ABSTRACT
As interest in antibody-based drug development continues to increase, the biopharmaceutical industry has begun to focus on complex multi-specific antibodies (MsAbs) as an up-and-coming class of biologic that differ from natural monoclonal antibodies through their ability to bind to more than one type of antigen. As techniques to generate such molecules have diversified, so have their formats and the need for standard notation. Previous efforts to develop a notation language for macromolecule drugs have been insufficient, or too complex, for MsAbs. Here, we present Antibody Markup Language (AbML), a new notation language specifically for antibody formats that overcomes the limitations of existing languages and can annotate all current antibody formats, including fusions, fragments, standard antibodies and MsAbs, as well as all currently conceivable future formats. AbML V1.1 also provides explicit support for T-cell receptor domains. To assist users of this language we have also developed a tool, abYdraw, that can draw antibody schematics from AbML strings or generate an AbML string from a drawn antibody schematic. AbML has the potential to become a standardized notation for describing new MsAb formats entering clinical trials.
Abbreviations: AbML: Antibody Markup Language; ADC: Antibody-drug conjugate; CAS: Chemical Abstracts Service; CH: Constant heavy; CL: Constant light; Fv: Variable fragment; HELM: Hierarchical Editing Language for Macromolecules; HSA: Human serum albumin; INN: International Nonproprietary Names; KIH: Knobs-into-holes; mAbs: Monoclonal antibodies; MsAb: Multi-specific antibody; WHO: World Health Organization; PEG: Poly-ethylene glycol; scFv: Single-chain variable fragment; SMILES: Simplified Molecular-Input Line-Entry System; VH: Variable heavy; VHH: Single-domain (Camelid) variable heavy; VL: Variable light
Introduction
Immunoglobulins, otherwise known as antibodies, are useful tools in biology and medicine owing to their natural ability to bind a specific antigen. When clonally expanded, monoclonal antibodies (mAbs) have clinical applications as molecular diagnostics, medical imaging agents, and therapeutics.Citation1 Multi-specific antibodies (MsAbs) are engineered proteins that differ from naturally occurring mAbs in their ability to bind to more than one type of antigen. Generally, this is achieved through multiple different antigen combining sites, and the popularity of these formats has been recognized by the World Health Organization’s International Nonproprietary Names (INN) Expert Group, which now gives the suffix stem ‘-mig’ to such proteins (https://cdn.who.int/media/docs/default-source/international-nonproprietary-names-(inn)/new_mab_-nomenclature-_2021.pdf). An exception to the use of multiple combining sites is bimekizumab, which is a conventional IgG with identical binding sites on both arms that binds to both IL-17A and IL-17F through a single type of combining site.Citation2 This would not be given the ‘-mig’ stem in the new INN scheme. While the majority of MsAbs are bispecific (binding to two epitopes though different combining sites), trispecific and tetraspecific antibodies have also been developed. This versatile class of molecules has become a keen focus of therapeutic clinical development because multi-specificity allows two molecules (as is the case with emicizumab) or two cells (as is the case with blinatumomab and catumaxomab) to be brought into close proximity.Citation3 There is a particular interest in immunomodulatory cancer treatment,Citation4 in which two of the approved drugs mentioned above (blinatumomab and catumaxomab – the latter now withdrawn) are used.Citation5,Citation6 Other approved MsAbs include emicizumab (also mentioned above) for Factor VIII deficiency hemophilia.Citation7 It binds activated factor IX and factor X to restore function of missing activated factor VIII in patients with hemophilia. More recently FDA-approved bispecifics include amivantamab and faricimab.
