designed variants of ACE2-Fc that decouple anti-SARS-CoV-2 activities from unwanted cardiovascular effects. Int J Biol Macromol. 2020;165:1626–33. doi:10.1016/j.ijbiomac.2020.10.120.
- Lei C, Qian K, Li T, Zhang S, Fu W, Ding M, Hu S. Neutralization of SARS-CoV-2 spike pseudotyped virus by recombinant ACE2-Ig. Nat Commun. 2020;11:2070. doi:10.1038/s41467-020-16048-4.
- Iwanaga N, Cooper L, Rong L, Maness NJ, Beddingfield B, Qin Z, Crabtree J, Tripp RA, Yang H, Blair R, et al. ACE2-IgG1 fusions with improved in vitro and in vivo activity against SARS-CoV-2. iScience. 2022;25:103670. doi:10.1016/j.isci.2021.103670.
- Vickers C, Hales P, Kaushik V, Dick L, Gavin J, Tang J, Godbout K, Parsons T, Baronas E, Hsieh F, et al. Hydrolysis of biological peptides by human angiotensin-converting enzyme-related carboxypeptidase. J Biol Chem. 2002;277:14838–43. doi:10.1074/jbc.M200581200.
- Glasgow A, Glasgow J, Limonta D, Solomon P, Lui I, Zhang Y, Nix MA, Rettko NJ, Zha S, Yamin R, et al. Engineered ACE2 receptor traps potently neutralize SARS-CoV-2. Proc Natl Acad Sci U S A. 2020;117:28046–55. doi:10.1073/pnas.2016093117.
- Fridrich S, Karmilin K, Stocker W. Handling metalloproteinases. Curr Protoc Protein Sci. 2016;83(21). doi:10.1002/0471140864.ps2116s83.
- Namuswe F, Berg JM. Secondary interactions involving zinc-bound ligands: roles in structural stabilization and macromolecular interactions. J Inorg Biochem. 2012;111:146–49. doi:10.1016/j.jinorgbio.2011.10.018.
- McCall KA, Huang C, Fierke CA. Function and mechanism of zinc metalloenzymes. J Nutr. 2000;130:1437S–46S. doi:10.1093/jn/130.5.1437S.
- Miao X, Luo Y, Huang X, Lee SMY, Yuan Z, Tang Y, Chen L, Wang C, Wu F, Xu Y, et al. A novel biparatopic hybrid antibody-ACE2 fusion that blocks SARS-CoV-2 infection: implications for therapy. MAbs. 2020;12:1804241. doi:10.1080/19420862.2020.1804241.
- Xu C, Wang Y, Liu C, Zhang C, Han W, Hong X, Wang Y, Hong Q, Wang S, Zhao Q, et al. Conformational dynamics of SARS-CoV-2 trimeric spike glycoprotein in complex with receptor ACE2 revealed by cryo-EM. Sci Adv. 2021;7. doi:10.1126/sciadv.abe5575.
- Walls AC, Park YJ, Tortorici MA, Wall A, McGuire AT, Veesler DS. Function, and antigenicity of the SARS-CoV-2 spike glycoprotein. Cell. 2020;181:281–92 e6. doi:10.1016/j.cell.2020.02.058.
- Lan J, Ge J, Yu J, Shan S, Zhou H, Fan S, Zhang Q, Shi X, Wang Q, Zhang L, et al. Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor. Nature. 2020;581:215–20. doi:10.1038/s41586-020-2180-5.
- Bertoglio F, Meier D, Langreder N, Steinke S, Rand U, Simonelli L, Heine PA, Ballmann R, Schneider KT, Roth KDR, et al. SARS-CoV-2 neutralizing human recombinant antibodies selected from pre-pandemic healthy donors binding at RBD-ACE2 interface. Nat Commun. 2021;12:1577. doi:10.1038/s41467-021-21609-2.
