ABSTRACT
The intense international focus on the COVID-19 pandemic has provided a unique opportunity to use a wide array of novel tools to carry out scientific studies on the SARS-CoV-2 virus. The value of these comparative studies extends far beyond their consequences for SARS-CoV-2, providing broad implications for health-related science. Here we specifically discuss the impacts of these comparisons on advances in vaccines, the analysis of host humoral immunity, and antibody discovery. As an extension, we also discuss potential synergies between these areas.
Abbreviations: CoVIC: The Coronavirus Immunotherapeutic Consortium; EUA: Emergency Use Authorization
KEYWORDS:
One silver lining of the COVID-19 pandemic has been the intense scientific focus on solving problems related to it, which have far broader implications beyond the pandemic itself. These problems provided a unique opportunity to directly compare many different technologies with an intensity far exceeding the average pace of research, with perhaps the most striking advances exhibited in vaccine development efforts. Since 1913, when the first vaccine was licensed (rabies), only 103 vaccines against 32 different pathogens (including SARS-CoV-2) have been approved for use in the US (https://www.fda.gov/vaccines-blood-biologics/vaccines/vaccines-licensed-use-united-states). Furthermore, for only a few pathogens (e.g., Herpes Zoster, Polio, Typhoid, Influenza), is more than one type of vaccine available (e.g., attenuated virus and recombinant protein), each reflecting years of research. Vaccine development is typically a slow, expensive process, requiring the production and testing of multiple candidates over many years, with safety at a premium as vaccines are usually administered to healthy individuals.
The advent of COVID-19 completely upended this slow development paradigm, with the pandemic adding urgency to spur development on a global scale. In January 2020, the genomic sequence of the SARS-CoV-2 virus was posted on the internet, and the race was on to develop new vaccines starting from that sequence alone. The wealth of prior knowledge on coronaviruses, particularly SARS-CoV-1, including early genome sequencing and structural studies, allowed the scientific community to hit the ground running. The development of a deep understanding of the virus, its mechanisms of infection, and neutralization rapidly accumulated in the first year.Citation1–12 For the first time in history, reverse vaccinology, synthetic biology, structural virology, cryogenic electron microscopy, vaccine adjuvants, and lipid nanoparticle technology were combined to engineer and develop 409 vaccines (198 preclinical, 170 clinical, and 41 approved). These 409 vaccines comprised eight different vaccine classes (protein subunit, replicating and non-viral vectors, DNA, RNA, live and attenuated virus, and virus-like particles) against this single virus (https://www.who.int/publications/m/item/draft-landscape-of-COVID-19-candidate-vaccines). The expertise accumulated in years of prior research made swift development of these vaccines possible once the genomic sequence was available. Although the jury is still out on the long-term side effects, by most measures, RNA vaccines have proved to be the most successful. They were the fastest to obtain approval, garnering Emergency Use Authorization (EUA) within a year of the genomic sequence being available and full approval eight months later. They were the most effective in preventing infection and symptomatic disease without an increase in short-term side effects.Citation13 Furthermore, RNA vaccines have been the fastest to update for new strains, with the BA-5 version approved within five months of its first appearance in South Africa. Only in the reduction of mortality does another format (viral vector) have a possible edge.Citation13
Most SARS-CoV-2 vaccines have been based on unmodified versions of the virus, or proteins derived from it, delivered as proteins, viral particles, DNA, or RNA. Beyond these straightforward virus-like immunogens, the pandemic has also stimulated the development of novel nanoparticle-like fusion protein vaccines, which elicit particularly robust immune responses by virtue of their multimeric nature. The nanoparticle-like fusion protein vaccines are presently delivered as proteinsCitation14 or DNA,Citation15 although there is no reason RNA could not also be used.
The efforts invested in the fast-paced development of new technologies to produce effective vaccines against SARS-CoV-2 will also pay future dividends by providing answers to new therapeutic challenges. These include emerging infections, drug-resistant bacteria, chronic infection, and cancer, where it may be possible to engineer RNA vaccines to encode genes and molecules to (re)activate the immune system. If the overall safety of RNA vaccines is unequivocally demonstrated, rapid approval of different RNA therapeutics is likely to follow, with the era of customized treatment for individuals targeting patient-specific tumor antigens becoming a potential reality.