The engineering techniques to produce MsAbs that were first developed in the 1970s have evolved substantially. At first, the ‘quadroma’ was created by fusing two hybridoma cell lines used for generating mAbs, which would then result in some cases where two halves with different antigen-binding fragments (Fabs) form heterodimers, yielding a molecule with two specificities.Citation8,Citation9 This technique offered poor yield owing to the disfavored formation of the desired heterodimers. For example, given one hybridoma producing VHa/VLa and another producing VHb/VLb, accounting for symmetry, 10 possible antibodies could be produced by the quadroma: VHa/VLa–VHa/VLa, VHa/VLa–VHa/VLb, VHa/VLa–VHb/VLa, VHa/VLb–VHa/VLb, VHa/VLb–VHb/VLa, VHa/VLb–VHb/VLb, VHb/VLa–VHb/VLa, VHb/VLa–VHb/VLb, VHb/VLb–VHb/VLb and finally VHa/VLa–VHb/VLb, the desired product. Consequently, efforts for more scalable synthesis have led to new techniques of MsAb generation.Citation10
DNA recombination has allowed greater flexibility in designing MsAbs with IgG-like formats, which can be done by appending additional variable fragments (Fv) at the N-termini of the light and heavy chains.Citation11 On dimerization, this approach generates a symmetrical MsAb. Recombination also allows linking of VH and VL domains to form an scFv, which may be sequentially added via engineered linkers onto the N- or C-termini of both light and heavy chains.Citation12 Camelid single domain VHH fragments, also called nanobodies, may be added in the same way. All of these give rise to symmetrical antibodies.
Alternatively, asymmetric antibodies can be produced by introducing mutations that encourage heterodimerization of heavy chains or specific pairings of light and heavy chains. Additional residue mutations for knobs-into-holes (KIH) formatsCitation13 are typically used to form heavy-chain heterodimers by introducing mutations in the CH3 domains, while introduction of positively and negatively charged residues in the CH1 and CL domains of one armCitation14 assist in the correct pairing of light and heavy chains to make the desired asymmetric antibody format more favorable.Citation10
Protein engineering also allows the generation of smaller fragment-based MsAbs, including 2-chained diabodies or a single chain consisting of a sequence of scFvs. These non-IgG-like molecules are advantageous because they are easier to produce (requiring no glycosylation), but they are limited by short half-lives, which can be extended through human serum albumin (HSA) fusion or PEGylation (addition of polyethylene glycol), or the addition of disulfide bonds.Citation1,Citation15
Antibody-drug conjugates (ADCs) have become a popular approach for delivering small-molecule drugs to an intended target.Citation16 Most recently chemical conjugation has also been exploited to allow modular combination of protein domains, which has given rise to great diversity in structures and presentation of these molecules.Citation10 Ligating antibody fragments in this way has been seen in the ‘Dock and Lock’ format, while the potential of chemical ligation has also been demonstrated through production of MsAbs by ligating two IgG molecules to give IgG-IgG molecules.Citation17
Molecules based around T-cell receptors and fusions of these with scFvs (such as the ImmTAC format), including the recently FDA-approved tebentafusp, are also becoming popular. Therefore, being able to describe and draw these formats is becoming more important.
While only a few MsAbs have thus-far been approved (all bispecifics), many more are in development, including many in clinical trials. Given the huge diversity of possible MsAb formats, a standardized format for description and annotation would be advantageous, for example, when they are submitted for an INN or for regulatory approval. For small-molecule drugs, ‘Simplified Molecular-Input Line-Entry System’ (SMILES) stringsCitation18 have been adopted as a standard for describing organic molecules. As yet, no such standard has been widely adopted for biologics.
The Hierarchical Editing Language for Macromolecules (HELM)Citation19 was introduced in 2012 as a general tool for describing biologics (including antibodies) and is promoted by the Pistoia Alliance (https://www.pistoiaalliance.org/projects/current-projects/hierarchical-editing-language-for-macromolecules/). It provides a visual editor and has the support of a number of large pharmaceutical companies, including GSK, Merck, Roche, and Pfizer. Nonetheless, it has only gained limited traction in the annotation of antibodies and is not currently used by regulatory authorities, the INN Expert Group or the Chemical Abstracts Service for description of antibody-based drugs. Current limitations that make HELM less suitable for MsAbs are as follows: 1) its necessary complexity to be able to annotate other kinds of biologics, 2) while the editor allows changing colors of domains, the markup language does not allow for notation of Fv specificities, and 3) it does not allow comments or notes about additional fused domains that can be added to an antibody. Furthermore, rather than allowing the user to draw a schematic for an MsAb using simple domain blocks, the HELM editor requires amino acid sequences in an attempt to draw a schematic automatically. In addition, the editor only saves XML versions of the HELM notation, which is less suitable for text embedding to propagate the HELM string.