- Bracken CJ, Lim SA, Solomon P, Rettko NJ, Nguyen DP, Zha BS, Schaefer K, Byrnes JR, Zhou J, Lui I, et al. Bi-paratopic and multivalent VH domains block ACE2 binding and neutralize SARS-CoV-2. Nat Chem Biol. 2021;17:113–21. doi:10.1038/s41589-020-00679-1.
- Hwang YC, Lu RM, Su SC, Chiang PY, Ko SH, Ke FY, Liang KH, Hsieh TY, Wu HC. Monoclonal antibodies for COVID-19 therapy and SARS-CoV-2 detection. J Biomed Sci. 2022;29:1. doi:10.1186/s12929-021-00784-w.
- Shi R, Shan C, Duan X, Chen Z, Liu P, Song J, Song T, Bi X, Han C, Wu L, et al. A human neutralizing antibody targets the receptor-binding site of SARS-CoV-2. Nature. 2020;584:120–24. doi:10.1038/s41586-020-2381-y.
- Wu Y, Wang F, Shen C, Peng W, Li D, Zhao C, Li Z, Li S, Bi Y, Yang Y, et al. A noncompeting pair of human neutralizing antibodies block COVID-19 virus binding to its receptor ACE2. Science. 2020;368:1274–78. doi:10.1126/science.abc2241.
- Wang Q, Zhang Y, Wu L, Niu S, Song C, Zhang Z, Lu G, Qiao C, Hu Y, Yuen KY, et al. Structural and functional basis of SARS-CoV-2 entry by using human ACE2. Cell. 2020;181:894–904 e9. doi:10.1016/j.cell.2020.03.045.
- Gonzalez-Perez D, Garcia-Ruiz E, Alcalde M. Saccharomyces cerevisiae in directed evolution: an efficient tool to improve enzymes. Bioeng Bugs. 2012;3:172–77. doi:10.4161/bbug.19544.
- Li Y, Wang H, Tang X, Fang S, Ma D, Du C, Wang Y, Pan H, Yao W, Zhang R, et al. SARS-CoV-2 and three related coronaviruses utilize multiple ACE2 orthologs and are potently blocked by an improved ACE2-Ig. J Virol. 2020;94. doi:10.1128/JVI.01283-20.
- Guy JL, Jackson RM, Jensen HA, Hooper NM, Turner AJ. Identification of critical active-site residues in angiotensin-converting enzyme-2 (ACE2) by site-directed mutagenesis. FEBS J. 2005;272:3512–20. doi:10.1111/j.1742-4658.2005.04756.x.
- Liu P, Xie X, Gao L, Jin J. His345 mutant of angiotensin-converting enzyme 2 (ACE2) remains enzymatically active against angiotensin II. Proc Natl Acad Sci U S A. 2021:118. doi:10.1073/pnas.2023648118.
- Glasgow J, Glasgow A, Kortemme T, Wells JA , et al. Reply to Liu et al.: Specific mutations matter in specificity and catalysis in ACE2. Proc Natl Acad Sci U S A. 2021;118. doi:10.1073/pnas.2024450118.
- Higuchi Y, Suzuki T, Arimori T, Ikemura N, Mihara E, Kirita Y, Ohgitani E, Mazda O, Motooka D, Nakamura S, et al. Engineered ACE2 receptor therapy overcomes mutational escape of SARS-CoV-2. Nat Commun. 2021;12:3802. doi:10.1038/s41467-021-24013-y.
- Swiech K, de Freitas Mc, Covas DT, Picanco-Castro V. Recombinant glycoprotein production in human cell lines. Methods Mol Biol. 2015;1258:223–40. doi:10.1007/978-1-4939-2205-5_12.
- Gemmill TR, Trimble RB. Overview of N- and O-linked oligosaccharide structures found in various yeast species. Biochim Biophys Acta. 1999;1426:227–37. doi:10.1016/s0304-4165(98)00126-3.