Another sphere touched by COVID-19 is that of antibodies, the fastest growing class of therapeutics, representing nine of the top 20 best-selling drugs,Citation16 and a global market projected to more than double to over $450 billion by 2028 (https://www.fortunebusinessinsights.com/monoclonal-antibody-therapy-market-102734). While vaccines are the best option to prevent COVID-19, monoclonal antibodies provided the first-line treatment during the pandemic’s early phase and are still a valid option for certain vulnerable populations before or after exposure to SARS-CoV-2, such as the unvaccinated or recently vaccinated high-risk patients.Citation17
No other virus has had host immunity studied as intensively as SARS-CoV-2. This scrutiny has led to the development, validation, improvement, and high throughput screening application of many innovative tools for COVID-19 infection that will be invaluable in the general study of other infections and immune responses. These include microfluidics,Citation18,Citation19 single B cell antibody testing and gene cloning techniques,Citation20,Citation21 direct identification of antibody genes encoding neutralizing antibodies,Citation22 sewage based epidemiological viral detection,Citation23,Citation24 detailed dissection of antibody reactivity using variant yeast antigen display librariesCitation11,Citation25–27 and single-cell multi-omics (single-cell transcriptome, cell-surface protein, and lymphocyte antigen receptor repertoire) analyses.Citation28 Beyond understanding the immune response, some of these techniques also focus on isolating or analyzing antibodies with potential therapeutic value, which may offer improvements over the use of traditional methods based on human or mouse B cell hybridoma technology from immunized in-bred and/or transgenic mice or infected/immunized human subjects to obtain antibodies.Citation29,Citation30
In contrast to using immune sources, antibodies can also be selected from vast “naïve” in vitro antibody libraries, an approach developed over 30 years ago,Citation31–33 and for which the Nobel Prize was awarded in 2018. These libraries comprise billions of different human antibodies, and upon their development in 1991, were touted as a technology that would rapidly replace the use of animals. While significant numbers of therapeutic antibodies are derived from such libraries, the majority still come from immune sources since antibodies from naïve libraries generally tend to have lower affinities and can have inferior biophysical characteristics (known in the field as poor “developability”), complicating their advancement to successful drugs.Citation34
Before the pandemic, it was impossible to directly compare different antibody discovery platforms, as all were validated on different targets by different groups using different parameters. This situation changed when the international spotlight on COVID-19 led to essentially every capable group isolating antibodies against a single target: the SARS-CoV-2 spike protein. This singular focus provided a unique opportunity to directly compare different discovery platforms for their efficiency and speed, as well as the properties of the derived antibodies for their affinities, neutralizing potencies, and developability.Citation35 The Coronavirus Immunotherapeutic Consortium (CoVIC), established to analyze candidate antibody therapeutics in standardized side-by-side assays,Citation36 compared over 350 antibodies against the spike protein (from 56 groups worldwide) derived from COVID-19 survivors, naïve phage display libraries, in silico methods, and other strategies. Although the antibodies were blinded, making it impossible to determine the most effective way to generate antibodies against this single target, this global study provides a benchmark against which different antibody discovery strategies can now be assessed. This approach was recently adopted by our group to validate a new naïve antibody library design.Citation35
Anti-spike protein antibodies selected from this libraryCitation37 were compared to all antibodies published worldwide, including CoVIC, in terms of affinity and viral neutralization and found to be comparable to the very best immune SARS-CoV-2 antibodies. Furthermore, antibodies selected from this new naïve library design had developability properties equivalent to the best-behaved clinical antibodies, reflecting the value of using developable clinical antibodies as library scaffolds. Other naïve libraries obtained directly from healthy nonimmune human donors have also been able to generate potent neutralizing antibodies.Citation38 This approach contrasts with identifying convalescent patients with high serum titers and isolating antibody genes from their B cells using different approaches.Citation36 SARS-CoV-2 originated in Wuhan, China, and months passed before convalescent patients from which antibodies could be isolated in the US were identified. The ability to directly and rapidly (less than four weeks) generate antibodies from naïve libraries as potent as those from the immune system heralds a new approach that bypasses the need for immune responses altogether and allows for the generation of potent antibodies suitable for diagnostics, therapeutics, and prevention, within weeks of having the antigen in hand.