Here, we present a new antibody annotation language, Antibody Markup Language (AbML), designed specifically to address the needs of the antibody community in describing the ever-increasing diversity of antibody-based drugs (including fusions, fragments, standard antibodies and MsAbs) in a simple and effective manner. As of AbML V1.1, there is also explicit support for T-cell receptor domains. We have also developed a graphical editor, abYdraw, which uses AbML to render schematics of antibody-based drugs, as well as producing AbML expressions from drawn antibody schematics.
Results
Antibody markup language
AbML is based on describing antibody domains, arranged in a string and separated by connectors, representing antibody chains from N-terminus to C-terminus. The aim is to provide as simple a format as possible while conveying all necessary information.
Each domain is separated by a ‘-’ character and is numbered sequentially in order of its appearance in the expression. In this respect, hinges and artificial linkers can be considered more like domains as they are numbered and are separated from neighboring domains with a ‘-’ character. Whitespace, including line breaks is ignored in AbML except for comments given in square brackets.
Chains are separated by ‘|’ characters. Chains that are part of the antibody molecule can be presented in any order, but any additional chains that interact with antibody chains (e.g., via a disulfide or a domain pairing with a domain conjugated to the antibody) are placed last. In a multi-chain structure, every chain must have at least one domain that interacts with a domain on a different chain.
Domains
A domain annotation always begins with the domain type. The following domain types are permitted: ‘VH’, ‘VL’, ‘VHH’, ‘CH1’, ‘CH2’, ‘CH3’, ‘CH4’, ‘CL’, ‘X’, ‘C’, ‘H’ and ‘L’ as explained in the AbML Guidesheet () and AbML Format Description (Supplementary File 1). In addition, domain types for T-cell receptor domains are allowed: ‘VA’, ‘CA’, ‘VB’, ‘CB’, ‘VG’, ‘CG’, ‘VD’, ‘CD’. Domains of type ‘X’ are ‘extra’ protein domains that are not part of a standard immunoglobulin and will usually be described by associated comments; ‘C’ domains are chemical conjugation moieties, while ‘H’ and ‘L’ refer to hinge regions and artificial linkers, respectively.
Figure 1. AbML Guidesheet explaining the properties of the language. All possible domain types, modifications, connectors and comment types as well as how to notate pairings and disulfide bonds are given in a color-coded fashion relating to the example antibody domain highlighted in red. The antibody schematic was rendered with abYdraw and numbers represent the numbering of each domain given in the AbML and labeled on the schematic. A dagger in table headings indicates optional information that may be omitted from domain information.
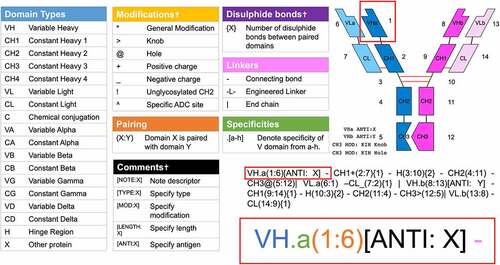
For the Fv (i.e., the VH and VL domains), the specificity is indicated by appending a ‘.’ followed by a letter corresponding to the specificity (e.g., VH.a); specificities may be omitted when the antibody is monospecific.
Where a VH/VL can bind multiple antigens (as is the case with bimekizumab, as noted above), this can be indicated with multiple letters (e.g., VH.ab, VL.ab). Typically, an interacting pair of VH and VL domains would both be assigned identical specificity descriptors, but exceptions apply when two different heavy chains share a common light chain. In this case one heavy chain would be VH.a and the other would be VH.b, while the light chain would be VL.ab.
Each domain is given a unique identifying number in parentheses (e.g., VH.a(1)) and this notation can be extended to indicate a domain with which it interacts by following the domain number with a colon and the identifying number of another domain (e.g., VH.a(1:6)). In the case of multiple interactions, multiple interacting domains may be specified (e.g., X(1:2,3,4)), as is the case in protein multimers.
If the interacting domains have disulfide bonds between them, these are indicated in curly brackets to indicate the number of disulfide bonds. (e.g., CH1(2:7){1}).