- Benton DJ, Wrobel AG, Xu P, Roustan C, Martin SR, Rosenthal PB, Skehel JJ, Gamblin SJ. Receptor binding and priming of the spike protein of SARS-CoV-2 for membrane fusion. Nature. 2020;588:327–30. doi:10.1038/s41586-020-2772-0.
- Shimon A, Shani O, Diskin R. Structural basis for receptor selectivity by the whitewater arroyo mammarenavirus. J Mol Biol. 2017;429:2825–39. doi:10.1016/j.jmb.2017.07.011.
- Wacharapluesadee S, Tan CW, Maneeorn P, Duengkae P, Zhu F, Joyjinda Y, Kaewpom T, Chia WN, Ampoot W, Lim BL, et al. Evidence for SARS-CoV-2 related coronaviruses circulating in bats and pangolins in Southeast Asia. Nat Commun. 2021;12:972. doi:10.1038/s41467-021-21240-1.
- Cohen-Dvashi H, Weinstein J, Katz M, Eilon M, Mor Y, Shimon A, Strobelt R, Shemesh M, Fleishman SJ, Diskin R. Coronacept – a potent immunoadhesin against SARS-CoV-2. bioRxiv. 2020. doi:10.1101/2020.08.12.247940.
- Mou H, Quinlan BD, Peng H, Liu G, Guo Y, Peng S, Zhang L, Davis-Gardner ME, Gardner MR, Crynen G, et al. Mutations derived from horseshoe bat ACE2 orthologs enhance ACE2-Fc neutralization of SARS-CoV-2. PLoS Pathog. 2021;17:e1009501. doi:10.1371/journal.ppat.1009501.
- Xiao K, Zhai J, Feng Y, Zhou N, Zhang X, Zou JJ, Li N, Guo Y, Li X, Shen X, et al. Isolation of SARS-CoV-2-related coronavirus from Malayan pangolins. Nature. 2020;583:286–89. doi:10.1038/s41586-020-2313-x.
- Letko M, Marzi A, Munster V. Functional assessment of cell entry and receptor usage for SARS-CoV-2 and other lineage B betacoronaviruses. Nat Microbiol. 2020;5:562–69. doi:10.1038/s41564-020-0688-y.
- Mallapaty S. The search for animals harbouring coronavirus - and why it matters. Nature. 2021;591:26–28. doi:10.1038/d41586-021-00531-z.
- Patterson EI, Elia G, Grassi A, Giordano A, Desario C, Medardo M, Smith SL, Anderson ER, Prince T, Patterson GT, et al. Evidence of exposure to SARS-CoV-2 in cats and dogs from households in Italy. bioRxiv. 2020. doi:10.1101/2020.07.21.214346.
- Sit THC, Brackman CJ, Ip SM, Tam KWS, Law PYT, Emw T, Vyt Y, Sims LD, Tsang DNC, Chu DKW, et al. Infection of dogs with SARS-CoV-2. Nature. 2020;586:776–78. doi:10.1038/s41586-020-2334-5.
- Eroshenko N, Gill T, Keaveney MK, Church GM, Trevejo JM, Rajaniemi H. Implications of antibody-dependent enhancement of infection for SARS-CoV-2 countermeasures. Nat Biotechnol. 2020;38:789–91. doi:10.1038/s41587-020-0577-1.
- Jafarzadeh A, Chauhan P, Saha B, Jafarzadeh S, Nemati M. Contribution of monocytes and macrophages to the local tissue inflammation and cytokine storm in COVID-19: lessons from SARS and MERS, and potential therapeutic interventions. Life Sci. 2020;257:118102. doi:10.1016/j.lfs.2020.118102.
- Manickam C, Sugawara S, Reeves RK. Friends or foes? The knowns and unknowns of natural killer cell biology in COVID-19 and other coronaviruses in July 2020. PLoS Pathog. 2020;16:e1008820. doi:10.1371/journal.ppat.1008820.