These results validate the original promise of naïve antibody libraries to eliminate animal use in therapeutic antibody discovery. This is true not just for diseases in which antibodies have proved particularly useful (cancer and autoimmunity), but also in swift responses to future pandemic threats: the present one has made clear the importance of preparation for the inevitable next one. While highly potent antibodies, comparable to some EUA-approved antibodies, have been generated from naïve libraries, all approved anti-SARS-CoV-2 antibodies to date, including broadly neutralizing antibodies, such as sotrovimab,Citation39 have been derived from immune responses. It remains to be seen whether similar broadly neutralizing antibodies can also be selected from naïve libraries.
The CoVIC study focused mainly on the biological functions of antibodies obtained from different antibody discovery strategies without considering downstream drug development aspects such as timeline, the scale of discovery, the proportion of neutralizing antibodies obtained with each approach, the genetic/structural diversity (i.e., different paratopes and epitope binding) of different antibodies obtained, and an estimate of the cost to obtain clinical candidate molecules.
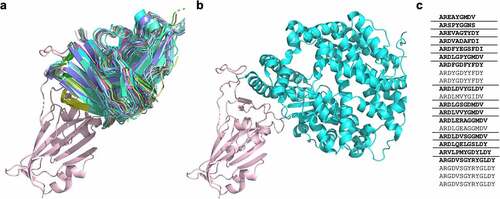
Another aspect of antibody science touched by responses to SARS-CoV-2 has been the analysis of the modalities by which antibodies with different sequences bind to the same target. While it is known that antibody responses often target similar epitopes, without the opportunity for the extensive structural analysis the pandemic provided, it has been less clear whether this reflects binding to identical epitopes or merely overlap, resulting in similar biological effects. A recent analysis of the crystal structures of antibodies binding the SARS-CoV-2 receptor binding domainCitation40 indicates that, at least for this target, numerous antibodies with diverse HCDR3s (but all derived from IGHV3–53/IGHV3–66 germline genes) bind the neutralizing epitope essentially identically (). This indicates greater structural convergence in the humoral immune response than suggested by sequence-based analysis of the HCDR3 alone, as well as the importance of the germline gene and other CDRs in directing epitope binding activity.Citation41
Figure 1. a) A cartoon representation of 22 antibodies binding to the “neck” of SARS-CoV-2 RBD (salmon), with b) showing ACE-2 (turquoise) binding at the same site. c) The CDRH3 sequences represented across the 22 RBD ‘neck’-binding antibodies. Lenient VH-clonotypes are separated with solid lines, with the cluster representative highlighted in bold font (derived from Robinson et al.Citation39).
If antibody generation, particularly using in vitro methods, could be routinely combined with mRNA vaccination, which has already been tested in clinical trials for in situ antibody expression within an infectious context,Citation42 the world of antibody therapy may be revolutionized as dramatically as has the world of vaccination. By avoiding the need to establish cell banks, bioreactors, and antibody purification, mRNA injection could lead to antibody development and administration as rapidly as is now seen for mRNA vaccines. Furthermore, mixing mRNAs encoding vaccines with mRNAs encoding neutralizing antibodies may provide simultaneous and immediate active and passive vaccination with a single injection.
Finally, these unprecedented worldwide efforts to expedite the development and production of vaccines and antibodies occurred in parallel to the streamlining of regulatory procedures. This demonstrates such regulations can also be accelerated without increased risk to patients, in anticipation of the argument that approval for treatments for other diseases could also benefit from this time advantage.
Without the COVID-19 pandemic spurring international focus on SARS-CoV-2, we likely would have waited decades for these insights.
Disclosure statement
AART and CLL received financing from Specifica Inc. SD, MFE, FF, LPS, LN, EM, TM, ADA, KP, and ARMB are employed by Specifica Inc.