Thus a normal IgG antibody could be described by the AbML string:
VH.a(1:6)-CH1(2:7){1}-H(3:10){2}-CH2(4:11)-CH3(5:12)|
VL.a(6:1)-CL(7:2){1}|
VH.a(8:13)-CH1(9:14){1}-H(10:3){2}-CH2(11:4)-CH3(12:5)|
VL.a(13:8)-CL(14:9){1}
Note that line breaks and spacing are ignored and that for a mono-specific antibody the ‘a’ to indicate specificity is optional.
Consequently, the first line shows the first heavy chain consisting of domains VH, CH1, Hinge, CH2 and CH3 and these are numbered as domains 1–5. The end of the chain is indicated by the |. The second line shows the first light chain (domains VL and CL, numbered as domains 6 and 7). The third line shows the second heavy chain consisting of domains VH, CH1, Hinge, CH2 and CH3 (domain numbers 8–12) while the fourth line shows the second light chain (domains VL and CL, numbered as domains 13 and 14). Domain interactions are shown after colons (e.g., VH.a(1:6) indicates that this domain interacts with domain 6, which is VL.a(6:1)). Hinge region H(3:10){2} shows the interaction with hinge region H(10:3){2} and the {2} indicates that there are two disulfide bonds.
Modifications
Modifications to domains are indicated by characters immediately following the domain type. Seven such characters are currently supported. ‘>’ and ‘@’ are used to indicate knobs and holes, respectively, for KIH heterodimer pairing. ‘+’ and ‘_’ are used to indicate positive or negative mutations for charge pairing. Note that ‘_’ is used instead of ‘-’ for a negative charge since ‘-’ is used between domains.
Other general modifications (e.g., mutations to enhance or abrogate effector functions) can be indicated with a ‘*’, which can be elaborated by a comment. The carat (^) is used to indicate specific ADC conjugation sites. Currently, nonspecific ADC conjugation sites are more common and these are indicated by adding a pseudo-chain at the end of the AbML annotation: ‘|[ADC]’. Finally, ‘!’ can only appear in CH2 as it specifies that this domain is not glycosylated.
If there are multiple modification symbols, they can appear in any order. However, ‘@’ cannot be combined with ‘>’, and ‘+’ cannot be combined with ‘_’ since they are mutually exclusive opposite modifications.
Each domain may be followed by an optional comma-separated list of comments within a set of square brackets. These comments can denote the nature of modifications or ‘extra’ non-antibody protein domains, as well as antigen specificities or the length of a domain or linker. A full list of keywords and modifications can be found in the AbML Format Description (Supplementary File 1).
abYdraw
abYdraw is a graphical program written in Python3 where users may input expressions in AbML to obtain a schematic of their designed antibody by clicking the ‘Get Structure’ button. However, the user is also able to draw antibodies, and derivatives, by arranging standard domains and connecting them with connectors to obtain the appropriate expression for their design by using the ‘Get AbML’ button. Once the AbML is obtained for the drawing, using ‘Get Structure’ will re-render the schematic automatically. Both functions can be run in sequence using the ‘Tidy’ button. The program will also print out comments made in the AbML string and highlight the domain linked to those comments. abYdraw can be used to export these schematics as figures for publication and to generate a standardized expression that may be used in MsAb annotations.
The interface draws domains as blocks labeled with their domain type and any specified modifications. In the case of the negative charge modification, the ‘_’ is replaced with a minus sign in the rendered image. For KIH modifications, the ‘@’ and ‘>’ characters are omitted, as these modifications are used to affect the shape of the rendered domain. KIH adaptations are displayed by constant domains with either a cutout or an extension to their side that slots together to demonstrate how these domains are paired.
By default, domains are colored according to their specificity descriptor. Consequently, it is possible that chains will have blocks of different colors when domains of different specificities are given in the same chain. Normal connections between each domain are given by black lines that are drawn from the bottom of one domain to the top of the next domain. Artificial linkers are shown as purple lines, disulfide bonds are shown as red lines and hinges are shown in dark green. Default colors for all domain and bond types may be changed in the settings menu.
Variable domains appear with a cutout at the top of the domain referring to their antigen-combining site, which pairs with another to give a complete Fv. Nanobody domains (i.e., a VH domain that does not interact with anything else and indicated in AbML as ‘VHH’) have a unique domain shape reflecting their single-domain binding site.