- de Taeye Sw, Bentlage AEH, Mebius MM, Meesters JI, Lissenberg-Thunnissen S, Falck D, Senard T, Salehi N, Wuhrer M, Schuurman J, et al. FcgammaR binding and ADCC Activity of human igg allotypes. Front Immunol. 2020;11:740. doi:10.3389/fimmu.2020.00740.
- van Erp Ea, Luytjes W, Ferwerda G, van Kasteren PB, van Erp EA. Fc-Mediated antibody effector functions during respiratory syncytial virus infection and disease. Front Immunol. 2019;10:548. doi:10.3389/fimmu.2019.00548.
- Svilenov HL, Sacherl J, Reiter A, Wolff LS, Cheng CC, Stern M, Grass V, Feuerherd M, Wachs FP, Simonavicius N, et al. Picomolar inhibition of SARS-CoV-2 variants of concern by an engineered ACE2-IgG4-Fc fusion protein. Antiviral Res. 2021;196:105197. doi:10.1016/j.antiviral.2021.105197.
- Correia IR. Stability of IgG isotypes in serum. MAbs. 2010;2(3):221–32. doi:10.4161/mabs.2.3.11788.
- Dumet C, Pottier J, Gouilleux-Gruart V, Watier H. Insights into the IgG heavy chain engineering patent landscape as applied to IgG4 antibody development. MAbs. 2019;11(8):1341–50. doi:10.1080/19420862.2019.1664365.
- Aalberse RC, Schuurman J. IgG4 breaking the rules. Immunology. 2002;105(1):9–19. doi:10.1046/j.0019-2805.2001.01341.x.
- Handlogten MW, Peng L, Christian EA, Xu W, Lin S, Venkat R, Dall’Acqua W, Ahuja S. Prevention of Fab-arm exchange and antibody reduction via stabilization of the IgG4 hinge region. MAbs. 2020;12:1779974. doi:10.1080/19420862.2020.1779974.
- Silva JP, Vetterlein O, Jose J, Peters S, Kirby H. The S228P mutation prevents in vivo and in vitro IgG4 Fab-arm exchange as demonstrated using a combination of novel quantitative immunoassays and physiological matrix preparation. J Biol Chem. 2015;290:5462–69. doi:10.1074/jbc.M114.600973.
- Ferrari M, Mekkaoui L, Ilca FT, Akbar Z, Bughda R, Lamb K, Ward K, Parekh F, Karattil R, Allen C, et al. Characterization of a Novel ACE2-Based therapeutic with enhanced rather than reduced activity against SARS-CoV-2 variants. J Virol. 2021;95:e0068521. doi:10.1128/JVI.00685-21.
- Zhang H, Penninger JM, Li Y, Zhong N, Slutsky AS. Angiotensin-converting enzyme 2 (ACE2) as a SARS-CoV-2 receptor: molecular mechanisms and potential therapeutic target. Intensive Care Med. 2020;46:586–90. doi:10.1007/s00134-020-05985-9.
- Tovey MG, Lallemand C. Immunogenicity and other problems associated with the use of biopharmaceuticals. Ther Adv Drug Saf. 2011;2:113–28. doi:10.1177/2042098611406318.
- Alfaleh MA, Alsaab HO, Mahmoud AB, Alkayyal AA, Jones ML, Mahler SM, Hashem AM. Phage display derived monoclonal antibodies: from bench to bedside. Front Immunol. 2020;11:1986. doi:10.3389/fimmu.2020.01986.
- Salazar-Fontana LI, Desai DD, Khan TA, Pillutla RC, Prior S, Ramakrishnan R, Schneider J, Joseph A. Approaches to mitigate the unwanted immunogenicity of therapeutic proteins during drug development. AAPS J. 2017;19:377–85. doi:10.1208/s12248-016-0030-z.