Additional information
Funding
References
- Wrapp D, Wang N, Corbett KS, Goldsmith JA, Hsieh C-L, Abiona O, Graham BS, McLellan JS. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science. 2020;367(6483):1260–5. doi:10.1126/science.abb2507
- Amanat F, Stadlbauer D, Strohmeier S, Nguyen THO, Chromikova V, McMahon M, Jiang K, Arunkumar GA, Jurczyszak D, Polanco J, et al. A serological assay to detect SARS-CoV-2 seroconversion in humans. Nat Med. 2020;26(7):1033–36. doi:10.1038/s41591-020-0913-5
- Baum A, Fulton BO, Wloga E, Copin R, Pascal KE, Russo V, Giordano S, Lanza K, Negron N, Ni M, et al. Antibody cocktail to SARS-CoV-2 spike protein prevents rapid mutational escape seen with individual antibodies. Science. 2020;369(6506):1014–18. doi:10.1126/science.abd0831
- Brouwer PJM, Caniels TG, van der Straten K, Snitselaar JL, Aldon Y, Bangaru S, Torres JL, Okba NMA, Claireaux M, Kerster G, et al. Potent neutralizing antibodies from COVID-19 patients define multiple targets of vulnerability. Science. 2020;369(6504):643–50. doi:10.1126/science.abc5902
- Hoffmann M, Kleine-Weber H, Schroeder S, Kruger N, Herrler T, Erichsen S, Schiergens TS, Herrler G, Wu N-H, Nitsche A, et al. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020;181(2):271–80 e8. doi:10.1016/j.cell.2020.02.052
- Jackson LA, Anderson EJ, Rouphael NG, Roberts PC, Makhene M, Coler RN, McCullough MP, Chappell JD, Denison MR, Stevens LJ, et al. An mRNA vaccine against SARS-CoV-2 — preliminary report. N Engl J Med. 2020;383(20):1920–31. doi:10.1056/NEJMoa2022483
- Muruato AE, Fontes-Garfias CR, Ren P, Garcia-Blanco MA, Menachery VD, Xie X, Shi P-Y. A high-throughput neutralizing antibody assay for COVID-19 diagnosis and vaccine evaluation. Nat Commun. 2020;11(1):4059. doi:10.1038/s41467-020-17892-0
- Robbiani DF, Gaebler C, Muecksch F, Lorenzi JCC, Wang Z, Cho A, Agudelo M, Barnes CO, Gazumyan A, Finkin S, et al. Convergent antibody responses to SARS-CoV-2 in convalescent individuals. Nature. 2020;584(7821):437–42. doi:10.1038/s41586-020-2456-9
- Sariol A, Perlman S. Lessons for COVID-19 immunity from other coronavirus infections. Immunity. 2020;53(2):248–63. doi:10.1016/j.immuni.2020.07.005
- Shi R, Shan C, Duan X, Chen Z, Liu P, Song J, Song T, Bi X, Han C, Wu L, et al. A human neutralizing antibody targets the receptor-binding site of SARS-CoV-2. Nature. 2020;584(7819):120–24. doi:10.1038/s41586-020-2381-y
- Starr TN, Greaney AJ, Hilton SK, Ellis D, Crawford KHD, Dingens AS, Navarro MJ, Bowen JE, Tortorici MA, Walls AC, et al. Deep mutational scanning of SARS-CoV-2 receptor binding domain reveals constraints on folding and ACE2 binding. Cell. 2020;182(5):1295–310 e20. doi:10.1016/j.cell.2020.08.012
- Yan R, Zhang Y, Li Y, Xia L, Guo Y, Zhou Q. Structural basis for the recognition of SARS-CoV-2 by full-length human ACE2. Science. 2020;367(6485):1444–48. doi:10.1126/science.abb2762
- Korang SK, von Rohden E, Veroniki AA, Ong G, Ngalamika O, Siddiqui F, Juul S, Nielsen EE, Feinberg JB, Petersen JJ, et al. Vaccines to prevent COVID-19: a living systematic review with trial sequential analysis and network meta-analysis of randomized clinical trials. PLoS One. 2022;17(1):e0260733. doi:10.1371/journal.pone.0260733
- Cohen AA, van Doremalen N, Greaney AJ, Andersen H, Sharma A, Starr TN, Keeffe JR, Fan C, Schulz JE, Gnanapragasam PNP, et al. Mosaic RBD nanoparticles protect against challenge by diverse sarbecoviruses in animal models. Science. 2022;377(6606):eabq0839. doi:10.1126/science.abq0839