Users may draw antibody-based drugs from scratch or begin with a template design of common formats (including MsAbs) that may be manipulated by the user. To draw domains, a user must select a specificity and any modifications for that domain and then place it on the canvas. Both specificities and modifications can be updated whilst on the canvas by selecting a specificity or modification, but not a domain type. Once drawn, domains may be moved to a space where they interact with other domains to be paired. VH and VL domains must face each other to be considered as interacting. Users can right-click newly drawn domains to change the direction they are facing. Nanobody domains cannot be paired with other domains, as these are single-domain VHH fragments. Standard connectors between domains are drawn by starting on the N-terminal domain of the pair and ending on the C-terminal domain of the pair. Disulfide bonds can be drawn starting from either of the interacting domains (including linkers and hinges). To insert a comment (e.g., NOTE, TYPE, ANTI, MOD), the appropriate comment-type button is clicked and, in the case of TYPE and MOD, which have restricted allowed values, the required value is selected from a drop-down list. If the desired comment is not available, comment text is typed into the text entry box and the required domain is clicked to associate the comment with that domain. Clicking the ‘Tidy’ button will then relocate the comment to the bottom of the canvas, but the comment will still be associated with the specified domain.
While AbML is designed as a simple markup language to describe domain connectivity and interactions, AbML V1.1 also allows sequence information to be associated with each domain using ASEQ and DSEQ keywords for amino acid and DNA sequences, respectively. These are provided after the main AbML annotation. The current version of abYdraw does not display this information.
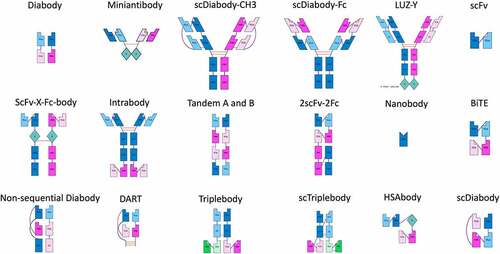
demonstrate the rendering abilities of abYdraw from AbML strings (Supplementary File 2) describing numerous antibody formats mentioned by Spiess et al.Citation10
Figure 2. Schematics of 4-chained antibodies rendered in abYdraw. The AbML to generate these images is included in Supplementary File 2.
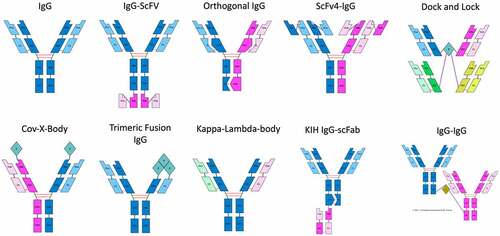
Figure 3. Schematics of 2-chained and single-chained antibodies rendered in abYdraw. The AbML to generate these images is included in Supplementary File 2.
Discussion
By addressing the pitfalls of currently available annotation languages, we have developed AbML, which is loosely based on the established HELM notation for macromolecule biologics, but simplified and adapted specifically to describe antibody formats in a straightforward manner. AbML has been carefully designed to allow annotation of future possible formats and we have demonstrated that it can be applied to all existing MsAbs described by Spiess et al.Citation10 as well as newer antibodies listed by the WHO’s INN Expert Group.
The simplicity of AbML over HELM allows greater accessibility and the potential to extend the language in future by inserting additional modification symbols and domain types. This future-proofing of the language will allow it to cope with the inevitably expanding formats of recombinant and chemically conjugated antibody-based drugs. In general, the ‘X’ and ‘C’ domains can be used to describe a multitude of possible fusion proteins, drug conjugates and chemical bonds using the comments system, and consequently we do not expect the language to require constant updating.
We hope that abYdraw, which is able both to generate and render AbML, will make AbML more accessible and will promote its use as a standard method for describing antibody formats. We are also providing compiled application versions for Linux, Mac OS and Windows environments, avoiding the need to install Python and required libraries, and to run the program from the command line.
abYdraw includes a library of commonly used MsAb formats complete with their AbML strings and diagrams that can be used as starting points for researchers to draw and describe newly designed drugs.
Currently, abYdraw has some minor limitations that may need to be addressed in future. It only supports eight specificities (i.e., letters a–h), but this should be enough for all currently conceivable constructs. abYdraw also limits domain pairings to those normally seen, i.e., VH/VL, CH1/CL, CH2/CH2, CH3/CH3, CH4/CH4, Vα/Vβ, Cα/Cβ, Vγ/Vδ, Cγ/Cδ and hinge-hinge. In addition, interactions may be specified between ‘extra’ (non-antibody) domains and chemical conjugation moieties. This works best when specifying interactions between identical domains, such as X/X in the case of protein multimers and L/L where pairs of linkers are joined by disulfide bonds. This could be improved by allowing better rendering when linking two non-identical domains, e.g., X/L pairings, which it is possible to specify in AbML.
While we support drug conjugation (random or site-specific) in AbML, these are not currently rendered or supported in abYdraw and we foresee the need to support associated features, including spacers and specific payloads.Citation20
We have recently added a command-line interface that allows abYdraw to be used for automatically rendering AbML strings without use of the graphical user interface. This will be useful, for example, in the context of generating images on the fly in web pages. In future, porting abYdraw to JavaScript would allow the full graphical user interface to be used via a web page with no need to install software locally.
To conclude, our annotation language AbML is a new descriptor language for MsAb formats and its ability to annotate all existing MsAb formats has been demonstrated. We expect this language and its corresponding tool abYdraw to become useful in the development of future MsAb drugs, allowing for standardization of MsAb description as part of ushering in a new era of MsAb drug development. Improved descriptions of their formats will demonstrate the most popular formats and potentially those that are likely to work as drugs, therefore prompting greater development in the multi-specific antibody-based drug field.
Materials and methods
Development of antibody markup language (AbML)
The requirements for AbML were as follows:
The language needed to be simple to encourage its use, but sufficiently flexible to describe all current MsAb formats and all those that could be envisioned in future.
As well as standard antibody domains, it needed to be able to describe modified domains (e.g., KIH), non-antibody domains and chemical conjugation.
Interactions between domains and (multiple) disulfides linking domains needed to be described.
The specificity of different VH/VL domains needed to be indicated.
Three types of connection between domains needed to be allowed: normal peptide connections between domains, natural (or engineered) hinge regions and artificial (engineered) peptide linkers.
AbML needed to support additional optional comments including general notes, types of additional domains, modifications, and region lengths.
With these requirements in mind, the formats of over 60 MsAbs described by Spiess et al.Citation10 were used as a starting point to ensure all such formats could be described. New INN annotations of MsAbs (post 2016) were also examined to ensure that they could all be annotated.
It was decided that AbML should have a similar structure to HELM,Citation19 but simplified and adapted specifically for MsAbs. For example, HELM would require one to specify a constant heavy (‘CH’) domain and add a comment to specify the CH type (e.g., CH1, CH2). To simplify this, AbML adopts separate domain types (e.g., ‘CH1’, ‘CH2’).
As described in the requirements above, to improve the description of antibodies, we provide three types of peptide connectors between domains: 1) natural short peptide connectors (as seen, for example, joining VH and CH1 domains). The standard definitions of the boundaries of antibody domains include these linking peptides and consequently they do not need to be indicated as separate regions of the structure; 2) natural hinge regions (as seen between CH1 and CH2 domains); and 3) engineered linkers (for example, between the VH and VL domains of an scFv). Hinges and engineered linkers differ from simple connectors in that they can be considered as connector-based ‘domains’ that can interact with one another and be joined via disulfide bonds.
Development of abYdraw
abYdraw was initially developed to render AbML strings as images, but it was then extended to make AbML more accessible by providing a graphical editor. abYdraw allows an AbML string to be entered via the graphical user interface and rendered as an image; alternatively, an image can be created or manipulated to generate an AbML string. abYdraw was implemented in Python3 using TKinter (a standard Python package) for the interface.
Software availability
Compiled apps for Linux, Mac OS and Windows are freely downloadable from http://www.bioinf.org.uk/software/abydraw/ while an introduction to AbML and the latest AbML Format Description (i.e., any updates to Supplementary File 2) are available at: http://www.bioinf.org.uk/abs/abml/ Source code for abYdraw, released under GPL3, is available at https://github.com/JamesSweetJones/abYdraw
Supplemental Material
Download Zip (60.5 KB)Disclosure statement
The authors report there are no competing interests to declare.