- Konrath KM, Liaw K, Wu Y, Zhu X, Walker SN, Xu Z, Schultheis K, Chokkalingam N, Chawla H, Du J, et al. Nucleic acid delivery of immune-focused SARS-CoV-2 nanoparticles drives rapid and potent immunogenicity capable of single-dose protection. Cell Rep. 2022;38(5):110318. doi:10.1016/j.celrep.2022.110318
- Mullard A. FDA approves 100th monoclonal antibody product. Nat Rev Drug Discov. 2021;20(7):491–95. doi:10.1038/d41573-021-00079-7
- Sun Y, Ho M. Emerging antibody-based therapeutics against SARS-CoV-2 during the global pandemic. Antib Ther. 2020;3(4):246–56. doi:10.1093/abt/tbaa025
- Jamiruddin MR, Meghla BA, Islam DZ, Tisha TA, Khandker SS, Khondoker MU, Haq MA, Adnan N, Haque M, et al. Microfluidics technology in SARS-CoV-2 diagnosis and beyond: a systematic review. Life (Basel). 2022;12(5):649. doi:10.3390/life12050649
- Swank Z, Michielin G, Yip HM, Cohen P, Andrey DO, Vuilleumier N, Kaiser L, Eckerle I, Meyer B, Maerkl SJ, et al. A high-throughput microfluidic nanoimmunoassay for detecting anti-SARS-CoV-2 antibodies in serum or ultralow-volume blood samples. Proc Natl Acad Sci U S A. 2021;118:18. doi:10.1073/pnas.2025289118
- Zost SJ, Gilchuk P, Chen RE, Case JB, Reidy JX, Trivette A, Nargi RS, Sutton RE, Suryadevara N, Chen EC, et al. Rapid isolation and profiling of a diverse panel of human monoclonal antibodies targeting the SARS-CoV-2 spike protein. Nat Med. 2020;26(9):1422–27. doi:10.1038/s41591-020-0998-x
- Pedrioli A, Oxenius A. Single B cell technologies for monoclonal antibody discovery. Trends Immunol. 2021;42(12):1143–58. doi:10.1016/j.it.2021.10.008
- Shiakolas AR, Kramer KJ, Johnson NV, Wall SC, Suryadevara N, Wrapp D, Periasamy S, Pilewski KA, Raju N, Nargi R, et al. Efficient discovery of SARS-CoV-2-neutralizing antibodies via B cell receptor sequencing and ligand blocking. Nat Biotechnol. 2022;40(8):1270–75. doi:10.1038/s41587-022-01232-2
- Coulliette-Salmond AD, Alleman MM, Wilnique P, Rey-Benito G, Wright HB, Hecker JW, Miles S, Peñaranda S, Lafontant D, Corvil S, et al. Haiti poliovirus environmental surveillance. Am J Trop Med Hyg. 2019;101(6):1240–48. doi:10.4269/ajtmh.19-0469
- Brumfield KD, Leddy M, Usmani M, Cotruvo JA, Tien C-T, Dorsey S, Graubics K, Fanelli B, Zhou I, Registe N, et al. Microbiome analysis for wastewater surveillance during COVID-19. mBio. 2022;13(4):e0059122. doi:10.1128/mbio.00591-22
- Francino-Urdaniz IM, Steiner PJ, Kirby MB, Zhao F, Haas CM, Barman S, Rhodes ER, Leonard AC, Peng L, Sprenger KG, et al. One-shot identification of SARS-CoV-2 S RBD escape mutants using yeast screening. Cell Rep. 2021;36(9):109627. doi:10.1016/j.celrep.2021.109627
- Starr TN, Greaney AJ, Addetia A, Hannon WW, Choudhary MC, Dingens AS, Li JZ, Bloom JD. Prospective mapping of viral mutations that escape antibodies used to treat COVID-19. Science. 2021;371(6531):850–54. doi:10.1126/science.abf9302
- Greaney AJ, Starr TN, Gilchuk P, Zost SJ, Binshtein E, Loes AN, Hilton SK, Huddleston J, Eguia R, Crawford KHD, et al. Complete mapping of mutations to the SARS-CoV-2 spike receptor-binding domain that escape antibody recognition. Cell Host Microbe. 2021;29(1):44–57 e9. doi:10.1016/j.chom.2020.11.007
- Stephenson E, Reynolds G, Botting RA, Calero-Nieto FJ, Morgan MD, Tuong ZK, Bach K, Sungnak W, Worlock KB, Yoshida M, et al. Single-cell multi-omics analysis of the immune response in COVID-19. Nat Med. 2021;27(5):904–16. doi:10.1038/s41591-021-01329-2