Supplementary material
Supplemental data for this article can be accessed online at https://doi.org/10.1080/19420862.2022.2101183
Correction Statement
This article has been corrected with minor changes. These changes do not impact the academic content of the article.
Additional information
Funding
References
- Ma J, Mo Y, Tang M, Shen J, Qi Y, Zhao W, Huang Y, Xu Y, Qian C. Bispecific antibodies: from research to clinical application. Front Immunol. 2021;12:626616.
- Adams R, Maroof A, Baker T, G LAD, Oliver R, Paveley R, Rapecki S, Shaw S, Vajjah P, West S, et al. Bimekizumab, a novel humanized IgG1 antibody that neutralizes both IL-17A and IL-17F. Front Immunol. 2020;11. doi:10.3389/fimmu.2020.01894.
- Fan G, Wang Z, Hao M, Li J. Bispecific antibodies and their applications. J Hematol Oncol. 2015;8:130.
- Labrijn AF, Janmaat ML, Reichert JM, Parren PWHI. Bispecific antibodies: a mechanistic review of the pipeline. Nat Rev Drug Discov. 2019;18:585–7.
- Wilke AC, Gökbuget N. Clinical applications and safety evaluation of the new CD19 specific T-cell engager antibody construct blinatumomab. Expert Opin Drug Saf. 2017;16:1191–202.
- Seimetz D. Novel monoclonal antibodies for cancer treatment: the trifunctional antibody catumaxomab (Removab). J Cancer. 2011;2:309–16.
- Schmitt C, Adamkewicz JI, Xu J, Petry C, Catalani O, Young G, Negrier C, Callaghan MU, Levy GG. Pharmacokinetics and pharmacodynamics of emicizumab in persons with hemophilia A with factor VIII inhibitors: HAVEN 1 study. Thromb Haemost. 2021;121:351–60.
- Milstein C, Cuello AC. Hybrid hybridomas and their use in immunohistochemistry. Nature. 1983;305:537–40.
- Kontermann RE, Brinkmann U. Bispecific antibodies. Drug Discov Today. 2015;20:838–47.
- Spiess C, Zhai Q, Carter PJ. Alternative molecular formats and therapeutic applications for bispecific antibodies. Mol Immunol. 2015;67(2 Pt A):95–106. doi:10.1016/j.molimm.2015.01.003.
- Brinkmann U, Kontermann RE. The making of bispecific antibodies. MAbs. 2017;9:182–212.
- Le Gall F, Kipriyanov SM, Moldenhauer G, Little M. Di-, tri- and tetrameric single chain Fv antibody fragments against human CD19: effect of valency on cell binding. FEBS Lett. 1999;453:164–68.
- Ridgway JBB, Presta LG, Carter PJ. ‘knobs-into-holes’ engineering of antibody CH3 domains for heavy chain heterodimerization. Protein Eng Des Sel. 1996;9:617–21.
- Gunasekaran K, Pentony M, Shen M, Garrett L, Forte C, Woodward A, B NS, Born T, Retter M, Manchulenko K, et al. Enhancing antibody Fc heterodimer formation through electrostatic steering effects: applications to bispecific molecules and monovalent IgG. J Biol Chem. 2010;285:19637–46.
- Kontermann RE. Strategies for extended serum half-life of protein therapeutics. Curr Opin Biotechnol. 2011;22:868–76.
- Sau S, Alsaab HO, Kashaw SK, Tatiparti K, Iyer AK. Advances in antibody-drug conjugates: a new era of targeted cancer therapy. Drug Discov Today. 2017;22:1547–56.
- Szijj P, Chudasama V. The renaissance of chemically generated bispecific antibodies. Nat Rev Chem. 2021;5:78–92.
- Weininger D. SMILES, a chemical language and information system. 1. introduction to methodology and encoding rules. J Chem Inf Comput Sci. 1988;28:31–36.
- Zhang T, Li H, Xi H, Stanton RV, Rotstein SH. HELM: a hierarchical notation language for complex biomolecule structure representation. J Chem Inf Model. 2012;52:2796–806.
- Dal Corso A, Pignataro L, Belvisi L, Gennari C. Innovative linker strategies for tumor-targeted drug conjugates. Chem Eur J. 2019;25:14740–57.