- Hartley GE, Edwards ESJ, Aui PM, Varese N, Stojanovic S, McMahon J, Peleg AY, Boo I, Drummer HE, Hogarth PM, et al. Rapid generation of durable B cell memory to SARS-CoV-2 spike and nucleocapsid proteins in COVID-19 and convalescence. Sci Immunol. 2020;5(54). doi:10.1126/sciimmunol.abf8891
- Rogers TF, Zhao F, Huang D, Beutler N, Burns A, He W-T, Limbo O, Smith C, Song G, Woehl J, et al. Isolation of potent SARS-CoV-2 neutralizing antibodies and protection from disease in a small animal model. Science. 2020;369(6506):956–63. doi:10.1126/science.abc7520
- Barbas CF 3rd, Kang AS, Lerner RA, Benkovic SJ. Assembly of combinatorial antibody libraries on phage surfaces: the gene III site. Proc Natl Acad Sci U S A. 1991;88(18):7978–82. doi:10.1073/pnas.88.18.7978
- Breitling F, Dübel S, Seehaus T, Klewinghaus I, Little M. A surface expression vector for antibody screening. Gene. 1991;104(2):147–53. doi:10.1016/0378-1119(91)90244-6
- Marks JD, Hoogenboom HR, Bonnert TP, McCafferty J, Griffiths AD, Winter G. By-passing immunization. Human antibodies from V-gene libraries displayed on phage. J Mol Biol. 1991;222(3):581–97. doi:10.1016/0022-2836(91)90498-U
- Jain T, Sun T, Durand S, Hall A, Houston NR, Nett JH, Sharkey B, Bobrowicz B, Caffry I, Yu Y, et al. Biophysical properties of the clinical-stage antibody landscape. Proc Natl Acad Sci U S A. 2017;114(5):944–49. doi:10.1073/pnas.1616408114
- Azevedo Reis Teixeira A, Erasmus MF, D’Angelo S, Naranjo L, Ferrara F, Leal-Lopes C, Durrant O, Galmiche C, Morelli A, Scott-Tucker A, et al. Drug-like antibodies with high affinity, diversity and developability directly from next-generation antibody libraries. MAbs. 2021;13(1):1980942. doi:10.1080/19420862.2021.1980942
- Hastie KM, Li H, Bedinger D, Schendel SL, Dennison SM, Li K, Rayaprolu V, Yu X, Mann C, Zandonatti M, et al. Defining variant-resistant epitopes targeted by SARS-CoV-2 antibodies: a global consortium study. Science. 2021;374(6566):472–78. doi:10.1126/science.abh2315
- Ferrara F, Erasmus MF, D’Angelo S, Leal-Lopes C, Teixeira AA, Choudhary A, Honnen W, Calianese D, Huang D, Peng L, et al. A pandemic-enabled comparison of discovery platforms demonstrates a naive antibody library can match the best immune-sourced antibodies. Nat Commun. 2022;13(1):462. doi:10.1038/s41467-021-27799-z
- Zhao S, Zhang H, Yang X, Zhang H, Chen Y, Zhan Y, Zhang X, Jiang R, Liu M, Liu L, et al. Identification of potent human neutralizing antibodies against SARS-CoV-2 implications for development of therapeutics and prophylactics. Nat Commun. 2021;12(1):4887. doi:10.1038/s41467-021-25153-x
- Pinto D, Park Y-J, Beltramello M, Walls AC, Tortorici MA, Bianchi S, Jaconi S, Culap K, Zatta F, De Marco A, et al. Cross-neutralization of SARS-CoV-2 by a human monoclonal SARS-CoV antibody. Nature. 2020;583(7815):290–95. doi:10.1038/s41586-020-2349-y
- Robinson SA, Raybould MIJ, Schneider C, Wong WK, Marks C, Deane CM, Fraternali F. Epitope profiling using computational structural modelling demonstrated on coronavirus-binding antibodies. PLoS Comput Biol. 2021;17(12):e1009675. doi:10.1371/journal.pcbi.1009675
- D’Angelo S, Ferrara F, Naranjo L, Erasmus MF, Hraber P, Bradbury ARM. Many routes to an antibody heavy-chain CDR3: necessary, yet insufficient, for specific binding. Front Immunol. 2018;9:395. doi:10.3389/fimmu.2018.00395
- August A, Attarwala HZ, Himansu S, Kalidindi S, Lu S, Pajon R, Han S, Lecerf J-M, Tomassini JE, Hard M, et al. A phase 1 trial of lipid-encapsulated mRNA encoding a monoclonal antibody with neutralizing activity against Chikungunya virus. Nat Med. 2021;27(12):2224–33. doi:10.1038/s41591-021-01573-6
